Abstract
Trypanosoma brucei causes human African trypanosomiasis (HAT). The pyrrolopyrimidine AEE788 (a hit for anti-HAT drug discovery) associates with three trypanosome protein kinases. Herein we delineate the effects of AEE788 on T. brucei using chemical biology strategies. AEE788 treatment inhibits DNA replication in the kinetoplast (mitochondrial nucleoid) and nucleus. In addition, AEE788 blocks duplication of the basal body and the bilobe without affecting mitosis. Thus, AEE788 prevents entry into the S-phase of the cell division cycle. To study the kinetics of early events in trypanosome division, we employed an “AEE788 block and release” protocol to stage entry into the S-phase. A time-course of DNA synthesis (nuclear and kinetoplast DNA), duplication of organelles (basal body, bilobe, kinetoplast, nucleus), and cytokinesis was obtained. Unexpected findings include the following: 1) basal body and bilobe duplication are concurrent; 2) maturation of probasal bodies, marked by TbRP2 recruitment, is coupled with nascent basal body assembly, monitored by localization of TbSAS6 at newly forming basal bodies; and 3) kinetoplast division is observed in G2 after completion of nuclear DNA synthesis. Prolonged exposure of trypanosomes to AEE788 inhibited transferrin endocytosis, altered cell morphology, and decreased cell viability. To discover putative effectors for the pleiotropic effects of AEE788, proteome-wide changes in protein phosphorylation induced by the drug were determined. Putative effectors include an SR protein kinase, bilobe proteins, TbSAS4, TbRP2, and BILBO-1. Loss of function of one or more of these effectors can, from published literature, explain the polypharmacology of AEE788 on trypanosome biology.
Introduction
Trypanosoma brucei is a protozoan parasite that causes human African trypanosomiasis (HAT) (reviewed by Lejon and Büscher, 2005; Kennedy, 2013). Current HAT chemotherapies are administered by injection and have toxic side effects (for review, see Babokhov et al., 2013), making them far from ideal. An attractive drug discovery approach for neglected tropical diseases, such as HAT, is chemical scaffold repurposing (Patel et al., 2013). In this strategy, drugs with proven efficacy against other diseases are screened for activity against HAT, reducing the time and cost associated with early-stage drug discovery (DiMasi et al., 2003). We identified a small-molecule kinase inhibitor, AEE788 (Traxler et al., 2004; Meco et al., 2010), as a “hit” (growth inhibitory concentration by 50% = 2.5 µM) for HAT drug discovery. Subsequently, AEE788 was established as an anti-trypanosomal lead drug (Behera et al., 2014). AEE788 forms complexes with three trypanosome protein kinases (Katiyar et al., 2013), suggesting that it is a multitargeted antagonist or agonist (Dar and Shokat, 2011) whose toxicity to trypanosomes is likely based on exerting pleiotropic biologic effects.
Stages of the trypanosome cell division cycle can be identified by enumeration of single-copy organelles, including the kinetoplast [mitochondrial nucleoid containing kinetoplast DNA (kDNA)], basal body, and nucleus (Sherwin and Gull, 1989; Woodward and Gull, 1990). In G1, trypanosomes have a single round kinetoplast and a single nucleus (1K1N) (Sherwin and Gull, 1989). As cells transition into the S-phase, synthesis of kDNA (for review, see Jensen and Englund, 2012) is associated with kinetoplast elongation (Gluenz et al., 2011), generating early S-phase cells with a single elongated kinetoplast (Ke) and one nucleus (1Ke1N) (Siegel et al., 2008). Division of the kinetoplast precedes mitosis forming a 2K1N population (Woodward and Gull, 1990). Mitosis produces 2K2N trypanosomes, which become two 1K1N cells after cytokinesis (Sherwin and Gull, 1989), completing the division cycle (for review, see Hammarton, 2007; Li, 2012; Zhou et al., 2014).
The basal body is the microtubule-organizing center for the flagellar axoneme. Additionally, the basal body is attached to the kinetoplast (Ogbadoyi et al., 2003) and has a role in inheritance of the mitochondrial genome (Robinson and Gull, 1991). Accordingly, basal body biogenesis is tightly coordinated with the cell division cycle (Sherwin and Gull, 1989; Woodward and Gull, 1990; Gluenz et al., 2011). Prior to duplication, trypanosomes have a mature basal body (mBB) adjacent to an immature probasal body (pBB) (Sherwin and Gull, 1989). Maturation of the pBB produces cells with two mBBs, each of which seed a new pBB (Sherwin and Gull, 1989; Lacomble et al., 2010; Gluenz et al., 2011). No quantitative time-course study of the conversion of putative intermediates into mBBs has been reported.
The flagellum exits the trypanosome cell body via the flagellar pocket (Lacomble et al., 2009). Duplication of the flagellar pocket depends on basal body duplication and separation (Lacomble et al., 2010). Outside the cell body, the flagellar membrane is conjoined to the plasma membrane by the flagellar attachment zone (FAZ) (Sherwin and Gull, 1989; Kohl et al., 1999). Cytokinesis requires duplication of the flagellum and its associated cytoskeletal structures (Robinson et al., 1995; Kohl et al., 2003). The bilobe is a cytoskeletal structure closely associated with the FAZ filament (Esson et al., 2012) and is implicated in FAZ formation (Zhou et al., 2010; Bangs, 2011). The flagellar pocket is the major site of endocytosis, a process needed for nutrient uptake (for review, Field et al., 2009). Bloodstream trypanosomes require host transferrin (Tf), as a source of iron, for proliferation (Schell et al., 1991). Interestingly, trypanosome glycogen synthase kinase (TbGSK3β), an AEE788-associated protein kinase (Katiyar et al., 2013), regulates Tf endocytosis (Guyett et al., 2016).
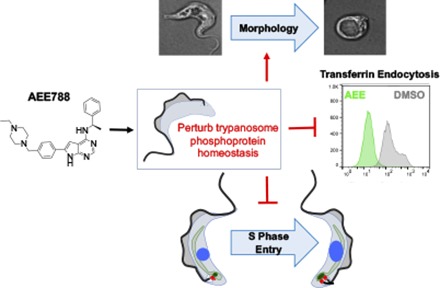
In our effort to understand the basis of AEE788 toxicity in T. brucei, we show that AEE788 blocks S-phase entry of bloodstream trypanosomes, inhibits Tf endocytosis, and alters cell morphology. Unexpectedly, we found that AEE788 could be used to enrich pre–S-phase trypanosomes. Using a novel “AEE788 block and release” protocol, we document the kinetics of DNA replication and subcellular organelle duplication in bloodstream trypanosomes. Finally, we show that AEE788 perturbs phosphoprotein homeostasis, offering insight into the putative effector proteins involved in AEE788-disrupted phospho-signaling pathways in the African trypanosome.
Materials and Methods
Parasite Cultures.
Bloodstream T. brucei, RUMP528 (Leal et al., 2001) or Lister 427, was cultured in HMI-9 medium supplemented with 10% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA), 10% Serum Plus (SAFC Biosciences, Lenexa, KS), and 1% antibiotic-antimycotic solution (Corning, Corning, NY) at 37°C, 5% CO2 (Hirumi and Hirumi, 1994). For all experiments, trypanosomes were harvested in logarithmic phase (i.e., less than 1 × 106 cells/ml).
Time-Dependent Inhibition of Trypanosome Proliferation at a Cytostatic Concentration of AEE788.
T. brucei were resuspended at 5 × 105 cells/ml (5 ml), in a Corning 25-cm2 culture flask, and treated with AEE788 (Novartis, Basel, Switzerland) to achieve a final concentration of 5 µM or equal volume (0.1%) of the drug solvent dimethylsulfoxide (DMSO) (Thermo Fisher, Waltham, MA). Cells were incubated at 37°C in 5% CO2. Trypanosome density was measured with a hemocytometer after 4, 9, and 16 hours of incubation. Both sides of the hemocytometer were counted twice and averaged for every time point. Biologic replicates were performed twice.
4′,6-Diamidino-2-Phenylindole Staining of DNA in the Kinetoplast and Nucleus After AEE788 Treatment.
T. brucei (5 × 105 cells/ml) was treated with AEE788 (5 µM), or an equal volume (0.1%) of DMSO (drug solvent) for 4 hours at 37°C in 5% CO2. Treated cells were pelleted (3000g for 5 minutes), resuspended in 1 ml of 4% paraformaldehyde (PFA) (Affymetrix, Santa Clara, CA) in phosphate-buffered saline (PBS) (Thermo Fisher), and incubated for 15 minutes at room temperature. Cells were pelleted by centrifugation, as described previously, and adhered to poly-l-lysine (Sigma-Aldrich, St. Louis, MO)–coated coverslips for 15 minutes. Coverslips were briefly washed with PBS before being mounted onto microscope slides with VectaShield Mounting Medium (Vector Laboratories, Burlingame, CA), containing 1.5 µM 4′,6-diamidino-2-phenylindole (DAPI) to stain nuclear and kinetoplast DNA. Trypanosomes were visualized with a high-sensitivity interline camera on an EVOS fluorescence (EVOS FL) microscope (Life Technologies, Grand Island, NY). The number of kinetoplasts and nuclei per cell, in 150 trypanosomes, were scored in four independent experiments.
Time-Course for Duplication of the Kinetoplast and Nucleus.
After a 4-hour treatment with AEE788 (5 µM), trypanosomes were washed twice and resuspended in drug-free HMI-9 medium (5 × 105 cells/ml). Cells were returned to an incubator (37°C, 5% CO2) for 1, 2, 3, 4, 5, or 6 hours. Cells were fixed and stained with DAPI as described above. Trypanosomes (150) were scored based on their number of kinetoplasts and nuclei (n = 3 for each time point).
Detection of DNA Synthesis with 5-Ethynyl-2′-Deoxyuridine.
Bloodstream trypanosomes (5 × 105 cells/ml) were treated with AEE788 (5 µM) or DMSO (0.1%) for 4 hours at 37°C in 5% CO2. 5-Ethynyl-2′-deoxyuridine (EdU) (300 µM) (Life Technologies) and 2′-deoxycytidine (200 µM) (Sigma-Aldrich) were added to both DMSO- and AEE788-treated samples 3.5 hours into the 4-hour treatment (i.e., 30-minute labeling period). After the 4-hour incubation, cells were washed once in PBS supplemented with 1% glucose (PBSG), fixed with 4% PFA in PBS (15 minutes), adhered to poly-l-lysine–coated coverslips, and permeabilized with 0.5% Triton X-100 (Thermo Fisher) in PBS for 25 minutes at room temperature. Permeabilized trypanosomes were washed with PBS and incubated in the dark for 30 minutes in a click-iT reaction cocktail, as follows: 4 mM copper sulfate (Sigma-Aldrich); 60 µM azide conjugated to Alexa Fluor 488 (Life Technologies); 1× Tris-buffered saline (20 mM Tris base; Genesee Scientific, San Diego, CA); 0.14 M NaCl (Sigma-Aldrich); and 300 mM ascorbic acid (Avantor Performance Materials, Center Valley, PA). Cells were washed three times in PBS (3 minutes each) before mounting with VectaShield Mounting Medium, containing DAPI (1.5 µM), onto microscope slides. Cells were visualized by fluorescence microscopy on the Applied Precision DeltaVision II Microscope System (GE Healthcare, Issaquah, WA) on an IX-71 inverted microscope (Olympus, Center Valley, PA). Images were captured with a cooled charge-coupled device (CCD) camera. The kinetoplast and nucleus of each trypanosome (n = 100-150) were characterized as EdU positive or EdU negative in three independent experiments.
Time-Course of DNA Synthesis.
Trypanosomes were treated for 4 hours with AEE788 (5 µM), washed twice, and resuspended in drug-free HMI-9 medium (5 × 105 cells/ml). Trypanosome aliquots (2 × 106 cells) were harvested every hour over a 3-hour time-course (1–4 hours after AEE788 washout) and incubated in medium containing EdU (300 µM) and 2′-deoxycytidine (200 µM) for 30 minutes (37°C, 5% CO2). Cells were subsequently processed as described above. Cells were first collected from 0 to 4 hours after AEE788 washout to identify the range of DNA synthesis. Subsequently, trypanosomes were harvested from 0 to 3 hours after AEE788 washoff (n = 3) to monitor the initiation of DNA synthesis. Additionally, cells were collected between 2 and 4 hours after AEE788 washout (n = 2) in attempts to detect termination of DNA synthesis. The kinetoplast and nucleus of 100–150 trypanosomes were scored as EdU positive or EdU negative at each time point for all experiments (0 and 2 hours, n = 6; 1 hour, n = 4; 3 and 4 hours, n = 3).
Immunofluorescence Detection of Basal Bodies and Bilobes.
Trypanosomes (5 × 105 cells/ml) were treated with AEE788 (5 µM) or an equal volume (0.1%) of DMSO (drug solvent) for 4 hours at 37°C, 5% CO2. Cells were washed once with PBSG, adhered to poly-l-lysine–coated coverslips for 5 minutes, quickly air dried, and fixed with methanol (Thermo Fisher) for 20 minutes at −20°C. Coverslips were briefly rinsed with PBS and rehydrated in blocking buffer [1% bovine serum albumin (BSA) (Sigma-Aldrich) in PBS] for 1 hour. Permeabilized trypanosomes were either costained with the primary antibodies YL1/2 (EMD Millipore, Billerica, MA) (Andre et al., 2014) and anti-TbSAS6 (Hu et al., 2015a) to detect basal bodies or stained with 20H5 (EMD Millipore) (He et al., 2005) for bilobes. The TbSAS6 antibody was a gift from Dr. Ziyin Li (University of Texas Health Science Center, San Antonio, TX). Antibodies were diluted (YL1/2 at 1:1000; anti-TbSAS6 and 20H5 at 1:500) in blocking buffer and incubated with cells for 1 hour at room temperature. Cells were rinsed three times, 5 minutes each, in PBS prior to exposure to the secondary antibody at a dilution of 1:2000 in blocking buffer for 1 hour at room temperature: Alexa Fluor 488 goat anti-rat and Alexa Fluor 594 goat anti-rabbit or Alexa Fluor 488 goat anti-mouse, respectively (Molecular Probes, Eugene, OR). Cells were rinsed three 2times, 5 minutes each, in PBS and mounted onto microscope slides with VectaShield Mounting Medium supplemented with DAPI (1.5 µM). Cells were then visualized with a DeltaVision Microscope System II, at the Biomedical Microscopy Core at the University of Georgia, and images were captured with a cooled CCD camera. The number of basal bodies and bilobes were quantitated in three independent experiments (100–150 trypanosome quantitated per experiment). Basal bodies were considered mature if they were colabeled with YL1/2 and anti-TbSAS6 or if they were labeled by YL1/2 alone. Basal bodies labeled solely by anti-TbSAS6 were counted as probasal bodies.
Time-Course of Basal Body and Bilobe Duplication.
Trypanosomes (5 × 105 cells/ml) were treated with AEE788 (5 µM), for 4 hours (37°C, 5% CO2), washed twice in drug-free HMI-9 medium, and resuspended in drug-free medium. Cells were returned to the incubator for 0, 2, 2.5, 3, or 3.5 hours, collected, and prepared for immunofluorescence assays as described above (YL1/2 and anti-TbSAS6 double labeling or 20H5 staining). The time-course was repeated in three independent experiments with the number of basal bodies (in YL1/2- and anti-TbSAS6–stained cells) and bilobes (in 20H5-stained cells) assessed in 100–150 trypanosomes at each time point for all experiments.
Analysis of Time-Course Studies Using Nonlinear Regression Curve Fitting.
Nonlinear regression curves were applied to time-course data documenting the recovery of DNA synthesis and organelle duplication (kinetoplast, basal body, bilobe, and nucleus) after an “AEE788 block and release” protocol (see above) using GraphPad Prism. GraphPad was used to calculate the time at which 50% of the observed maximum is reached (T50) (e.g., DNA synthesis) and was achieved based on a sigmoidal function. Calculations for kinetoplast elongation (measured by the percentage of 1Ke1N cells) and cytokinesis (based on the reappearance of 1K1N cells) were based on time points between 0 and 4 hours and 5 and 6 hours, respectively, when the minimum and maximum for these events were observed (a third-order polynomial nonlinear regression was used to show data trends for these events). Based on the T50 and the Hill slope (provided by GraphPad Prism), we calculated the time at which 10% (T10) and 90% (T90) of the maximum were achieved using the following equation provided by GraphPad Software: Tx = ((x/100 − x)1/H)T50, where H is the Hill slope and x is the desired percentage (of maximum).
Assessment of Cell Viability After AEE788 Treatment.
Trypanosomes (5 × 105 cells/ml) were treated with AEE788 (5 µM) or an equal volume (0.1%) of DMSO (drug solvent) for 4, 9, or 16 hours. Thereafter, cells from each treatment group (1 ml each) were aliquoted into 1.5 ml microcentrifuge tubes and treated with propidium iodide (PI) (3 µM) (Sigma Aldrich). Cells were immediately incubated on ice for 15 minutes and analyzed using a Beckman Coulter (Brea, CA) CyAn Flow Cytometer to measure PI fluorescence. FlowJo software (FlowJo, LLC, Ashland, OR) was used to gate live cell populations based on size and shape (forward and side scatter) and to quantitate the fluorescence intensity of PI in 10,000 trypanosomes (n = 2).
Evaluation of Trypanosome Endocytosis of Tf, BSA, and Tomato Lectin.
Trypanosomes were treated with AEE788 (5 µM) or equal volume (0.1%) of DMSO (drug solvent) for 9 hours (37°C, 5% CO2). Cells were washed and resuspended in serum-free HMI-9 medium devoid of AEE788 or DMSO (5 × 105 cells/ml). Trypanosomes were incubated with fluorescent endocytic cargo for 15 minutes at 37°C, 5% CO2: 25 µg Tf-Alexa Fluor 488 Conjugate (Thermo Fisher), 25 BSA labeled with Alexa Fluor 647 (Thermo Fisher), or 10 µg DyLight 488-tomato lectin (TL) (Vector Laboratories). Cells were subsequently transferred to an ice-water bath and washed with cold PBSG at 4°C (3000g for 5 minutes). Cells were resuspended in 1 ml PBSG, with PI (3 µM), as a marker for nonviable cells, and analyzed on the Beckman Coulter CyAn Flow Cytometer. FlowJo software (FlowJo, LLC) was used to gate viable trypanosome populations, based on size, shape (forward and side scatter), and PI exclusion. Fluorescence intensity of endocytic cargo was measured only in viable cell populations (negative for PI uptake). FlowJo was then used to determine the median fluorescence intensity of each endocytic cargo in trypanosome populations (15,000 events, n = 3).
Quantitation of Changes in Trypanosome Morphology.
Trypanosomes (5 × 105 cells/ml) were treated with AEE788 (5 µM) or an equal volume (0.1%) of DMSO (drug solvent) for 4, 9, or 16 hours. After each incubation period, cells were transferred to a hemocytometer and visualized (live) with an EVOS XL Core microscope (Thermo Fisher). Cells (100/incubation period) were scored based on morphology in two independent experiments.
Immunofluorescence Detection of the Paraflagellar Rod and Flagellum.
Trypanosomes (5 × 105 cells/ml) were treated with DMSO or AEE788 for 16 hours (37°C, 5% CO2), washed with PBSG and adhered to poly-l-lysine–coated coverslips (5 minutes). Once adhered, cells were quickly air dried and fixed with methanol for 20 minutes at −20°C. Cells were rehydrated in blocking buffer (PBS supplemented with 1% BSA) for 1 hour. Subsequently, trypanosomes were incubated with anti-paraflagellar rod (PFR) 2 (1:500) and 20H5 (1:500) in blocking buffer for 1 hour at room temperature. The polyclonal rabbit antibody against PFR2 was generated by GenScript (Piscataway Township, NJ). Trypanosomes were washed three times, 5 minutes each, in PBS before addition of fluorescent secondary antibodies (1:2000 in blocking buffer) for 1 hour at room temperature (Alexa Fluor 488 goat anti-rabbit or Alexa Fluor 594 goat anti-mouse, respectively). Cells were washed three times in PBS, 5 minutes each, prior to mounting onto microscope slides with VectaShield Mounting Medium containing DAPI (1.5 µM). Cells were visualized by fluorescence microscopy on an Applied Precision DeltaVision II Microscope System (GE Healthcare) with an Olympus IX-71 inverted microscope. Images were captured with a cooled CCD camera.
Scanning Electron Microscopy.
T. brucei (5 × 105 cells/ml) cells were treated with AEE788 (5 µM) for 12 hours in HMI-9 medium. Cells were centrifuged (1500g for 5 minutes) and washed with ice-cold PBSG. Cells were fixed with 2% glutaraldehyde in PBS for 1 hour at room temperature, washed with PBS, and adhered to poly-l-lysine–coated coverslips. Cells on coverslips were treated with OsO4 (1%) for 30 minutes at room temperature, washed three times in water, and dehydrated with increasing concentrations of ethanol by incubating them sequentially in 25%, 30%, 50%, 75%, 85%, 95%, and 100% ethanol for 5 minutes each. The samples were dried at a critical point with a Tousimis (Rockville, MD) Critical Point Dryer (Samdri-780 A), and sputter coated (gold) with an SPI Module Sputter Coater (SPI Supplies, West Chester, PA) following standard protocols. Samples were viewed using a Carl Zeiss (Jena, Germany) 1450EP variable pressure scanning electron microscope at the Georgia Electron Microscopy at the University of Georgia (Athens, GA).
Phosphopeptide Enrichment and Identification in AEE788-Treated Trypanosomes.
Trypanosomes (5 × 105 cells/ml) were treated with either AEE788 (5 µM) or equivalent volume (0.1%) DMSO (drug solvent) at 37°C (4 or 9 hours). Trypanosomes (2 × 108 cells) were moved to ice, washed with cold PBSG containing 1× HALT phosphatase inhibitor cocktail (PIC) (Thermo Fisher), lysed by sonication in 50 mM HEPES (Thermo Fisher), pH 7.6, 8 M urea (Thermo Fisher), 4 mM dithiothreitol (Sigma-Aldrich), 1× HALT PIC and alkylated with 9 mM iodoacetamide (Bio-Rad, Hercules, CA) for 30 minutes (away from light). The lysate was diluted five-fold with 50 mM HEPES, pH 7.6, and 1× HALT PIC (1.6 M urea final) followed by protein digestion with immobilized trypsin agarose (Thermo Fisher) for 48 hours at room temperature. After collecting the beads by centrifugation, the peptide supernatant was diluted 10-fold with 0.1% trifluoroacetic acid (TFA) (Thermo Fisher) and desalted over a Sep-Pak C18 column (Waters, Milford MA). A step gradient of acetonitrile (25% followed by 50%) (Thermo Fisher) was used to elute peptides. Eluates were dried via vacuum centrifugation. Phosphopeptides were then enriched by FeCl3-charged metal affinity chromatography (IMAC) made in-house (Proteomics Core at Fred Hutchinson Cancer Research Center). Briefly, peptide samples were resuspended in 80% acetonitrile and 0.1% TFA and were loaded onto FeCl3-charged IMAC resin (10 µl bed volume). The resin was washed three times with 150 µl of 80% acetonitrile in 0.1% TFA, then received a final wash with 1% TFA (150 µl). The peptides were eluted twice (3 minutes each) with 150 µl of 500 mM potassium phosphate (pH 7) and desalted using ZipTip C18 (EMD Millipore) before mass spectrometry analysis.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis was performed with an Easy-nLC 1000 (Thermo Scientific) coupled to an Orbitrap Elite Mass Spectrometer (Thermo Scientific). The liquid chromatography system (New Objective, Inc., Woburn, MA) was configured in a vented format (Licklider et al., 2002) consisting of a fused-silica nanospray needle (50 µm i.d.; PicoTip emitter; New Objective) packed in-house (Fred Hutchinson Proteomics Facility) with Magic C18 AQ 100-Å reverse-phase medium (Michrom BioResources, Auburn, CA) (25 cm), and a trap (100 µm i.d.; IntegraFrit Capillary; New Objective) containing Magic C18 AQ 200Å (2 cm). The peptide sample was diluted in 10 µl of 2% acetonitrile and 0.1% formic acid in water, and 8 µl was loaded onto the column for separation using a two-mobile phase system consisting of 0.1% formic acid in water (A) and 0.1% acetic acid in acetonitrile (B). A 60- or 90-minute gradient from 7% to 35% acetonitrile in 0.1% formic acid at a flow rate of 400 nl/min was used for chromatographic separation. The mass spectrometer was operated in a data-dependent MS/MS mode over the m/z range of 400–1800 at the 240,000 mass resolutions. For each cycle, the 20 most abundant ions from the scan were selected for MS/MS analysis using 35% normalized collision energy. Selected ions were dynamically excluded for 30 seconds.
Raw MS/MS data were analyzed with Proteome Discoverer software version 1.4 (Thermo Fisher) using SEQUEST (Eng et al., 1994) as a search engine against TriTrypDB database version 4.1 (from TritrypDB.org), which included common contaminants such as human keratin. The database contained 8614 protein entries including contaminants. The following modifications were considered: carbamidomethylation of cysteine as a fixed modification; and phosphorylation of serine, threonine, tyrosine, and oxidation of methionine as variable modifications. The enzyme was set to trypsin, allowing up to two missed cleavages. The precursor and fragment mass tolerances were set to 10 ppm and 0.6 Da, respectively. Search results were run through Percolator (Käll et al., 2007) for scoring. The results were filtered for peptides identified with a false discovery rate lower than 0.05. Phosphorylation sites were evaluated and probability values were calculated using PhosphoRS version 3.1 (Taus et al., 2011). Specific phosphorylation sites in Table 1 were assigned if the PhosphoRS probability for the site was 80% or greater.
TABLE 1.
Select examples of phosphoproteins affected by short-term (4 h) AEE8788 treatment
After treatment of trypanosomes with DMSO (0.1%) or AEE788 (5 µM) for 4 h, peptides were harvested and phosphopeptides were enriched over an IMAC column. LC-MS/MS was used to monitor the abundance of phosphopeptides in three independent experiments. Spectral counts indicate the combined number of times a phosphopeptide was observed over all experiments. The number in parenthesis indicates the total number of peptides observed for the parent protein over all experiments (summation of all peptides observed in the IMAC elution and flow through). (Taus et al., 2011) probability ≥80%]. A Student’s t test was used to determine whether the change in phosphopeptide abundance was statistically significant (P < 0.05).
| Gene ID | Production Description | Identified Phosphopeptide | Spectral Counts |
P Value | |
|---|---|---|---|---|---|
| DMSO | AEE788 | ||||
| Decreased | |||||
| Tb427.06.4970 | SRPK | HsASTNGPSQPAHQR | 6 (15) | 1 (3) | 0.038 |
| Tb427.10.3010 | Bilobe protein | sRISTGISFLSK | 5 (18) | 0 (7) | 0.038 |
| Tb427tmp.02.0810 | TbSAS4 | LAVGDANHSESIGDKSVstK | 8 (12) | 2 (3) | 0.013 |
| Increased | |||||
| Tb427.01.2100 | Calpain-like cysteine peptidase | AEEASPAPSPAGEsDEKAsKSEHESEAK | 20 (88) | 44 (99) | 0.03 |
SRPK, serine-arginine protein kinase.
Statistical Analysis.
To quantitate the effect of AEE788 on organelle (basal body, bilobe, nucleus, kinetoplast) duplication and trypanosome morphology, the distribution of cells was grouped according to organelle content per trypanosome or trypanosome shape after treatment with drug or DMSO. To determine whether AEE788 caused statistically significant changes in these distributions, we compared the distribution obtained after exposure to AEE788 to that observed after treatment with DMSO (i.e., control) using the Pearson χ2 test of independence (⍺ = 0.0005).
A two-sample Student’s t test was used to compare the median fluorescence of endocytosed cargo (measure of internalization) between DMSO- and AEE788-treated cells (⍺ = 0.005).
Results
AEE788 Inhibits Kinetoplast Duplication.
Our primary objective in these studies was to characterize pharmacological effects of AEE788 on bloodstream trypanosomes. To achieve this goal, it was necessary to work with higher cell densities, and therefore higher drug concentrations, than previously used in proliferation inhibition assays (Behera et al., 2014) to provide adequate numbers of trypanosomes for follow-up phenotypic evaluation. We first identified the optimal AEE788 concentration and treatment time for “mode of action” studies (conditions that inhibit proliferation without death, thereby providing an opportunity to characterize disrupted pathways in living cells). We found that AEE788 (5 µM) arrested proliferation between 4 and 9 hours of treatment, but beyond 9 hours the drug caused cell density to decrease (Supplemental Fig. 1). These data indicate that AEE788 halts bloodstream trypanosome division within a single duplication cycle (∼6–7 hours) (Hesse et al., 1995; Ajoko and Steverding, 2015).
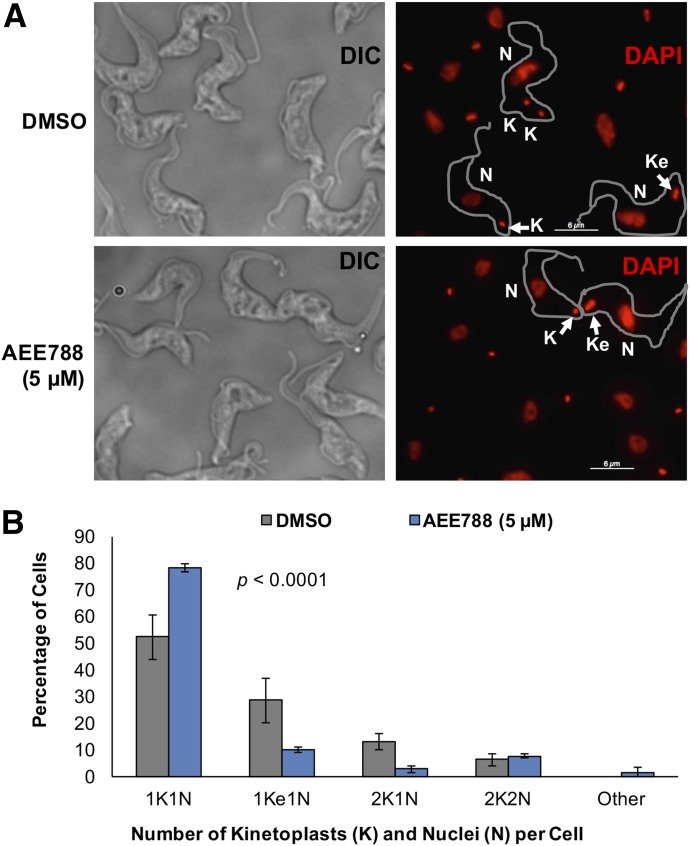
One hypothesis to explain the inability of cells to proliferate in the presence of AEE788 (Supplemental Fig. 1) is that trypanosomes fail to progress through a specific point in the division cycle. To determine whether AEE788 interfered with the cell division cycle, we used DAPI to quantitate the number of kinetoplasts and nuclei per cell. After a 4-hour incubation with AEE788 the proportion of trypanosomes with one round kinetoplast (1K) and a single nucleus (1N) increased, compared with control cells treated with DMSO (drug vehicle) (Fig. 1A). Quantitation of the percentage of cells with each “karyotype” (i.e., number of kinetoplasts and nuclei) demonstrated that AEE788 caused a statistically significant change in the cell type distribution compared with the control population (p = 7.4 × 10−19). The proportion of cells with a 1K1N configuration (i.e., G1 trypanosomes) increased from 52.2% to 78.2% (Fig. 1B). Concomitantly, the percentage of cells in S-phase (i.e., 1Ke1N cells) (Siegel et al., 2008; Kaufmann et al., 2012) dropped from 28.6% to 9.8% (Fig. 1B). A decrease in the percentage of 2K1N cells, from 13% in the control (DMSO-treated) to 2.7%, after AEE788 treatment indicated that kinetoplast duplication was blocked (Fig. 1B). In contrast, the proportion of postmitotic trypanosomes (2K2N) was unchanged during the 4-hour AEE788 treatment, implying that mitosis was not affected (Fig. 1B).
Fig. 1.
AEE788 blocks kinetoplast elongation and division. Trypanosomes (5 × 105 cells/ml) were treated with AEE788 (5 µM) or DMSO (0.1%), in HMI-9 medium, for 4 hours. Cells were fixed in PFA, and the kinetoplast and nuclear DNA were stained with DAPI. The number of kinetoplasts and nuclei in 150 trypanosomes were quantitated. (A) Representative images of DAPI-stained trypanosomes after treatment with DMSO (top) or AEE788 (bottom). Scale bar, 6 µm. (B) The average percentage of trypanosomes within each kinetoplast (K) and nucleus (N) configuration are shown. Error bars represent S.D. among four biologic replicates. The distribution of kinetoplasts and nuclei (per trypanosome) in DMSO-treated and AEE788-treated cells was compared using a Pearson χ2 test (p = 7.4 × 10−19).
AEE788 Prevents DNA Synthesis in the Kinetoplast and Nucleus.
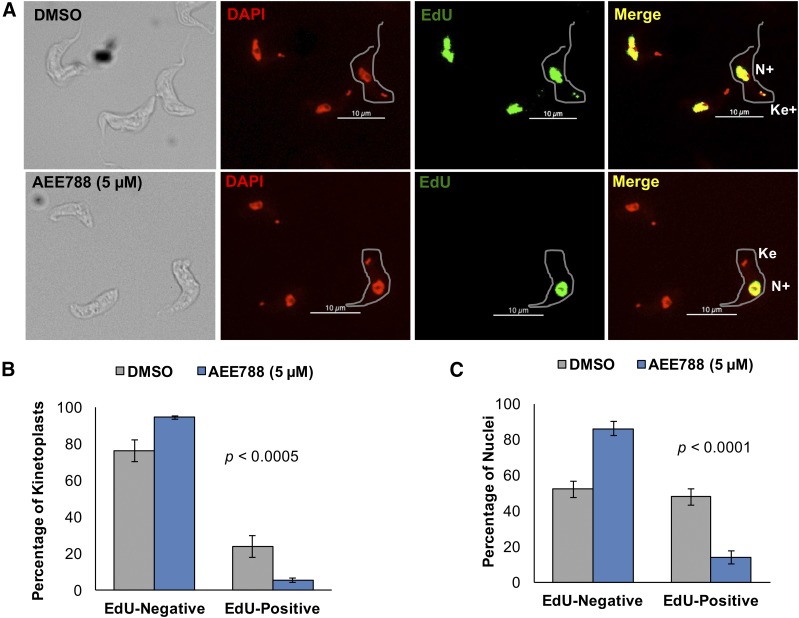
Failure of the kinetoplast to elongate after AEE788 treatment (Fig. 1) led us to hypothesize that the drug impairs kDNA synthesis. We tested this hypothesis by labeling kinetoplast and nuclear DNA with a thymidine analog, EdU (Cavanagh et al., 2011), in the absence or presence of AEE788 (Fig. 2A). EdU labeling was performed for 30 minutes to detect newly synthesized DNA. Unlike nuclear incorporation of EdU, which can be visualized throughout the nucleus, kDNA incorporation of EdU is limited to the ends of the kinetoplast DNA network (Fig. 2A) where newly synthesized minicircles are attached (for review, see Liu et al., 2005).
Fig. 2.
AEE788 decreases DNA synthesis in the kinetoplast and nucleus. Trypanosomes were treated with AEE788 (5 µM) or DMSO (0.1%) for 4 hours. EdU and 2′-deoxycytidine were added to both cultures during the last 30 minutes of treatment. Incorporated EdU was detected in a click-iT reaction with a fluorescent azide. (A) Kinetoplasts (K) and nuclei (N) were scored as EdU-positive (K+ or N+) or EdU-negative after treatment with DMSO (top) or AEE788 (bottom). Scale bar, 10 µm. (B) Quantitation of the average percentage of trypanosomes (n = 125) with EdU-negative or EdU-positive kinetoplasts after AEE788 or DMSO treatment. (C) Quantitation of the average proportion of cells (n = 125) with EdU-negative or EdU-positive nuclei after AEE788 or DMSO treatment. Error bars denote the S.D. in three independent experiments. Differences in the distribution of trypanosomes with EdU-positive and EdU-negative kinetoplasts or nuclei in cell populations treated with DMSO or AEE788 were assessed with a Pearson χ2 test (p = 4.9 × 10−4 for the kinetoplast, and p = 3.1 × 10−19 for the nucleus).
In control trypanosomes (treated with DMSO), 23.8% incorporated EdU into the kDNA network (proportional to the number of 1Ke1N cells) (Fig. 1B), while only 5.5% of kinetoplasts in AEE788-treated trypanosomes incorporated EdU (Fig. 2B). The distribution of replicating and nonreplicating kinetoplasts was significantly altered, compared with control trypanosomes, by AEE788 treatment (p = 4.9 × 10−4). Nuclear DNA synthesis was also inhibited by AEE788 treatment. Only 14% of AEE788-treated trypanosomes incorporated EdU in the nucleus compared with 52.2% in the control (Fig. 2C), leading to a statistically significant difference in the distribution of S-phase nuclei (p = 3.1 × 10−19). Inhibition of DNA synthesis in both trypanosome DNA-containing organelles suggests that AEE788 impairs entry into the S-phase of the cell cycle, as the protein factors and DNA origins needed for DNA replication in the nucleus and kinetoplast differ (for review, see Jensen and Englund, 2012; Povelones, 2014; Tiengwe et al., 2014). In DMSO-treated populations, the percentage of cells synthesizing kDNA is approximately 50% of the proportion synthesizing nuclear DNA (Fig. 2, B and C). This observation may be explained by the fact that: 1) the time-course of DNA synthesis differs between kDNA and chromosomal DNA (Fig. 5, B and C) (Woodward and Gull, 1990); and 2) the sensitivity of EdU detection is higher in the nucleus, which contains more DNA (Borst et al., 1982).
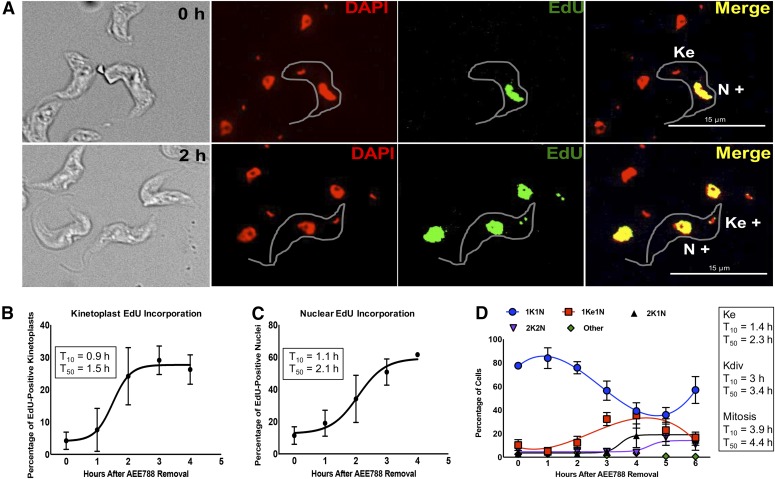
Fig. 5.
Time-course of DNA replication and division in the kinetoplast and nucleus after withdrawal of AEE788. Trypanosomes were treated with AEE788 (5 µM, 4 hours), rinsed, and placed in drug-free HMI-9 medium. The time-course of DNA synthesis was monitored by EdU incorporation, and DAPI was used to visualize the division of the kinetoplast and nucleus. (A) Representative images of trypanosomes directly after AEE788 treatment (0 hour, top) or 2 hours after AEE788 washout (bottom). Kinetoplasts and nuclei, in 100–150 trypanosomes, were scored as EdU-positive (K+ or N+) or EdU-negative. Scale bar, 15 µm. The average proportion of cells with EdU-positive kinetoplasts (B) or nuclei (C), are indicated at every hour after AEE788 withdrawal. S.D.s between independent experiments are shown (n = 6 for 0 and 2 hours; n = 4 for 1 hour; n = 3 for 3 and 4 hours). A sigmoidal nonlinear regression curve was fit to the data points using GraphPad Prism, and the time at which 10% (T10) or 50% (T50) of the population became EdU positive, compared with the observed maximum (4 hours), was calculated for kinetoplast (B) and nuclear (C) EdU incorporation. (D) The average percentage of cells (n = 115) with the indicated kinetoplast (K) and nucleus (N) configuration is shown for every hour after AEE788 withdrawal. The S.D. represents variation among three independent experiments. Data trends are represented by nonlinear regression curves using a third-order polynomial equation for 1K1N and 1Ke1N data, and a sigmoidal nonlinear regression curve for 2K1N and 2K2N populations. T10 and T50 values (calculated by GraphPad software) for each event are listed [T10 and T50 for the appearance of 1Ke1N cells is based on a sigmoidal nonlinear regression curve from 0 to 4 hours (maximum)].
Effect of AEE788 on Duplication of the Basal Body and Bilobe.
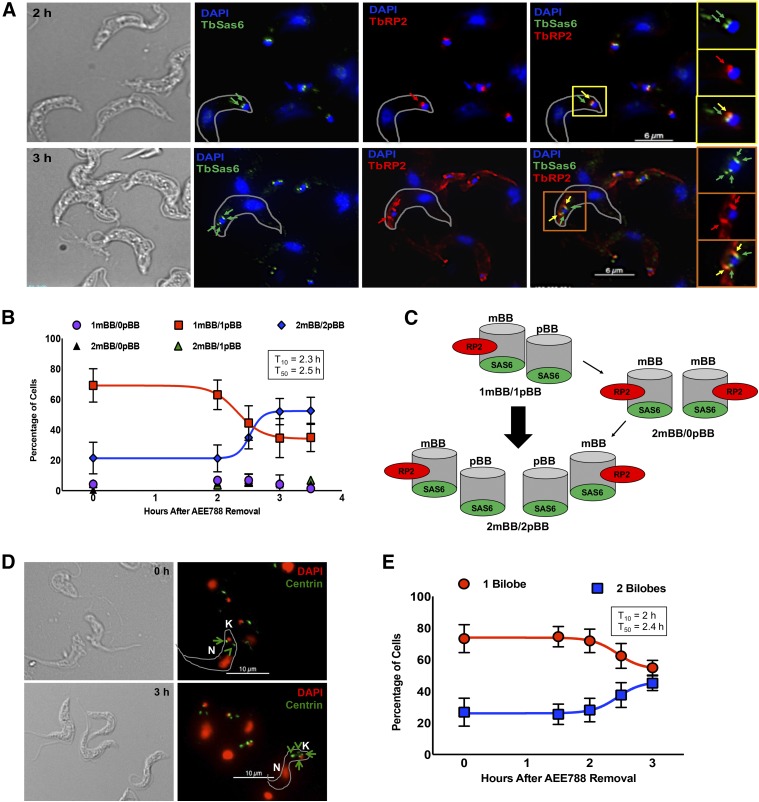
Trypanosomes in G1 have a single mBB paired with an immature pBB, with each containing TbSAS6. TbRP2 (recognized by the antibody YL1/2) (Andre et al., 2014) is localized to transitional fibers, found only on mBBs (Andre et al., 2014). Maturation of the pBB is thought to precede assembly of new ones (Sherwin and Gull, 1989; Lacomble et al., 2010; Gluenz et al., 2011; Ikeda and de Graffenried, 2012). Thus, trypanosomes with two mBBs lacking adjacent pBBs (2 mBBs/0 pBB) are thought to arise first as intermediates in the biogenesis of the organelle. Subsequently, a new pBB is assembled adjacent to each mBB to form 2 mBB/2 pBB trypanosomes. Migration of each mBB/pBB pair away from each other correlates with scission of the kinetoplast (Sherwin and Gull, 1989; Robinson and Gull, 1991; Lacomble et al., 2010; Gluenz et al., 2011). Given that AEE788 blocked division of the kinetoplast (Fig. 1), we hypothesized that the drug inhibited basal body duplication. We tested this possibility using immunofluorescence to detect the number of mBBs and pBBs per trypanosome (Fig. 3A).
Fig. 3.
AEE788 prevents basal body duplication. Anti-TbSAS6 and YL1/2 were used to quantitate the number of mBBs and immature pBBs per trypanosome after treatment with AEE788 (5 µM) or DMSO (0.1%). (A) Representative staining pattern of YL1/2 (red) and anti-TbSAS6 (green) after DMSO (top) or AEE788 (bottom) treatment. Cells are counterstained with DAPI (1.5 µM). Red arrows indicate TbRP2+ foci (mBB), green arrows indicate TbSAS6+ foci (pBB), and yellow arrows indicate colocalization of TbRP2 and TbSAS6 (mBB). Scale bar, 6 µm. The basal bodies and associated kinetoplast (K) are enlarged in a single trypanosome for both treatment groups: DMSO (yellow boxes) and AEE788 (orange boxes). (B) Average percentage of trypanosomes (n = 125) with the indicated number of mBBs and pBBs after treatment with AEE788 or DMSO. Error bars represent the S.D. in three independent experiments. Statistical significance of changes in the distribution of the number of basal bodies (per cell) in the trypanosome population was determined with a Pearson χ2 test (p = 0.3 × 10−18).
The distribution of basal bodies (i.e., number of mBBs or pBBs per cell) was skewed toward trypanosomes with unduplicated basal bodies (1 mBB/1 pBB) after AEE788 treatment (p = 1.3 × 10−18). In a control population (exposed to DMSO), 35.5% of cells had one mBB and one pBB (1 mBB/1 pBB) (Fig. 3B). This population doubled to 73.5% after a 4-hour treatment with AEE788 (Fig. 3B). Additionally, the fraction of trypanosomes with 2 mBBs/2 pBBs dropped from 54.2% in the control to 21.5% in AEE788-treated trypanosomes (Fig. 3B). Infrequently, trypanosomes with 1 mBB/0 pBB or 2 mBBs/1 pBB were detected, likely a staining artifact, and these populations remained the same after DMSO or AEE788 treatment (Fig. 3B). The data indicate that, in the presence of AEE788, targeting of TbRP2 to the second basal body fails, possibly due to: 1) the absence of new transitional fibers and/or 2) an inability to deliver TbRP2 to new mBBs. Further, AEE788 prevents the assembly of new TbSAS6-positive pBBs in the absence of TbRP2 recruitment (Fig. 3B). Together, these data indicate that AEE788 inhibits basal body duplication by interfering with the recruitment of proteins to the organelle.
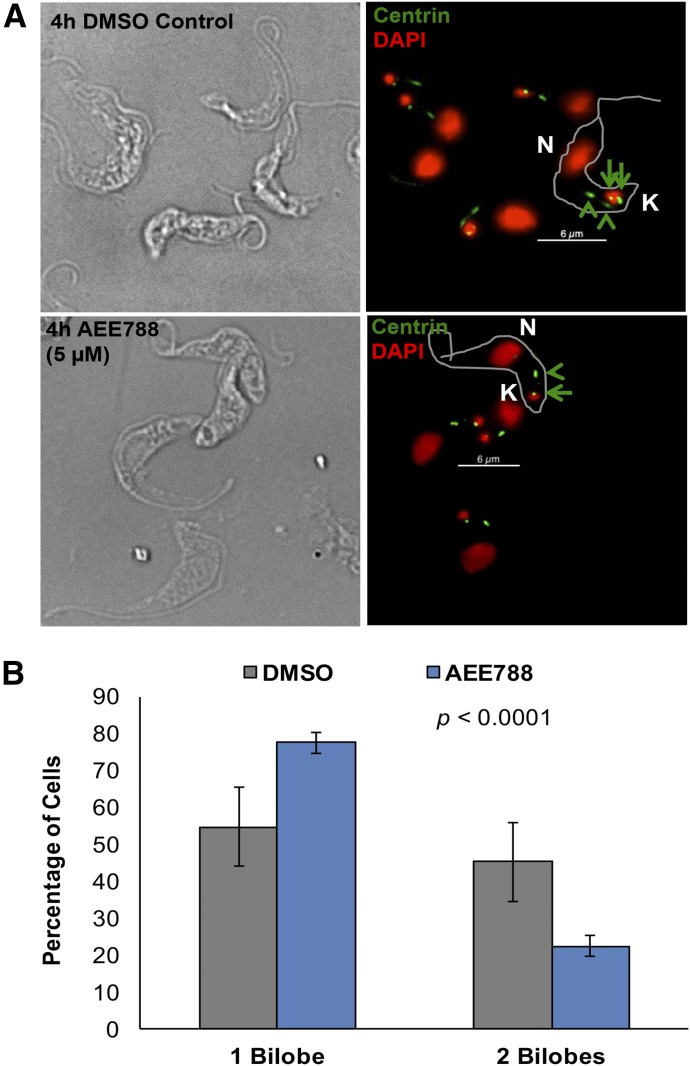
Failure of AEE788-treated trypanosomes to synthesize DNA (Fig. 2) indicated that the drug blocked the entry of trypanosomes into the S-phase. The bilobe, a centrin-containing cytoskeletal structure at the base of the flagellum (Esson et al., 2012), is duplicated in S-phase (Zhou et al., 2014). We postulated that because AEE788 prevented S-phase entry (Fig. 2) the drug would inhibit bilobe duplication. We tested this hypothesis by evaluating the effect of AEE788 on bilobe biogenesis using the antibody 20H5, which detects centrins at the bilobe and basal body (He et al., 2005) (Fig. 4A). AEE788 increased the fraction of trypanosomes with one bilobe from 54.7% to 77.5% and decreased the proportion of trypanosomes with two bilobes from 45.3% to 22.5% (Fig. 4B), a statistically significant change in the distribution of cells with unduplicated and duplicated bilobes (p = 3.6 × 10−9). We conclude that AEE788 prevents bilobe duplication.
Fig. 4.
Bilobe duplication is inhibited by AEE788. After a 4-hour treatment with AEE788 (5 µM) or DMSO (0.1%), trypanosomes were stained with the anti-centrin antibody 20H5 to detect the bilobe. (A) Representative images of 20H5-stained trypanosomes after treatment with DMSO (top) or AEE788 (bottom). Centrin is observed at the bilobe (green arrowheads) as well as the basal body (green arrows). DAPI was used to stain kinetoplast and nuclear DNA. K, kinetoplast; N, nucleus. Scale bar, 6 µm. (B) Average percentage of cells with one or two bilobes after AEE788 or DMSO treatment. Error bars represent S.D. among four biologic replicates. The distribution of trypanosomes with one or two bilobes in AEE788-treated cells was compared with the distribution observed in control cells (i.e., DMSO-treated) using a Pearson χ2 test (p = 3.6 × 10−9).
A Time-Course for DNA Synthesis and Duplication of Cytoskeletal Organelles during Trypanosome Division.
Experimental measurement of the kinetics of organelle duplication during bloodstream division has been hampered by the technical difficulties of enriching a pre–S-phase trypanosome population (Mutomba and Wang, 1996; Forsythe et al., 2009; Kabani et al., 2010; Archer et al., 2011). Discovery that AEE788 causes a buildup of pre–S-phase trypanosomes (Figs. 2–4) suggested that a “block-and-release” protocol using the drug might be valuable for time-course studies of organelle duplication during trypanosome division.
We first tested whether DNA synthesis would resume upon removal of AEE788 from the trypanosome culture, indicating re-entry into S-phase. For this objective, trypanosomes were treated with AEE788 (5 µM) for 4 hours, washed, and resuspended in drug-free HMI-9 medium. After AEE788 withdrawal, cell aliquots were obtained every hour and incubated with EdU (Cavanagh et al., 2011) for 30 minutes (1–4 hours after AEE788 washout). During the first hour after AEE788 removal, the percentage of trypanosomes that incorporated EdU into the kinetoplast (or nucleus) was similar to that observed immediately after AEE788 treatment (Fig. 5, A–C). However, by 2 hours the percentage of cells with EdU-positive kinetoplasts increased from 5%, immediately after AEE788 washout, to 25% (Fig. 5, A and B). Likewise, the number of nuclei that incorporated EdU increased from 12% to 35% (Fig. 5, A and C). Using a sigmoidal nonlinear regression curve, we estimated a time at which significant DNA synthesis [i.e., 10% of the observed maximum (4 hours) for EdU-positive kinetoplasts or nuclei] had occurred, designated as T10. Similarly, we defined the time by which the EdU-positive population increased to 50% (i.e., T50) or 90% (i.e., T90) compared with the observed maximum (4 hours). Initiation of nuclear DNA synthesis (T10 = 1.1 hours) and kDNA synthesis (T10 = 0.9 hour) occurred at similar times after AEE788 removal. However, the T50 for kinetoplast EdU incorporation (1.5 hours) was reached approximately 30 minutes earlier than that of nuclear incorporation (T50 = 2.1 hours), and it terminated an hour before nuclear DNA synthesis (T90 = 2.1 and 3 hours, respectively) (Fig. 5, B and C). These data are consistent with kinetoplast S-phase terminating prior to completion of nuclear DNA synthesis (Woodward and Gull, 1990). We next performed time-course experiments for the duplication of the kinetoplast (Fig. 5D), nucleus (Fig. 5D), basal body (Fig. 6, A and B), and bilobe (Fig. 6, D and E).
Fig. 6.
Kinetics of basal body and bilobe duplication. Trypanosomes were treated with AEE788 (5 µM, 4 hours) and then transferred to drug-free HMI-9 medium for up to 3 hours. Cells were retrieved every 30 minutes between 2 and 3 hours after AEE788 washout. (A) Basal body duplication was assessed with YL1/2 and anti-TbSAS6. YL1/2 recognizes mBBs (red arrows), and TbSAS6 localizes to immature pBBs (green arrows) and mBBs (yellow arrows). Scale bar, 6 µm. An unduplicated basal body (1 mBB/1 pBB) 1.5 hours after AEE788 washout (yellow boxes) and a duplicated basal body (2 mBBs/2 pBBs) 3 hours after AEE788 washout (orange boxes) are magnified. K, kinetoplast. (B) The average percentages of cells with the indicated number of mBBs and pBBs are shown at various times after AEE788 washout. Error bars represent the S.D. among three independent experiments. A sigmoidal nonlinear regression curve was fit to the data points in GraphPad Prism and the time by which 10% (T10) or 50% (T50) of the population, compared with the observed maximum, became 2 mBBs/2 pBBs is listed. (C) Schematic of nascent basal body duplication and pBB maturation (acquisition of TbRP2) occurring in the absence of intermediates with two mBBs and no pBBs (2 mBBs/0 pBB). (D) The anti-centrin antibody 20H5 was used to visualize bilobes. Green arrowheads indicate bilobes, green arrows point to basal bodies. K, kinetoplast; N, nucleus. Scale bar, 10 µm. (E) Quantitation of the average percentage of bilobes (BL) per cell after AEE788 withdrawal. Error bars represent the S.D. among three independent experiments. A sigmoidal nonlinear regression curve was fit to the data points in GraphPad Prism, and the T10 and T50 values, describing the formation of cells with duplicated bilobes, are provided.
Kinetoplast elongation (i.e., the appearance of 1Ke1N trypanosomes) was observed between 1 and 4 hours after AEE788 release (T10 = 1.4 hours; T50 = 2.3 hours; T90 = 3.3 hours) (Fig. 5D). From 1 to 4 hours, the fraction of 1Ke1N trypanosomes increased from 5% to 35% (Fig. 5D and Supplemental Fig. 2). Correspondingly, by 4 hours the 1K1N population was reduced from 77%, immediately after AEE788 withdrawal, to 39% (Fig. 5D). Kinetoplast division (defined as an increase in the percentage of 2K1N trypanosomes) was observed between 3 and 4 hours when the 2K1N population increased from 5% to 18.2% (T10 = 3 hours; T50 = 3.4 hours; T90 = 3.9 hours) (Fig. 5D and Supplemental Fig. 2). Mitosis was detectable between 4 and 5 hours with the number of 2K2N cells increasing from 2.7% to 17% (T10 = 3.9 hours; T50 = 4.4 hours; T90 = 5 hours), indicating that mitosis can be completed within 1 hour (Fig. 5D and Supplemental Fig. 2). Between 5 and 6 hours, the number of 1K1N trypanosomes increased (35.9 to 56.9%), with a simultaneous decrease in all other populations (Fig. 5D), demonstrating the completion of cytokinesis and the cell division cycle. These data are consistent with the 6- to 7-hour division time observed in bloodstream trypanosomes (Hesse et al., 1995; Ajoko and Steverding, 2015).
Basal bodies were costained using the antibody YL1/2 (for TbRP2-positive mBBs) (Andre et al., 2014) and anti-TbSAS6 (for mBBs and immature pBBs) (Hu et al., 2015a) (Fig. 6A). Immediately after AEE788 withdrawal, the majority of trypanosomes (73.4%) had 1 mBB/1 pBB, with 25.3% containing 2 mBBs/2 pBBs (Fig. 6B). Between 2 and 3 hours after AEE788 washout, the percentage of trypanosomes with 2 mBBs/2 pBBs increased from 24.4% to 56.6% (Fig. 6, A and B). A nonlinear regression analysis indicated that trypanosomes with 2 mBBs/2 pBBs emerged 2.3 hours (T10) after AEE788 removal and reached the observed maximum by 2.7 hours (T90) (Fig. 6B). Assuming that pBB maturation occurs prior to new pBB assembly (Sherwin and Gull, 1989; Lacomble et al., 2010; Gluenz et al., 2011; Ikeda and de Graffenried, 2012), one would expect to detect trypanosomes with two mBBs but no pBBs (2 mBBs/0 pBB). Surprisingly, we detected a small fraction of trypanosomes (<7%) with 2 mBBs/0 pBB (Fig. 6B). In fact, the kinetics of TbRP2 recruitment (a marker for pBB maturation) and assembly of new pBBs (monitored by TbSAS6) were remarkably similar (Supplemental Fig. 3). Our data suggest that TbRP2 recruitment to mBBs coincides with, and may be coordinated with, pBB assembly (Fig. 6C).
Bilobe duplication was examined using the anti-centrin antibody 20H5 (He et al., 2005) (Fig. 6D). During the first 2 hours after AEE788 washout, less than 30% of trypanosomes had two bilobes (Fig. 6E). By 3 hours, 45% of trypanosomes had two bilobes (T10 = 2 hours; T50 = 2.4 hours; T90 = 2.6 hours) (Fig. 6E). Thus, bilobe duplication occurs between 2 and 3 hours after release from an AEE788 block, coincident with basal body duplication (Fig. 7).
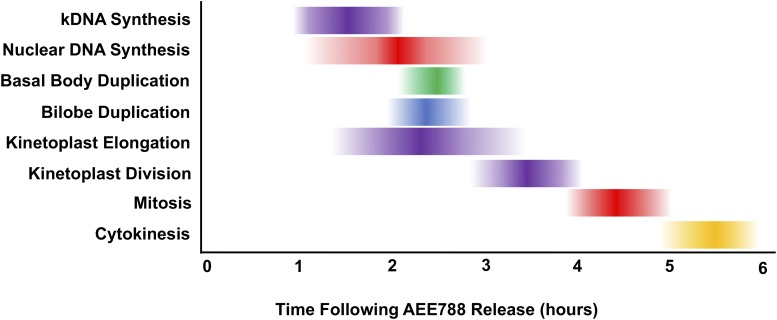
Fig. 7.
Time-course of major events in the trypanosome division cycle. AEE788 (5 µM, 4 hours) was used to block organelle duplication and DNA synthesis. After removing AEE788 from the medium, the onset and duration of organelle duplication and DNA synthesis were determined. The time at which, 10% (T10) (left border), 50% (T50), and 90% (T90) (right border) of the observed maximum was reached for each event was calculated, based on nonlinear regression curves (Figs. 5 and 6). The darkest shading corresponds to the T50 value (±S.E.).
A summary of the time-course for organelle duplication after AEE788 washout (Figs. 5 and 6) is presented in Fig. 7 based on the calculated T10, T50 and T90 values for each event. Briefly, kDNA synthesis and nuclear DNA synthesis begin at similar times after AEE788 removal (T10 = 0.9 hour and 1.1 hours, respectively). Kinetoplast elongation (T10 = 1.4 hours) is detected approximately 30 minutes after the start of kDNA synthesis and coincides with nuclear DNA synthesis (T50 = 2.3 and 2.1 hours, respectively). Basal body and bilobe duplication also occur during nuclear DNA synthesis (T50 = 2.5 and 2.4 hours, respectively). Termination of kDNA synthesis (T90 = 2.1 hours) is detected approximately 1 hour prior to the cessation of nuclear S-phase (T90 = 3 hours). The end of nuclear DNA synthesis marks the start of kinetoplast division (T10= 3 hours), which continues for 1 hour (T90 = 3.8 hours). Mitosis is completed within 1 hour (T10 = 3.9 hours; T90 = 5 hours). Last, cytokinesis occurs between 5 and 6 hours after trypanosomes have entered S-phase.
Trypanocidal Effects of AEE788 Are Associated with Endocytosis Defects and Changes in Cell Morphology.
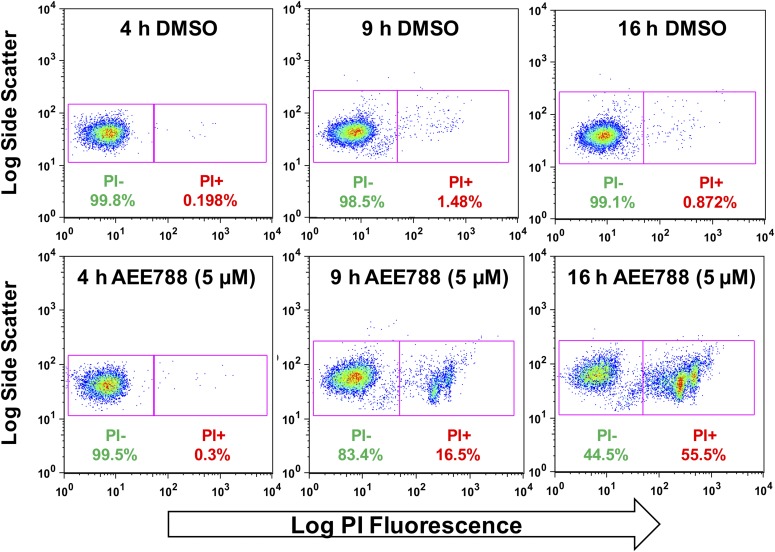
The ability of trypanosomes to resume division after a 4-hour treatment with AEE788 (Figs. 5 and 6) indicated that trypanosomes did not commit to death during that period of treatment. However, between 9 and 16 hours of AEE788 treatment trypanosome density decreases (Supplemental Fig. 1). Accordingly, we postulated that extended exposure to the drug was necessary to impair trypanosome viability. We tested this idea by staining trypanosomes with PI, which will not enter trypanosomes with an intact plasma membrane (Garner et al., 1986). By 4 hours, a small proportion of PI-positive trypanosomes (<0.4%) was observed in the control group (exposed to DMSO) as well as those treated with AEE788 (Fig. 8). After 9 hours of drug treatment, however, 16.5% of AEE788-treated trypanosomes (compared with 1.5% in the DMSO-treated controls) were positive for PI uptake, and by 16 hours half of the population stained with PI were positive (Fig. 8). We conclude that beyond 9 hours of treatment AEE788 (5 µM) decreases trypanosome viability.
Fig. 8.
Extended AEE788 exposure decreases trypanosome viability. Trypanosomes were treated with AEE788 (5 µM) or DMSO (0.1%) for 4, 9, or 16 hours, harvested, and treated with PI (3 µM) prior to analysis on a flow cytometer. Trypanosomes were gated based on size and shape (forward and side scatter) and the intensity of PI determined.
AEE788 associates with three trypanosome protein kinases (Katiyar et al., 2013). As such, the drug is likely to exert pleiotropic effects on trypanosome biology as a multitargeted kinase modulator. One AEE788-associated protein, TbGSK3β (Katiyar et al., 2013), regulates Tf endocytosis (Guyett et al., 2016). We therefore tested whether AEE788 (5 µM) affected trypanosome endocytic pathways. Ligands internalized through glycosylphosphatidylinositiol-anchored receptors, such as the Tf receptor (Steverding et al., 1994), follow a distinct endocytic pathway (Pal et al., 2002). We studied the effect of AEE788 treatment on the internalization of three endocytic cargos. Tf was used for receptor-mediated endocytosis; BSA was a marker for bulk-phase endocytosis (Morriswood and Schmidt, 2015; Guyett et al., 2016); and TL was used to evaluate the internalization of carbohydrate-binding proteins (Nolan et al., 1999; Field et al., 2004). Fluorescent cargo was used to monitor endocytosis after a 9-hour treatment with DMSO (drug solvent) or AEE788 (washed off prior to incubation with cargo). PI exclusion was used to gate for viable cells (Fig. 9A) before the fluorescence intensity of endocytic cargo was measured (Fig. 9, B–D). Based on the median fluorescence intensity, AEE788 decreased Tf endocytosis by 87% (Fig. 9B) (p = 2.8 × 10−3) but increased BSA internalization by 40% (Fig. 9C) (p = 3.1 × 10−3) without affecting TL uptake (Fig. 9D) (p = 0.9). Each cargo demonstrated a unique distribution of fluorescence intensity (proportional to the amount of cargo internalized) within the population (Fig. 9, B–D). For reasons that are unclear to us, AEE788 broadened the distribution of fluorescence associated with BSA or TL internalization (Fig. 9, C and D). These results demonstrate that trypanosomes are metabolically active after 9 hours of exposure to AEE788 and that Tf endocytosis is selectively inhibited.
Fig. 9.
Effect of AEE788 on endocytic pathways. Trypanosomes (5 × 105 cells/ml) were incubated with AEE788 (5 µM) or DMSO (0.1%) for 9 hours. Cells were subsequently washed and resuspended in serum-free medium (without drug or DMSO). Trypanosomes were incubated with fluorescent endocytic cargo (Tf, BSA, or TL) for 15 minutes (37°C). PI (3 µM) was used to stain dead cells. A flow cytometer was used to detect fluorescence intensity per cell. (A) FlowJo software was used to gate for live trypanosomes based on shape (forward and side scatter) and ability to exclude PI. Histograms depict fluorescence intensity for Tf (B), BSA (C), or TL (D) for every observed cell (n = 15,000 for each cargo). Bar graphs represent the average median fluorescence intensity (calculated with FloJo), with S.D. among three independent experiments shown, for Tf (B), BSA (C), or TL (D) after DMSO or AEE788 treatment. In statistical analysis, the median fluorescence of each cargo was compared between cells treated with DMSO or AEE788 using a Student’s t test (p = 0.002 for Tf, p = 0.003 for BSA, and p = 0.9 for TL).
AEE788 caused morphologic changes in trypanosomes in a time-dependent manner (Fig. 10A). Most trypanosomes had normal morphology after a 4-hour treatment with AEE788 (5 µM). However, by 9 hours the distribution of trypanosomes with altered morphology or normal shape shifted toward swollen and rounded cells compared with that found after 4 hours of AEE788 treatment (p = 6.6 × 10−17). By 16 hours, the majority of AEE788-treated trypanosomes had changed morphology compared with trypanosomes after the 4-hour treatment (p = 1.1 × 10−64) or after the 9-hour treatment (p = 5.6 × 10−25). Flagella of rounded trypanosomes were not observed by light microscopy (Fig. 10A). Despite this fact, no detached flagella were detected in the culture medium. This fact prompted us to use alternative methods to detect flagella on rounded trypanosomes. Employing markers for the flagellum (anti-centrin antibody 20H5) (de Graffenried et al., 2013) and paraflagellar rod (Portman and Gull, 2010) (anti-PFR2), we detected flagella juxtaposed to the periphery of rounded trypanosomes (Fig. 10B). The presence of flagella outside rounded trypanosomes was confirmed by scanning electron microscopy (Fig. 10C).
Fig. 10.
Prolonged AEE788 (5 micromolar) exposure changes trypanosome morphology. (A) The morphology of live trypanosomes (n = 100) was determined after different durations of AEE788 treatment (examples of trypanosome morphology are demonstrated by PFA-fixed cells). The S.D. of two experiments is shown. A Pearson χ2 test was used to compare the distribution of normal, swollen, and rounded cells among different treatment groups: 4–9 hours (p = 6.6 × 10−64); 4–16 hours (p = 1.1 × 10−64); 9–16 hours (p = 5.6 × 10−25). (B) After 16 hours of treatment with DMSO (top) or AEE788 (bottom), the PFR was visualized using an antibody against PFR2 (green) and the flagellum with the antibody 20H5 (red). 20H5 detects centrin at the basal body (arrow), bilobe (arrowhead), and the flagellum. K, kinetoplast; N, nucleus. Scale bar, 6 µm. (C) Trypanosomes were treated with AEE788 (5 µM) for 12 hours and visualized by S.E.M. The left panel demonstrates normal trypanosome morphology, the middle panel shows both rounded (left) and swollen (right) cells, and the right panel depicts a round trypanosome with flagellum at the cell periphery (white arrow). Scale bar, 2 µm.
Changes in Phosphoprotein Homeostasis in AEE788-Treated Trypanosomes.
The presence of AEE788 in complexes with trypanosome protein kinases (Katiyar et al., 2013) prompted us to determine whether AEE788 could alter phosphoprotein homeostasis in the parasite. We used IMAC enrichment of phosphopeptides, combined with LC-MS/MS, to identify changes in the abundance of protein phosphorylation after trypanosome exposure to AEE788. Because there are phenotypic differences associated with short-term (4 hours) compared with long-term (9 hours) AEE788 treatment, we examined trypanosome phosphopeptides obtained after both treatment times. A total of 244 trypanosome peptides (176 unique proteins) showed a 2-fold or greater change in phosphorylation after AEE788 treatment (Supplemental Table 1), confirming that AEE788 influences protein phosphorylation in T. brucei.
After 4 hours of AEE788 treatment, 56 unique trypanosome peptides showed decreased phosphorylation and 21 demonstrated increased phosphorylation (Supplemental Table 1). Proteins with decreased phosphorylation after 4 hours of AEE788 treatment include a serine-arginine protein kinase (for review, see Giannakouros et al., 2011), TbSAS4 (Hu et al., 2015b), and a bilobe protein (Morriswood et al., 2013) (Table 1). Proteins with increased phosphorylation include a protease (calpain-like cysteine peptidase) (Table 1).
After 9 hours of AEE788 (5 µM) treatment, 115 trypanosome peptides with decreased phosphorylation and 52 peptides with increased phosphorylation were identified (Supplemental Table 1). Thus, extended exposure to AEE788 affected more peptides (167) than the 4-hour treatment (77). Proteins with decreased phosphorylation after a 9-hour exposure to AEE788 include a NIMA (never in mitosis gene a)–related kinase (for review, see Fry et al., 2012), the basal body protein TbRP2 (Andre et al., 2014), a bilobe protein (Morriswood et al., 2013), and a flagellar pocket protein BILBO-1 (Bonhivers et al., 2008) (Table 2). Proteins with increased phosphorylation include a Tb14-3-3–associated kinase (TbAKB1) (Inoue et al., 2015), a bilobe protein (Morriswood et al., 2013), and a ubiquitin transferase (Table 2).
TABLE 2.
Select examples of phosphoproteins affected by long-term (9-h) AEE8788 treatment
After treatment of trypanosomes with DMSO (0.1%) or AEE788 (5 µM) for 4 h, trypanosome phosphopeptides were enriched over an IMAC column. Phosphopeptide abundance was monitored by LC-MS/MS in three independent experiments. Spectral counts indicate the combined number of times a phosphopeptide was observed over all three experiments. The number in parenthesis indicates the total number of peptides observed for the parent protein over all experiments (summation of all peptides observed in the IMAC elution and flow through). [phosphoRS (Taus et al., 2011) probability ≥80%]. A Student’s t test was used to determine whether the change in phosphopeptide abundance was statistically significant (P < 0.05).
| Gene ID | Production Description | Identified Phosphopeptide | Spectral Counts |
P Value | |
|---|---|---|---|---|---|
| DMSO | AEE788 | ||||
| Decreased | |||||
| Tb427.03.3080 | NEK | ADTsDIsLSHEDLsR | 12 (16) | 0 (7) | 0.02 |
| Tb427.10.14010 | TbRP2 | EATPPEsASRSDSSAPTTPHSR | 8 (25) | 1 (7) | 0.05 |
| Tb427.10.8820 | Bilobe protein | TIGTTSGHSTTNLssHTPEK | 6 (17) | 0 (5) | 0.03 |
| Tb427tmp.01.3960 | BILBO-1 | LMSEASsFLGNLR | 5 (41) | 0 (26) | 0.04 |
| Increased | |||||
| Tb427.10.14770 | Associated kinase of Tb14-3-3 | LANSsLPVsHTSTR | 7 (13) | 15 (18) | 0.02 |
| Tb427.07.7000 | Bilobe protein | TSSHIsEHGLDR | 0 (38) | 10 (67) | 0.02 |
| Tb427.04.310 | Ubiquitin-transferase | TTLSKsAHVsHER | 3 (3) | 8 (9) | 0.04 |
NEK, NIMA (never in mitosis gene a)–related kinase.
In some cases, the abundance of the phosphorylated peptide as well as the abundance of the parent protein (number in parentheses of Tables 1 and 2) changed. In most cases, the magnitude of change in phosphopeptide abundance exceeds that observed for total protein abundance [e.g., Tb427.01.2100 (Table 1) and Tb427.03.3080 (Table 2)]. These data may indicate that phosphorylation influences the stability of some trypanosome proteins, as observed in other eukaryotes (Yamamoto et al., 1999; Ishida et al., 2000; Vazquez et al., 2000; Ulery et al., 2006; Wang et al., 2014). Additionally, the altered phosphorylation of proteins involved in protein degradation (Tables 1 and 2 and Supplemental Table 1) may influence protein abundance.
Discussion
A New Tool for Identification of S-Phase Regulators in Bloodstream Trypanosomes.
S-phase is the period of DNA synthesis by the replisome (for review, see Machida and Dutta, 2005). DNA replication is restricted to S-phase to ensure that the genome is replicated only once per division cycle (Nishitani and Lygerou, 2002). Kinetoplastids are early-branching eukaryotes with a divergent genome (Berriman et al., 2005; Parsons et al., 2005), and signaling pathways responsible for entry into S-phase are not fully defined in bloodstream trypanosomes (for review, see Tiengwe et al., 2014). In higher eukaryotes, the Dbf4-dependent kinase (Zou and Stillman, 2000; Sheu and Stillman, 2006, 2010) and S-phase cyclin-dependent kinase (Zou and Stillman, 2000; Tanaka et al., 2007) promote the initiation of DNA synthesis. Trypanosomes lack homologs of the Dbf4-dependent kinase complex, and trypanosome homologs to cyclin-dependent kinases (TbCRKs) do not regulate DNA replication in bloodstream trypanosomes; knockdown of TbCRK1 and TbCRK2 arrests procyclic (insect stage) trypanosomes in G1 but does not prevent DNA synthesis in bloodstream trypanosomes (Tu and Wang, 2004, 2005; Tu et al., 2005).
AEE788 prevents trypanosome entry into S-phase by inhibiting DNA synthesis in the kinetoplast and nucleus (Fig. 2). Accordingly, by combining our phenotypic analysis with the identification of AEE788-affected phosphoproteins (Supplemental Table 1), we envision the use of AEE788 as a small-molecule tool to identify novel proteins (from effectors of the drug's action at 4 hours) (Supplemental Tables 1A–C) that regulate S-phase entry in bloodstream trypanosomes. In this strategy, proteins that are dephosphorylated (or hyperphosphorylated) will be knocked down (or overexpressed) genetically to determine their effect on DNA synthesis.
Kinetics of Organelle Duplication and Protein Recruitment to the Basal Body during Trypanosome Division.
A novel strategy using AEE788 in a “block and release” protocol was used to enrich pre–S-phase trypanosomes and to study the time-course of organelle duplication in bloodstream stage parasites (Figs. 5–7). Previous studies of basal body duplication in insect stage trypanosomes identified two groups of 1Ke1N cells based on pBB formation: 1) 1Ke1N cells with two mBBs each lacking a pBB (i.e., 2 mBBs/0 pBB); and 2) 1Ke1N cells with two mBBs paired with adjacent pBBs (i.e., 2 mBBs/2 pBBs) (Sherwin and Gull, 1989; Lacomble et al., 2010; Gluenz et al., 2011; Ikeda and de Graffenried, 2012). In our quantitation of SAS6/RP2 double-labeled basal bodies, we found less than 7% of trypanosomes with 2 mBBs/0 pBB (Fig. 6B). The data indicate that 2 mBBs/0 pBB is not a major intermediate for basal body duplication in bloodstream trypanosomes (Fig. 6C). This conclusion is reinforced by our observation that during duplication of basal bodies, TbRP2 is recruited to mBBs with the same kinetics as TbSAS6 localization at nascent pBBs (Fig. 6B and Supplemental Fig. 3). Hence, the recruitment of TbRP2 to mBBs is concurrent with new pBB formation. Our observations establish the utility of AEE788 as a small-molecule tool for monitoring the order of protein recruitment during basal body biogenesis (and perhaps of other cytoskeletal organelles).
Our data provide new insight into the sequence of S-phase events (DNA replication with respect to organelle duplication) in bloodstream trypanosomes. We found that kinetoplast elongation occurs throughout nuclear DNA synthesis, consistent with the annotation of 1Ke1N trypanosomes as S-phase cells (Siegel et al., 2008) (Fig. 7). Second, duplication of the basal body and bilobe are coincident (consistent with the idea of a continuous cytoskeletal network containing both organelles) (Gheiratmand et al., 2013). Duplication of these cytoskeletal structures occurs after kDNA synthesis but is concurrent with nuclear S-phase and kinetoplast elongation (both of which initiate approximately 30 minutes prior to duplication of the basal body and bilobe) (Fig. 7). Third, kinetoplast division does not occur immediately after kDNA synthesis but is observed 1 hour after termination of kDNA replication. During the intervening period, the basal body is duplicated. This lag between kDNA replication and kinetoplast division may reflect: 1) a requirement of two basal bodies to facilitate kinetoplast fission (Robinson and Gull, 1991; Lacomble et al., 2010), 2) a slow assembly of factors needed for kinetoplast division, or 3) both. Nuclear DNA synthesis was completed before kinetoplast division (Fig. 7), revealing that 2K1N trypanosomes are most likely in G2, which is in accord with previous work (Woodward and Gull, 1990; Siegel et al., 2008). Mitosis was observed 1 hour after replication of the nuclear genome, implying that the trypanosome G2 lasts 1 hour, during which kinetoplast division occurs.
Selective Inhibition of Endocytosis by AEE788.
Extended AEE788 treatment (9 hours) of trypanosomes inhibited Tf endocytosis (Fig. 9B). Interestingly, not all trypanosome endocytic pathways were affected by AEE788 treatment. Internalization of BSA, a marker for fluid-phase endocytosis (Morriswood and Schmidt, 2015), was increased after AEE788 treatment (Fig. 9C). A similar effect was observed after knockdown of TbGSK3β (Guyett et al., 2016). Future studies will address the basis of the ability of AEE788 to selectively inhibit Tf endocytosis (Fig. 9) by knocking down (or overexpressing) putative effectors of the drug's action (9 h) (Table 2 and Supplemental Table 1B and D).
Putative Effectors of AEE78 Action.
AEE788 treatment of trypanosomes caused dephosphorylation of some proteins but resulted in hyperphosphorylation of others (Tables 1 and 2 and Supplemental Table 1). Small-molecule kinase inhibitors can paradoxically lead to hyperphosphorylation of proteins (Okuzumi et al., 2009; Holderfield et al., 2013) through a variety of mechanisms including protection of their target from protein phosphatases (Gould et al., 2011), increasing (Zhang et al., 2003) or decreasing (Holderfield et al., 2013) inhibitory autophosphorylation, and activation of negative-feedback loops (Wan et al., 2007).
Proteins with altered phosphorylation after 4 hours of AEE788 treatment (Table 1) may be involved in biologic pathways disrupted during short-term AEE788 exposure (4 hours). They might be effectors for S-phase entry (Fig. 2) or duplication of the basal body (Fig. 3) and bilobe (Fig. 4). Of note, a cytoskeletal protein (TbSAS4) and a bilobe protein (Tb427.10.3010) (Morriswood et al., 2013) were dephosphorylated (Table 1). In other organisms, SAS4 is a centriolar protein (Hodges et al., 2010) with essential roles in centriole duplication (Leidel and Gönczy, 2003; Pelletier et al., 2006; Schmidt et al., 2009; Gogendeau et al., 2011; Novak et al., 2016). The role of TbSAS4 in bloodstream trypanosomes remains to be explored.
Extended exposure (9 hours) of trypanosomes to AEE788 inhibited Tf endocytosis (Fig. 9) and distorted cell morphology (Fig. 10). These phenotypes may be explained by postulating that two proteins with altered phosphorylation, namely, Tb14-3-3–associated protein kinase (TbAKB1) (Inoue et al., 2015) (Table 2) and BILBO-1 (Bonhivers et al., 2008) (Table 2), are effectors of AEE788 action. Knockdown of Tb14-3-3 reduces the size of recycling endosomes (Benz et al., 2010), and knockdown of BILBO-1 causes rounding of bloodstream trypanosomes (Bonhivers et al., 2008), comparable to the morphology of T. brucei observed after prolonged AEE788 treatment (Fig. 10).
The relative abundance of phosphopeptides in DMSO-treated cells (drug vehicle control) and AEE788-treated trypanosomes was determined by spectral counting of LC-MS/MS data. Spectral counting (for review, see Lundgren et al., 2010) has been used to document changes in protein expression (Old et al., 2005; Kislinger et al., 2006; Takadate et al., 2013) and phosphorylation (Xie et al., 2010; Dammer et al., 2015; Singec et al., 2016). However, there are limitations associated with this method: the dynamic range is poor for proteins of low abundance (Old et al., 2005). Additionally, the reproducibility of data may be compromised by nonidentical sampling of peptides between instrument runs (e.g., control versus experimental). The latter issue is mitigated by replicate runs and statistical analysis to improve confidence in identifying changes in protein levels between controls and experimental samples. Zhang et al. (2006) showed that the Student’s t test offers the lowest false-positive rate (<1%) for triplicate replicates (used in our analysis) when the fold-change in spectra is greater than two (Zhang et al., 2006). We reported proteins that were observed in three independent experiments and showed a statistically significant change in levels of phosphorylation as determined by Student’s t tests.
The functions of many phosphoproteins affected by AEE788 are unknown in bloodstream trypanosomes. Hence, the correlation of their dephosphorylation with the disruption of essential physiologic trypanosome pathways generates hypotheses as to the function of these uncharacterized phosphoproteins. In the future, we will focus on determining the role of these unstudied proteins in: 1) AEE788-perturbed pathways (Figs. 2–4, 9, and 10); and 2) how their phosphorylation may modulate their biologic functions.
Acknowledgments
The authors thank Julie Nelson at the CTEGD Cytometry Shared Resource Laboratory for help with flow cytometry. In addition, the authors thank John Shields at Georgia Electron Microscopy, and Muthugapatti Kandasamy at the Biomedical Microscopy Core for their assistance with microscopy experiments. For help with statistical analysis, the authors thank Jaxk Reeves and Wenhao Pan from the Department of Statistics at the University of Georgia. Finally, thanks to Frank Hardin and Samiksha Katiyar, who first documented the effect of AEE788 on trypanosome morphology; and Paul Guyett for insightful discussion.
Abbreviations
- 1K1N
single nucleus in kinetoplast
- 1Ke1N
one nucleus in elongated kinetoplast
- BSA
bovine serum albumin
- CCD
charge-coupled device
- DAPI
4′,6-diamidino-2-phenylindole
- DMSO
dimethylsulfoxide
- EdU
5-ethynyl-2′-deoxyuridine
- FAZ
flagellar attachment zone
- HAT
human African trypanosomiasis
- IMAC
immobilized-metal affinity chromatography
- kDNA
kinetoplast DNA
- Ke
elongated kinetoplast
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- mBB
mature basal body
- pBB
probasal body
- PBS
phosphate-buffered saline
- PBSG
phosphate-buffered saline with 1% glucose
- PFA
paraformaldehyde
- PFR
paraflagellar rod
- PI
propidium iodide
- PIC
phosphatase inhibitor cocktail
- Tf
transferrin
- TFA
trifluoroacetic acid
- TL
tomato lectin
- T10
time at which 10% of the observed maximum is reached
- T50
time at which 50% of the observed maximum is reached
- T90
time at which 90% of the observed maximum is reached
Authorship Contributions
Participated in research design: Sullenberger, Piqué, and Mensa-Wilmot.
Conducted experiments: Sullenberger, Ogata.
Wrote or contributed to the writing of the manuscript: Sullenberger, Mensa-Wilmot.
Footnotes
This work was funded by the National Institutes of Health [Grant R01AI124046].
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Ajoko C, Steverding D. (2015) A cultivation method for growing bloodstream forms of Trypanosoma brucei to higher cell density and for longer time. Parasitol Res 114:1611–1612. [DOI] [PubMed] [Google Scholar]
- Andre J, Kerry L, Qi X, Hawkins E, Drizyte K, Ginger ML, McKean PG. (2014) An alternative model for the role of RP2 protein in flagellum assembly in the African trypanosome. J Biol Chem 289:464–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SK, Inchaustegui D, Queiroz R, Clayton C. (2011) The cell cycle regulated transcriptome of Trypanosoma brucei. PLoS One 6:e18425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babokhov P, Sanyaolu AO, Oyibo WA, Fagbenro-Beyioku AF, Iriemenam NC. (2013) A current analysis of chemotherapy strategies for the treatment of human African trypanosomiasis. Pathog Glob Health 107:242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs JD. (2011) Replication of the ERES:Golgi junction in bloodstream-form African trypanosomes. Mol Microbiol 82:1433–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera R, Thomas SM, Mensa-Wilmot K. (2014) New chemical scaffolds for human african trypanosomiasis lead discovery from a screen of tyrosine kinase inhibitor drugs. Antimicrob Agents Chemother 58:2202–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz C, Engstler M, Hillmer S, Clayton C. (2010) Depletion of 14-3-3 proteins in bloodstream-form Trypanosoma brucei inhibits variant surface glycoprotein recycling. Int J Parasitol 40:629–634. [DOI] [PubMed] [Google Scholar]
- Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, et al. (2005) The genome of the African trypanosome Trypanosoma brucei. Science 309:416–422. [DOI] [PubMed] [Google Scholar]
- Bonhivers M, Nowacki S, Landrein N, Robinson DR. (2008) Biogenesis of the trypanosome endo-exocytotic organelle is cytoskeleton mediated. PLoS Biol 6:e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst P, van der Ploeg M, van Hoek JF, Tas J, James J. (1982) On the DNA content and ploidy of trypanosomes. Mol Biochem Parasitol 6:13–23. [DOI] [PubMed] [Google Scholar]
- Cavanagh BL, Walker T, Norazit A, Meedeniya AC. (2011) Thymidine analogues for tracking DNA synthesis. Molecules 16:7980–7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammer EB, Lee AK, Duong DM, Gearing M, Lah JJ, Levey AI, Seyfried NT. (2015) Quantitative phosphoproteomics of Alzheimer’s disease reveals cross-talk between kinases and small heat shock proteins. Proteomics 15:508–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar AC, Shokat KM. (2011) The evolution of protein kinase inhibitors from antagonists to agonists of cellular signaling. Annu Rev Biochem 80:769–795. [DOI] [PubMed] [Google Scholar]
- de Graffenried CL, Anrather D, Von Raußendorf F, Warren G. (2013) Polo-like kinase phosphorylation of bilobe-resident TbCentrin2 facilitates flagellar inheritance in Trypanosoma brucei. Mol Biol Cell 24:1947–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMasi JA, Hansen RW, Grabowski HG. (2003) The price of innovation: new estimates of drug development costs. J Health Econ 22:151–185. [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5:976–989. [DOI] [PubMed] [Google Scholar]
- Esson HJ, Morriswood B, Yavuz S, Vidilaseris K, Dong G, Warren G. (2012) Morphology of the trypanosome bilobe, a novel cytoskeletal structure. Eukaryot Cell 11:761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MC, Allen CL, Dhir V, Goulding D, Hall BS, Morgan GW, Veazey P, Engstler M. (2004) New approaches to the microscopic imaging of Trypanosoma brucei. Microsc Microanal 10:621–636. [DOI] [PubMed] [Google Scholar]
- Field MC, Lumb JH, Adung’a VO, Jones NG, Engstler M.(2009) Macromolecular trafficking and immune evasion in African trypanosomes. Int Rev Cell Mol Biol 278:1–67. [DOI] [PubMed] [Google Scholar]
- Forsythe GR, McCulloch R, Hammarton TC. (2009) Hydroxyurea-induced synchronisation of bloodstream stage Trypanosoma brucei. Mol Biochem Parasitol 164:131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry AM, O’Regan L, Sabir SR, Bayliss R. (2012) Cell cycle regulation by the NEK family of protein kinases. J Cell Sci 125:4423–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner DL, Pinkel D, Johnson LA, Pace MM. (1986) Assessment of spermatozoal function using dual fluorescent staining and flow cytometric analyses. Biol Reprod 34:127–138. [DOI] [PubMed] [Google Scholar]
- Gheiratmand L, Brasseur A, Zhou Q, He CY. (2013) Biochemical characterization of the bi-lobe reveals a continuous structural network linking the bi-lobe to other single-copied organelles in Trypanosoma brucei. J Biol Chem 288:3489–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakouros T, Nikolakaki E, Mylonis I, Georgatsou E. (2011) Serine-arginine protein kinases: a small protein kinase family with a large cellular presence. FEBS J 278:570–586. [DOI] [PubMed] [Google Scholar]
- Gluenz E, Povelones ML, Englund PT, Gull K. (2011) The kinetoplast duplication cycle in Trypanosoma brucei is orchestrated by cytoskeleton-mediated cell morphogenesis. Mol Cell Biol 31:1012–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogendeau D, Hurbain I, Raposo G, Cohen J, Koll F, Basto R. (2011) Sas-4 proteins are required during basal body duplication in Paramecium. Mol Biol Cell 22:1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould CM, Antal CE, Reyes G, Kunkel MT, Adams RA, Ziyar A, Riveros T, Newton AC. (2011) Active site inhibitors protect protein kinase C from dephosphorylation and stabilize its mature form. J Biol Chem 286:28922–28930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyett PJ, Xia S, Swinney DC, Pollastri MP, Mensa-Wilmot K. (2016) Glycogen synthase kinase 3β promotes the endocytosis of transferrin in the African trypanosome. ACS Infect Dis 2:518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarton TC. (2007) Cell cycle regulation in Trypanosoma brucei. Mol Biochem Parasitol 153:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He CY, Pypaert M, Warren G. (2005) Golgi duplication in Trypanosoma brucei requires Centrin2. Science 310:1196–1198. [DOI] [PubMed] [Google Scholar]
- Hesse F, Selzer PM, Mühlstädt K, Duszenko M. (1995) A novel cultivation technique for long-term maintenance of bloodstream form trypanosomes in vitro. Mol Biochem Parasitol 70:157–166. [DOI] [PubMed] [Google Scholar]
- Hirumi H, Hirumi K. (1994) Axenic culture of African trypanosome bloodstream forms. Parasitol Today 10:80–84. [DOI] [PubMed] [Google Scholar]
- Hodges ME, Scheumann N, Wickstead B, Langdale JA, Gull K. (2010) Reconstructing the evolutionary history of the centriole from protein components. J Cell Sci 123:1407–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holderfield M, Merritt H, Chan J, Wallroth M, Tandeske L, Zhai H, Tellew J, Hardy S, Hekmat-Nejad M, Stuart DD, et al. (2013) RAF inhibitors activate the MAPK pathway by relieving inhibitory autophosphorylation. Cancer Cell 23:594–602. [DOI] [PubMed] [Google Scholar]
- Hu H, Liu Y, Zhou Q, Siegel S, Li Z. (2015a) The centriole cartwheel protein SAS-6 in Trypanosoma brucei is required for probasal body biogenesis and flagellum assembly. Eukaryot Cell 14:898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Zhou Q, Li Z. (2015b) SAS-4 protein in Trypanosoma brucei controls life cycle transitions by modulating the length of the flagellum attachment zone filament. J Biol Chem 290:30453–30463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda KN, de Graffenried CL. (2012) Polo-like kinase is necessary for flagellum inheritance in Trypanosoma brucei. J Cell Sci 125:3173–3184. [DOI] [PubMed] [Google Scholar]
- Inoue M, Okamoto K, Uemura H, Yasuda K, Motohara Y, Morita K, Hiromura M, Reddy EP, Fukuma T, Horikoshi N. (2015) Identification and characterization of a cell division-regulating kinase AKB1 (associated kinase of Trypanosoma brucei 14-3-3) through proteomics study of the Tb14-3-3 binding proteins. J Biochem 158:49–60. [DOI] [PubMed] [Google Scholar]
- Ishida N, Kitagawa M, Hatakeyama S, Nakayama K. (2000) Phosphorylation at serine 10, a major phosphorylation site of p27(Kip1), increases its protein stability. J Biol Chem 275:25146–25154. [DOI] [PubMed] [Google Scholar]
- Jensen RE, Englund PT. (2012) Network news: the replication of kinetoplast DNA. Annu Rev Microbiol 66:473–491. [DOI] [PubMed] [Google Scholar]
- Kabani S, Waterfall M, Matthews KR. (2010) Cell-cycle synchronisation of bloodstream forms of Trypanosoma brucei using Vybrant DyeCycle Violet-based sorting. Mol Biochem Parasitol 169:59–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. (2007) Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods 4:923–925. [DOI] [PubMed] [Google Scholar]
- Katiyar S, Kufareva I, Behera R, Thomas SM, Ogata Y, Pollastri M, Abagyan R, Mensa-Wilmot K. (2013) Lapatinib-binding protein kinases in the African trypanosome: identification of cellular targets for kinase-directed chemical scaffolds. PLoS One 8:e56150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann D, Gassen A, Maiser A, Leonhardt H, Janzen CJ. (2012) Regulation and spatial organization of PCNA in Trypanosoma brucei. Biochem Biophys Res Commun 419:698–702. [DOI] [PubMed] [Google Scholar]
- Kennedy PGE. (2013) Clinical features, diagnosis, and treatment of human African trypanosomiasis (sleeping sickness). Lancet Neurol 12:186–194. [DOI] [PubMed] [Google Scholar]
- Kislinger T, Cox B, Kannan A, Chung C, Hu P, Ignatchenko A, Scott MS, Gramolini AO, Morris Q, Hallett MT, et al. (2006) Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell 125:173–186. [DOI] [PubMed] [Google Scholar]
- Kohl L, Robinson D, Bastin P. (2003) Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. EMBO J 22:5336–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl L, Sherwin T, Gull K. (1999) Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J Eukaryot Microbiol 46:105–109. [DOI] [PubMed] [Google Scholar]
- Lacomble S, Vaughan S, Gadelha C, Morphew MK, Shaw MK, McIntosh JR, Gull K. (2009) Three-dimensional cellular architecture of the flagellar pocket and associated cytoskeleton in trypanosomes revealed by electron microscope tomography. J Cell Sci 122:1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomble S, Vaughan S, Gadelha C, Morphew MK, Shaw MK, McIntosh JR, Gull K. (2010) Basal body movements orchestrate membrane organelle division and cell morphogenesis in Trypanosoma brucei. J Cell Sci 123:2884–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal S, Acosta-Serrano A, Morita YS, Englund PT, Böhme U, Cross GA. (2001) Virulence of Trypanosoma brucei strain 427 is not affected by the absence of glycosylphosphatidylinositol phospholipase C. Mol Biochem Parasitol 114:245–247. [DOI] [PubMed] [Google Scholar]
- Leidel S, Gönczy P. (2003) SAS-4 is essential for centrosome duplication in C elegans and is recruited to daughter centrioles once per cell cycle. Dev Cell 4:431–439. [DOI] [PubMed] [Google Scholar]
- Lejon V, Büscher P. (2005) Review article: cerebrospinal fluid in human African trypanosomiasis: a key to diagnosis, therapeutic decision and post-treatment follow-up. Trop Med Int Health 10:395–403. [DOI] [PubMed] [Google Scholar]
- Li Z. (2012) Regulation of the cell division cycle in Trypanosoma brucei. Eukaryot Cell 11:1180–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licklider LJ, Thoreen CC, Peng J, Gygi SP. (2002) Automation of nanoscale microcapillary liquid chromatography-tandem mass spectrometry with a vented column. Anal Chem 74:3076–3083. [DOI] [PubMed] [Google Scholar]
- Liu B, Liu Y, Motyka SA, Agbo EE, Englund PT. (2005) Fellowship of the rings: the replication of kinetoplast DNA. Trends Parasitol 21:363–369. [DOI] [PubMed] [Google Scholar]
- Lundgren DH, Hwang SI, Wu L, Han DK. (2010) Role of spectral counting in quantitative proteomics. Expert Rev Proteomics 7:39–53. [DOI] [PubMed] [Google Scholar]
- Machida YJ, Dutta A. (2005) Cellular checkpoint mechanisms monitoring proper initiation of DNA replication. J Biol Chem 280:6253–6256. [DOI] [PubMed] [Google Scholar]
- Meco D, Servidei T, Zannoni GF, Martinelli E, Prisco MG, de Waure C, Riccardi R. (2010) Dual inhibitor AEE788 reduces tumor growth in preclinical models of medulloblastoma. Transl Oncol 3:326–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morriswood B, Havlicek K, Demmel L, Yavuz S, Sealey-Cardona M, Vidilaseris K, Anrather D, Kostan J, Djinovic-Carugo K, Roux KJ, et al. (2013) Novel bilobe components in Trypanosoma brucei identified using proximity-dependent biotinylation. Eukaryot Cell 12:356–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morriswood B, Schmidt K. (2015) A MORN repeat protein facilitates protein entry into the flagellar pocket of Trypanosoma brucei. Eukaryot Cell 14:1081–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutomba MC, Wang CC. (1996) Effects of aphidicolin and hydroxyurea on the cell cycle and differentiation of Trypanosoma brucei bloodstream forms. Mol Biochem Parasitol 80:89–102. [DOI] [PubMed] [Google Scholar]
- Nishitani H, Lygerou Z. (2002) Control of DNA replication licensing in a cell cycle. Genes Cells 7:523–534. [DOI] [PubMed] [Google Scholar]
- Nolan DP, Geuskens M, Pays E. (1999) N-linked glycans containing linear poly-N-acetyllactosamine as sorting signals in endocytosis in Trypanosoma brucei. Curr Biol 9:1169–1172. [DOI] [PubMed] [Google Scholar]
- Novak ZA, Wainman A, Gartenmann L, Raff JW. (2016) Cdk1 phosphorylates Drosophila Sas-4 to recruit polo to daughter centrioles and convert them to centrosomes. Dev Cell 37:545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbadoyi EO, Robinson DR, Gull K. (2003) A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol Biol Cell 14:1769–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuzumi T, Fiedler D, Zhang C, Gray DC, Aizenstein B, Hoffman R, Shokat KM. (2009) Inhibitor hijacking of Akt activation. Nat Chem Biol 5:484–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. (2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics 4:1487–1502. [DOI] [PubMed] [Google Scholar]
- Pal A, Hall BS, Nesbeth DN, Field HI, Field MC. (2002) Differential endocytic functions of Trypanosoma brucei Rab5 isoforms reveal a glycosylphosphatidylinositol-specific endosomal pathway. J Biol Chem 277:9529–9539. [DOI] [PubMed] [Google Scholar]
- Parsons M, Worthey EA, Ward PN, Mottram JC. (2005) Comparative analysis of the kinomes of three pathogenic trypanosomatids: Leishmania major, Trypanosoma brucei and Trypanosoma cruzi. BMC Genomics 6:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel G, Karver CE, Behera R, Guyett PJ, Sullenberger C, Edwards P, Roncal NE, Mensa-Wilmot K, Pollastri MP. (2013) Kinase scaffold repurposing for neglected disease drug discovery: discovery of an efficacious, lapatinib-derived lead compound for trypanosomiasis. J Med Chem 56:3820–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L, O’Toole E, Schwager A, Hyman AA, Müller-Reichert T. (2006) Centriole assembly in Caenorhabditis elegans. Nature 444:619–623. [DOI] [PubMed] [Google Scholar]
- Portman N, Gull K. (2010) The paraflagellar rod of kinetoplastid parasites: from structure to components and function. Int J Parasitol 40:135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povelones ML. (2014) Beyond replication: division and segregation of mitochondrial DNA in kinetoplastids. Mol Biochem Parasitol 196:53–60. [DOI] [PubMed] [Google Scholar]
- Robinson DR, Gull K. (1991) Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature 352:731–733. [DOI] [PubMed] [Google Scholar]
- Robinson DR, Sherwin T, Ploubidou A, Byard EH, Gull K. (1995) Microtubule polarity and dynamics in the control of organelle positioning, segregation, and cytokinesis in the trypanosome cell cycle. J Cell Biol 128:1163–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell D, Borowy NK, Overath P. (1991) Transferrin is a growth factor for the bloodstream form of Trypanosoma brucei. Parasitol Res 77:558–560. [DOI] [PubMed] [Google Scholar]
- Schmidt TI, Kleylein-Sohn J, Westendorf J, Le Clech M, Lavoie SB, Stierhof YD, Nigg EA. (2009) Control of centriole length by CPAP and CP110. Curr Biol 19:1005–1011. [DOI] [PubMed] [Google Scholar]
- Sherwin T, Gull K. (1989) The cell division cycle of Trypanosoma brucei brucei: timing of event markers and cytoskeletal modulations. Philos Trans R Soc Lond B Biol Sci 323:573–588. [DOI] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B. (2006) Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell 24:101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B. (2010) The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 463:113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel TN, Hekstra DR, Cross GA. (2008) Analysis of the Trypanosoma brucei cell cycle by quantitative DAPI imaging. Mol Biochem Parasitol 160:171–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singec I, Crain AM, Hou J, Tobe BT, Talantova M, Winquist AA, Doctor KS, Choy J, Huang X, La Monaca E, et al. (2016) Quantitative Analysis of Human Pluripotency and Neural Specification by In-Depth (Phospho)Proteomic Profiling. Stem Cell Rep 7:527–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steverding D, Stierhof YD, Chaudhri M, Ligtenberg M, Schell D, Beck-Sickinger AG, Overath P. (1994) ESAG 6 and 7 products of Trypanosoma brucei form a transferrin binding protein complex. Eur J Cell Biol 64:78–87. [PubMed] [Google Scholar]
- Takadate T, Onogawa T, Fukuda T, Motoi F, Suzuki T, Fujii K, Kihara M, Mikami S, Bando Y, Maeda S, et al. (2013) Novel prognostic protein markers of resectable pancreatic cancer identified by coupled shotgun and targeted proteomics using formalin-fixed paraffin-embedded tissues. Int J Cancer 132:1368–1382. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. (2007) CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445:328–332. [DOI] [PubMed] [Google Scholar]
- Taus T, Köcher T, Pichler P, Paschke C, Schmidt A, Henrich C, Mechtler K. (2011) Universal and confident phosphorylation site localization using phosphoRS. J Proteome Res 10:5354–5362. [DOI] [PubMed] [Google Scholar]
- Tiengwe C, Marques CA, McCulloch R. (2014) Nuclear DNA replication initiation in kinetoplastid parasites: new insights into an ancient process. Trends Parasitol 30:27–36. [DOI] [PubMed] [Google Scholar]
- Traxler P, Allegrini PR, Brandt R, Brueggen J, Cozens R, Fabbro D, Grosios K, Lane HA, McSheehy P, Mestan J, et al. (2004) AEE788: a dual family epidermal growth factor receptor/ErbB2 and vascular endothelial growth factor receptor tyrosine kinase inhibitor with antitumor and antiangiogenic activity. Cancer Res 64:4931–4941. [DOI] [PubMed] [Google Scholar]
- Tu X, Mancuso J, Cande WZ, Wang CC. (2005) Distinct cytoskeletal modulation and regulation of G1-S transition in the two life stages of Trypanosoma brucei. J Cell Sci 118:4353–4364. [DOI] [PubMed] [Google Scholar]
- Tu X, Wang CC. (2004) The involvement of two cdc2-related kinases (CRKs) in Trypanosoma brucei cell cycle regulation and the distinctive stage-specific phenotypes caused by CRK3 depletion. J Biol Chem 279:20519–20528. [DOI] [PubMed] [Google Scholar]
- Tu X, Wang CC. (2005) Pairwise knockdowns of cdc2-related kinases (CRKs) in Trypanosoma brucei identified the CRKs for G1/S and G2/M transitions and demonstrated distinctive cytokinetic regulations between two developmental stages of the organism. Eukaryot Cell 4:755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulery PG, Rudenko G, Nestler EJ. (2006) Regulation of DeltaFosB stability by phosphorylation. J Neurosci 26:5131–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Ramaswamy S, Nakamura N, Sellers WR. (2000) Phosphorylation of the PTEN tail regulates protein stability and function. Mol Cell Biol 20:5010–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, Harkavy B, Shen N, Grohar P, Helman LJ. (2007) Rapamycin induces feedback activation of Akt signaling through an IGF-1R-dependent mechanism. Oncogene 26:1932–1940. [DOI] [PubMed] [Google Scholar]
- Wang SA, Hung CY, Chuang JY, Chang WC, Hsu TI, Hung JJ. (2014) Phosphorylation of p300 increases its protein degradation to enhance the lung cancer progression. Biochim Biophys Acta 1843:1135–1149. [DOI] [PubMed] [Google Scholar]
- Woodward R, Gull K. (1990) Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J Cell Sci 95:49–57. [DOI] [PubMed] [Google Scholar]
- Xie X, Feng S, Vuong H, Liu Y, Goodison S, Lubman DM. (2010) A comparative phosphoproteomic analysis of a human tumor metastasis model using a label-free quantitative approach. Electrophoresis 31:1842–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Kishida S, Kishida M, Ikeda S, Takada S, Kikuchi A. (1999) Phosphorylation of axin, a Wnt signal negative regulator, by glycogen synthase kinase-3beta regulates its stability. J Biol Chem 274:10681–10684. [DOI] [PubMed] [Google Scholar]
- Zhang B, VerBerkmoes NC, Langston MA, Uberbacher E, Hettich RL, Samatova NF. (2006) Detecting differential and correlated protein expression in label-free shotgun proteomics. J Proteome Res 5:2909–2918. [DOI] [PubMed] [Google Scholar]
- Zhang F, Phiel CJ, Spece L, Gurvich N, Klein PS. (2003) Inhibitory phosphorylation of glycogen synthase kinase-3 (GSK-3) in response to lithium. Evidence for autoregulation of GSK-3. J Biol Chem 278:33067–33077. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Gheiratmand L, Chen Y, Lim TK, Zhang J, Li S, Xia N, Liu B, Lin Q, He CY. (2010) A comparative proteomic analysis reveals a new bi-lobe protein required for bi-lobe duplication and cell division in Trypanosoma brucei. PLoS One 5:e9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Hu H, Li Z. (2014) New insights into the molecular mechanisms of mitosis and cytokinesis in trypanosomes. Int Rev Cell Mol Biol 308:127–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Stillman B. (2000) Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol Cell Biol 20:3086–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]