Abstract
IMGT/GENE-DB is the comprehensive IMGT genome database for immunoglobulin (IG) and T cell receptor (TR) genes from human and mouse, and, in development, from other vertebrates. IMGT/GENE-DB is the international reference for the IG and TR gene nomenclature and works in close collaboration with the HUGO Nomenclature Committee, Mouse Genome Database and genome committees for other species. IMGT/GENE-DB allows a search of IG and TR genes by locus, group and subgroup, which are CLASSIFICATION concepts of IMGT-ONTOLOGY. Short cuts allow the retrieval gene information by gene name or clone name. Direct links with configurable URL give access to information usable by humans or programs. An IMGT/GENE-DB entry displays accurate gene data related to genome (gene localization), allelic polymorphisms (number of alleles, IMGT reference sequences, functionality, etc.) gene expression (known cDNAs), proteins and structures (Protein displays, IMGT Colliers de Perles). It provides internal links to the IMGT sequence databases and to the IMGT Repertoire Web resources, and external links to genome and generalist sequence databases. IMGT/GENE-DB manages the IMGT reference directory used by the IMGT tools for IG and TR gene and allele comparison and assignment, and by the IMGT databases for gene data annotation. IMGT/GENE-DB is freely available at http://imgt.cines.fr.
INTRODUCTION
IMGT/GENE-DB, part of IMGT, the international ImMunoGeneTics information system®, http://imgt.cines.fr (1–4) is the comprehensive IMGT genome database, which has been developed to classify the immunoglobulin (IG) and the T cell receptor (TR) genes from vertebrate species, and to standardize and manage the complex IG and TR gene data knowledge (5) (http://www.bioinfo.de/isb/2003/04/0004/). The molecular genetics of the IG and TR genes is so complex and unique in the genome of vertebrates (6,7) that a specific gene database was required to manage all their characteristics. Indeed, the synthesis of IG and TR chains involves multigene families from four different gene types: variable (V), diversity (D), joining (J) and constant (C), each one with unique characteristics. These genes are organized in hundreds of cassettes, as in fish, or in large clusters from several hundred kilobases to one (or more) megabase(s), as in mouse and human (6,7). IG and TR genes that belong to same subgroup may be highly similar in their coding sequence, but at the same time, highly polymorphic (e.g. 13 allelic forms have been sequenced for the human IGHV2-70 gene) (6), with alleles displaying different functionalities. The presence of many pseudogenes in the loci, and the frequency of the polymorphisms by gene insertion and deletion in these multigene families, add an additional level of complexity (6,7). Although most human IG and TR genes were sequenced and characterized independently from and before the completion of the Human Genome Project, the classification and the characterization of the IG and TR genes remain a big challenge in the analysis of the genome. Indeed, the annotations of the IG and TR loci, which represent for instance, in human, ∼6 Mb on chromosomes 2, 7, 14 and 22, are not available through classical genome software, owing to the unique IG and TR gene structure (6,7). At the level of gene expression analysis (e.g. cDNAs), data are even more difficult to interpret as the mechanisms involved in the IG and TR synthesis include DNA rearrangements with large DNA deletion of several hundred kilobases, and recombinations, nucleotide deletions and insertions at the rearranged junctions and, for IG, somatic hypermutations. Such somatic mechanisms create an extraordinary diversity of 1012 different IG and TR per individual (6,7). Thus, most IG and TR expressed sequences, available in IMGT/LIGM-DB (8) (http://www3.oup.co.uk/nar/database/summary/504), the IMGT sequence database, and in IMGT/3Dstructure-DB, the IMGT 3D structure database (9) show significant nucleotide and amino acid differences, respectively, by comparison with the germline (not rearranged) sequences. IMGT/GENE-DB has been implemented to provide an easy and common access to standardized and expertly annotated IG and TR gene and allele data and knowledge. The first task of IMGT was to define a reference sequence for each individual gene and allele (6,7), based on the IMGT ‘gene’ and ‘allele’ concepts. IMGT/GENE-DB has been developed using Java and cgi programs and has been available on the Web since January 2003. IMGT/GENE-DB, which currently contains human and mouse IG and TR genes, is the international reference for the IG and TR gene nomenclature.
IMGT ‘GENE’ AND ‘ALLELE’ CONCEPTS
The IMGT ‘gene’ and ‘allele’ concepts represent the cornerstone of the IMGT-ONTOLOGY ‘CLASSIFICATION’ concept (10) and of the IMGT/GENE-DB implementation. A gene is a DNA sequence that can be potentially transcribed and/or translated (this definition includes the regulatory elements in 5′ and 3′, and the introns, if present). Instances of the ‘gene’ concept are gene names (10). By extension, orphons and pseudogenes are also instances of the ‘gene’ concept (6,7). The IMGT gene names integrate the main CLASSIFICATION concepts of IMGT-ONTOLOGY: the group, the subgroup, the locus and the chromosomal orphon set (10). All IMGT gene names for human IG and TR genes were approved by the Human Genome Organisation (HUGO) Gene Nomenclature Committee (HGNC) (11) in 1999, and entered in the Genome DataBase GDB (Canada) (12), LocusLink and Entrez Gene at NCBI (USA) (13). An allele is a polymorphic variant of a gene, which is characterized by the mutations of its sequence compared to the gene reference sequence designated as allele *01. An IMGT gene or allele name is systematically associated to a species. Each allele is characterized by its functionality and by an IMGT reference sequence (10). The allele functionality, part of the IDENTIFICATION concept of IMGT-ONTOLOGY, has three instances: functional (F), open reading frame (ORF) and pseudogene (P) (10). These instances refer to the V, D and J alleles in their ‘germline’ (non-rearranged) configuration (6,7), and to the C alleles (the configuration of the C genes that do not rearrange is ‘undefined’) (10). An IMGT/GENE-DB allele reference sequence is identified by the IMGT/LIGM-DB accession number, the IMGT gene and allele name, the species, the allele functionality, and the gene core (V-REGION, D-REGION, J-REGION and C-REGION) (10). The sequences of the gene core are extracted from the IMGT/LIGM-DB reference sequences. The IMGT/GENE-DB allele reference sequences are provided in FASTA format with a complete header, for example:
![]()
For C-REGION encoded by several exons, each exon is provided separately with, in addition, the complete artificially spliced C-REGION.
IMGT/GENE-DB CONTENT
As on July 2004, IMGT/GENE-DB contained 1375 genes and 2204 alleles from human and mouse (673 IG and TR genes and 1208 alleles from Homo sapiens, and 702 IG and TR genes and 996 alleles from mouse (most entries from Mus musculus, a few entries from Mus cookii, Mus minutoides, Mus pahari, Mus saxicola and Mus spretus) (Tables 1 and 2). This represents the complete set of human IG and TR genes, for all the seven loci (the three IG loci: IGH, IGK and IGL; and the four TR loci: TRA, TRB, TRG and TRD) and for the chromosomal orphon sets (6,7). The mouse entries are complete, except for the mouse IGHV group, which still has a provisional IMGT nomenclature but is near completion.
Table 1. IMGT/GENE-DB statistics: number of human and mouse IG genes, and within parentheses, number of alleles.
| Locus | IGH | IGK | IGL | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Group | IGHV | IGHD | IGHJ | IGHC | IGKV | IGKJ | IGKC | IGLV | IGLJ | IGLC | |
| Human | 164 (387) | 37 (44) | 9 (16) | 12 (45) | 108 (131) | 5 (9) | 1 (5) | 79 (129) | 7 (10) | 9 (22) | 431 (798) |
| Mousea | 216 (239) | 17 (19) | 4 (9) | 9 (26) | 176 (203) | 5 (10) | 6 (8) | 12 (19) | 7 (7) | 7 (10) | 459 (550) |
| Total | 380 (626) | 54 (63) | 13 (25) | 21 (71) | 284 (334) | 10 (19) | 7 (13) | 91 (148) | 14 (17) | 16 (32) | 890 (1348) |
aMus cookii, Mus minutoides, Mus musculus, Mus pahari, Mus saxicola and Mus spretus.
Table 2. IMGT/GENE-DB statistics: number of human and mouse TR genes, and within parentheses, number of alleles.
| Locus | TRA | TRB | TRG | TRD | Total | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group | TRAV | TRAJ | TRAC | TRBV | TRBD | TRBJ | TRBC | TRGV | TRGJ | TRGC | TRDV | TRDD | TRDJ | TRDC | |
| Human | 54 (112) | 61 (63) | 1 (1) | 76 (162) | 2 (3) | 14 (16) | 2 (4) | 14 (22) | 5 (6) | 2 (7) | 3 (6) | 3 (3) | 4 (4) | 1 (1) | 242 (410) |
| Mousea | 98 (233) | 60 (67) | 1 (2) | 35 (61) | 2 (2) | 14 (19) | 7 (9) | 7 (28) | 4 (4) | 4 (5) | 6 (10) | 2 (2) | 2 (3) | 1 (1) | 243 (446) |
| Total | 152 (345) | 121 (130) | 2 (3) | 111 (223) | 4 (5) | 28 (35) | 9 (13) | 21 (50) | 9 (10) | 6 (12) | 9 (16) | 5 (5) | 6 (7) | 2 (2) | 485 (856) |
aMus minutoides, Mus musculus, Mus pahari and Mus spretus.
IMGT/GENE-DB QUERY PAGE
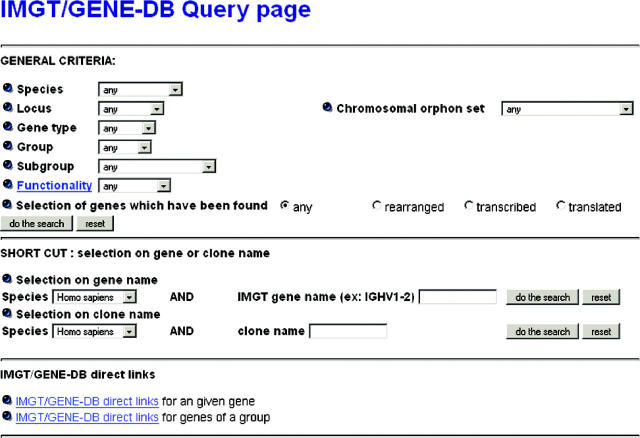
The IMGT/GENE-DB Query page comprises three types of search (Figure 1): (i) ‘GENERAL CRITERIA’ allows a search of IG and TR genes, for a given species, by locus or chromosomal orphon set, by gene type, group or subgroup, or functionality. The user can select genes that have been found rearranged, transcribed or translated. (ii) ‘SHORT CUT’ allows a selection, for a given species, on gene name or clone name. (iii) ‘IMGT/GENE-DB direct links’ gives access to a set of links, which allow the retrieval of the information related to either one given gene, or to genes of a group using configurable URL, which can be used by humans or programs.
Figure 1.
The IMGT/GENE-DB Query page.
IMGT/GENE-DB RESULT PAGE
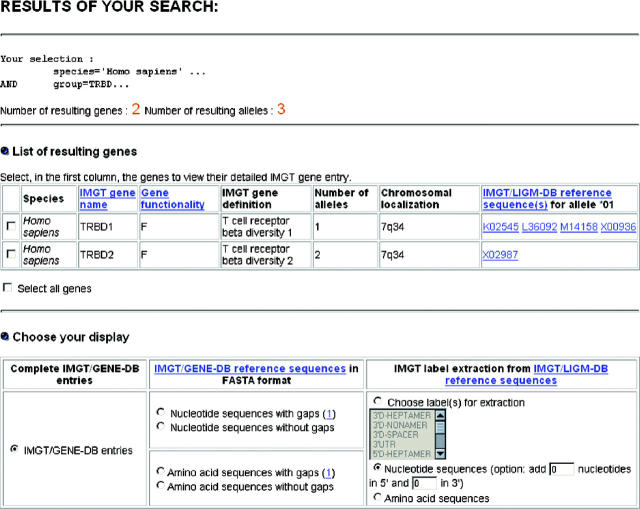
Following a ‘GENERAL CRITERIA’ or a ‘SHORT CUT’ selection, the IMGT/GENE-DB result page (Figure 2) shows, at the top, the user selection, the number of resulting genes and the number of resulting alleles, then the list of resulting genes with, for each gene, the species, IMGT gene name, gene functionality, IMGT gene definition, number of alleles, chromosomal localization and IMGT/LIGM-DB reference sequence(s) for the allele *01 (Figure 2). In the ‘Choose your display’ section, the user can select between three types of display: (i) the complete individual IMGT/GENE-DB entries for the genes selected in the list of resulting genes (an IMGT/GENE-DB entry is described in the next paragraph); (ii) the IMGT/GENE-DB allele reference sequences in FASTA format: nucleotide or amino acid sequences, either with gaps according to the IMGT unique numbering (14–16), or without gaps; (iii) the IMGT label sequences in FASTA format, extracted from expertly annotated IMGT/LIGM-DB reference sequences. This allows to retrieve any label sequence (V-EXON, V-HEPTAMER, etc.), the core regions of out-of-frame pseudogenes, which are not available in the IMGT/GENE-DB allele reference sequences, and the artificially spliced L-PART1+L-PART2 and L-PART1+V-EXON. For nucleotide sequences, the user has the possibility to extend the limits in 5′ or 3′ by typing the number of nucleotides of one's choice.
Figure 2.
The IMGT/GENE-DB result page and the three types of choice in ‘Choose your display’.
IMGT/GENE-DB ENTRY
An individual IMGT/GENE-DB entry provides a full characterization of a gene and of its alleles: IMGT name and definition, chromosomal localization, number of alleles, IMGT reference alleles and other sequences from the literature (as defined in IMGT Gene tables), and for each sequence, allele functionality, clone name, accession number, molecule type. The IMGT/GENE-DB entry gives also access (i) to the IMGT/GENE-DB allele reference sequences in FASTA format [nucleotide and amino acid sequences with gaps according to the IMGT unique numbering (14–16), or without gaps], (ii) to the IMGT Repertoire standardized resources (Chromosomal localization, Locus representation, Tables of alleles, Alignments of alleles, IMGT Protein displays, IMGT Colliers de Perles, etc.) via internal links (‘Locus and genes’, ‘Proteins and alleles’, ‘2D and 3D structures’, ‘Probes and RFLP’, ‘Gene regulation and expression’, ‘Genes and clinical entities’ sections), (iii) to the known IMGT/LIGM-DB cDNA sequences of the gene with a direct IMGT/LIGM-DB query, which then allows the choice of the nine different IMGT/LIGM-DB displays including IMGT/V-QUEST results (17,18), (iv) to the IMGT tools for genome analysis (IMGT/GeneSearch, IMGT/GeneView, IMGT/LocusView, IMGT/GeneInfo) (3,5,19), and (v) to the external links on genome databases LocusLink and Entrez Gene at NCBI, GDB, GeneCards (20), OMIM, MGD (21), sequence databases EMBL (22)/GenBank (23)/DDBJ (24) and nomenclature database HGNC Genenew (11).
CONCLUSION AND PERSPECTIVES
The central management of gene-related data in IMGT/GENE-DB improves the dynamic generation of knowledge resources from data, which are extracted from the IMGT sequence database IMGT/LIGM-DB, from HTML pages in IMGT Repertoire and from the IMGT tools for genome analysis. Reciprocally, the IMGT/GENE-DB data are used by other IMGT databases (IMGT/PRIMER-DB, IMGT/3D structure-DB) and tools (IMGT/V-QUEST, IMGT/JunctionAnalysis, etc.). The dynamic interactions are currently implemented through IMGT-Choreography (29) based on IMGT-ONTOLOGY and using IMGT-ML Web services. All the mouse IG and TR genes from IMGT/GENE-DB with IMGT reference sequences were provided by IMGT to HGNC and MGD in July 2002. IG and TR genes from genomes of other species (chimpanzee, rat, etc.), as well as members of the immunoglobulin superfamily (IgSF) and of the major histocompatibility complex superfamily (MhcSF) (currently described in the IMGT Repertoire ‘RPI’ section, for the related proteins of the immune system), will be added in IMGT/GENE-DB following the exhaustive analysis of the corresponding genes in IMGT.
CITATION
Users of IMGT/GENE-DB are requested to cite this article in their publications and to quote the IMGT® home page URL, http://imgt.cines.fr.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Tasuku Honjo, Leroy Hood, Gérard Lefranc, Fumihiko Matsuda and Hans Zachau for helpful discussion. We thank Richard Baldarelli, Judith Blake, Janan Eppig, Scott Federhen, Melissa Landrum, Ruth Lovering, Loïs Maltais, Donna Maglott, Chris Porter, Sue Povey, Marilyn Safran, Robert Sinclair and Hester Wain for their collaboration. We are deeply grateful to the IMGT team for its expertise and constant motivation, and specially to our curators for their hard work and enthusiasm. IMGT is funded by the European Union's 5th PCRDT programme (QLG2-2000-01287), the Centre National de la Recherche Scientifique (CNRS), and the Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche (Université Montpellier II Plan-Pluri-Formation, BIOSTIC-LR2004 and ACI-IMPBIO IMP82-2004).
REFERENCES
- 1.Lefranc M.-P. (2003) IMGT, the international ImMunoGeneTics database. Nucleic Acids Res., 31, 307–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lefranc M.-P. (2003) IMGT® databases, web resources and tools for immunoglobulin and T cell receptor sequence analysis, http://imgt.cines.fr. Leukemia, 17, 260–266. [DOI] [PubMed] [Google Scholar]
- 3.Lefranc M.-P. (2004) IMGT-ONTOLOGY and IMGT databases, tools and web resources for immunogenetics and immunoinformatics. Mol. Immunol., 40, 647–659. [DOI] [PubMed] [Google Scholar]
- 4.Lefranc M.-P. (2003) IMGT, the international ImMunoGeneTics information system®, http://imgt.cines.fr. In Bock,G. and Goode,J. (eds), Immunoinformatics: Bioinformatics Strategies for Better Understanding of Immune Function. Novartis Foundation Symposium 254. John Wiley and Sons, Chichester, pp. 126–136, discussion pp. 136–142, 216–222, 250–252. [PubMed] [Google Scholar]
- 5.Lefranc M.-P., Giudicelli,V., Ginestoux,C., Bosc,N., Folch,G., Guiraudou,D., Jabado-Michaloud,J., Magris,S., Scaviner,D., Thouvenin,V., Combres,K., Girod,D., Jeanjean,S., Protat,C., Yousfi Monod,M., Duprat,E., Kaas,Q., Pommié,C., Chaume,D. and Lefranc,G. (2004) IMGT-ONTOLOGY for Immunogenetics and Immunoinformatics (http://imgt.cines.fr). Epub In Silico Biology, 4, 0004. In Silico Biology, 4, 17–29. [PubMed] [Google Scholar]
- 6.Lefranc M.-P. and Lefranc,G. (2001) The Immunoglobulin FactsBook. Academic Press, London, UK. [Google Scholar]
- 7.Lefranc M.-P. and Lefranc,G. (2001) The T Cell Receptor FactsBook. Academic Press, London, UK. [Google Scholar]
- 8.Chaume D., Giudicelli,V. and Lefranc,M.-P. (2004) IMGT/LIGM-DB. In Galperin,M. (ed.), The Molecular Biology Database Collection. Nucleic Acids Res., 32, D3–D22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaas Q., Ruiz,M. and Lefranc,M.-P. (2004) IMGT/3Dstructure-DB and IMGT/StructuralQuery, a database and a tool for immunoglobulin, T cell receptor and MHC structural data. Nucleic Acids Res., 32, D208–D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giudicelli V. and Lefranc,M.-P. (1999) Ontology for immunogenetics: IMGT-ONTOLOGY. Bioinformatics, 15, 1047–1054. [DOI] [PubMed] [Google Scholar]
- 11.Wain H.M., Bruford,E.A., Lovering,R.C., Lush,M.J., Wright,M.W. and Povey,S. (2002) Guidelines for human gene nomenclature. Genomics, 79, 464–470. [DOI] [PubMed] [Google Scholar]
- 12.Letovsky S.I., Cottingham,R.W., Porter,C.J. and Li,P.W. (1998) GDB: the Human Genome Database. Nucleic Acids Res., 26, 94–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lefranc M.-P., Pommié,C., Ruiz,M., Giudicelli,V., Foulquier,E., Truong,L., Thouvenin-Contet,V. and Lefranc,G. (2003) IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol., 27, 55–77. [DOI] [PubMed] [Google Scholar]
- 15.Pommié C., Levadoux,S., Sabatier,R., Lefranc,G. and Lefranc,M.-P. (2004) IMGT standardized criteria for statistical analysis of immunoglobulin V-REGION amino acid properties. J. Mol. Recognit., 17, 17–32. [DOI] [PubMed] [Google Scholar]
- 16.Lefranc M.-P., Pommié,C., Kaas,Q., Duprat,E., Bosc,N., Guiraudou,D., Jean,C., Ruiz,M., Da Piédade,I., Rouard,M., Foulquier,E., Thouvenin,V. and Lefranc,G. (2003) IMGT unique numbering for immunoglobulin and T cell receptor constant domains and Ig superfamily C-like domains. Dev. Comp. Immunol., doi:10.1016/j.dci.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Giudicelli V., Chaume,D. and Lefranc,M.-P. (2004) IMGT/V-QUEST, an integrated software program for immunoglobulin and T cell receptor V-J and V-D-J rearrangement analysis. Nucleic Acids Res., 32, W435–W440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefranc M.-P. (2003) IMGT, the international ImMunoGeneTics information system®, http://imgt.cines.fr. Methods Mol. Biol., 248, 27–49. [DOI] [PubMed] [Google Scholar]
- 19.Baum P., Pasqual,N., Thuderoz,F., Hierle,V., Chaume,D., Lefranc,M.-P., Jouvin-Marche,E., Marche,N. and Demongeot,J. (2004) IMGT/GeneInfo: enhancing V(D)J recombination database accessibility. Nucleic Acids Res., 32, D51–D54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Safran M., Chalifa-Caspi,V., Shmueli,O., Olender,T., Lapidot,M., Rosen,N., Shmoish,M., Peter,Y., Glusman,G., Feldmesser,E., Adato,A., Peter,I., Khen,M., Atarot,T., Groner,Y. and Lancet,D. (2003) Human Gene-Centric databases at the Weizmann Institute of Science: GeneCards, UDB, CroW21 and HORDE. Nucleic Acids Res., 31, 142–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blake J.A., Richardson,J.E., Bult,C.J., Kadin,J.A., Eppig,J.T.; Mouse Genome Database Group. (2003) MGD: the Mouse Genome Database. Nucleic Acids Res., 31, 193–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulikova T., Aldebert,P., Althorpe,N., Baker,W., Bates,K., Browne,P., van den Broek,A., Cochrane,G., Duggan,K., Eberhardt,R. et al. (2004) The EMBL Nucleotide Sequence Database. Nucleic Acids Res., 32, D27–D30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J. and Wheeler,D.L. (2004) GenBank: update. Nucleic Acids Res., 32, D23–D26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyazaki S., Sugawara,H., Ikeo,K., Gojobori,T. and Tateno,Y. (2004) DDBJ in the stream of various biological data. Nucleic Acids Res., 32, D31–D34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yousfi Monod M., Giudicelli,V., Chaume,D. and Lefranc,M.-P. (2004) IMGT/JunctionAnalysis: the first tool for the analysis of the immunoglobulin and T cell receptor complex V-J and V-D-J JUNCTIONs. Bioinformatics, 20, I379–I385. [DOI] [PubMed] [Google Scholar]
- 26.Elemento O. and Lefranc,M.-P. (2003) IMGT/PhyloGene: an on-line tool for comparative analysis of immunoglobulin and T cell receptor genes. Dev. Comp. Immunol., 27, 763–779. [DOI] [PubMed] [Google Scholar]
- 27.Giudicelli V., Protat,C. and Lefranc,M.-P. (2003) The IMGT strategy for the automatic annotation of IG and TR cDNA sequences: IMGT/Automat. ECCB′2003, European Conference on Computational Biology. Ed DISC/Spid DKB-31, 103–104.
- 28.Folch G., Bertrand,J., Lemaitre,M. and Lefranc,M.-P. (2004) IMGT/PRIMER-DB. In Galperin,M. (ed.), The Molecular Biology Database Collection. Nucleic Acids Res., 32, D3–D22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaume D., Giudicelli,V., Combres,K., Ginestoux,C. and Lefranc,M.-P. IMGT-Choreography: processing of complex immunogenetics knowledge. Computational Methods in Systems Biology (Paris, France, May 26–28, 2004). Lecture Notes in BioInformatics. LNBI, Springer, in press. [Google Scholar]