Abstract
Successful completion of animal development is fundamentally reliant on nutritional cues. Surviving periods of nutritional insufficiency requires adaptations that are coordinated, in part, by neural circuits. As neuropeptides secreted by neuroendocrine (NE) cells modulate neural circuits, we investigated NE cell function during development under nutrient stress. Starved Drosophila larvae exhibited reduced pupariation if either insulin signaling or IP3/Ca2+ signaling were downregulated in NE cells. Moreover, an IP3R (inositol 1,4,5-trisphosphate receptor) loss-of-function mutant displayed reduced protein synthesis, which was rescued by overexpression of either InR (insulin receptor) or IP3R in NE cells of the mutant, suggesting that the two signaling pathways might be functionally compensatory. Furthermore, cultured IP3R mutant NE cells, but not neurons, exhibited reduced protein translation. Thus cell-specific regulation of protein synthesis by IP3R in NE cells influences protein metabolism. We propose that this regulation helps developing animals survive in poor nutritional conditions.
KEY WORDS: Insulin signaling, ER Ca2+ stores, dILP5, Starvation, Pupariation
Summary: Intracellular Ca2+ signaling regulates protein translation and can compensate for insulin signaling in specialized neuro-hormonal cells, thus enabling Drosophila larval to pupal development under acute starvation.
INTRODUCTION
Nutritional poverty during development has long-lasting effects on the growth and behavior of an animal. Although under-nutrition causes overall body size to decrease, the brain grows to near-normal size, a process termed ‘brain sparing’ (Dobbing and Sands, 1971). This suggests unique mechanisms in neuronal tissues to weather nutritional stress. Drosophila is an attractive model system to uncover these mechanisms because larvae subjected to nutrient restriction exhibit ‘brain sparing’ (Cheng et al., 2011) and nutritional effects on larval-to-pupal development are easily monitored. Additionally, growth signaling pathways activated by dietary cues such as insulin receptor (InR) and TOR signaling, are conserved in Drosophila (Padmanabha and Baker, 2014).
When starved, larval neural stem cells (NSCs) continue to proliferate by using an InR ortholog, Alk (Anaplastic lymphoma kinase) (Cheng et al., 2011). This study focuses on neuroendocrine (NE) cells, which, unlike NSCs, are differentiated and non-dividing. Importantly, neuropeptides released by NE cells modulate neural circuits that regulate processes associated with animal physiology and behavior (Nässel and Winther, 2010; Taghert and Nitabach, 2012), eventually influencing how animals adapt to external or internal stimuli. Crucially, NE cells produce peptide hormones that regulate feeding behavior and metabolism (Nässel and Winther, 2010), processes required for larvae to complete development successfully.
IP3R (Itp-r83A in Drosophila) is an endoplasmic reticulum (ER) channel that releases stored Ca2+ and acts downstream of G protein-coupled receptor activation. The ER-resident protein STIM (Stromal interaction molecule) conveys loss of stored Ca2+ to Orai (Olf186-F in Drosophila), a plasma membrane Ca2+ channel, thereby enabling store-operated Ca2+ entry (SOCE) from the extracellular milieu. SOCE occurs in both mammals (Prakriya and Lewis, 2015) and flies (Agrawal et al., 2010; Venkiteswaran and Hasan, 2009). Thus, all three molecules – IP3R, STIM and Orai – function during stimulus-dependent elevation of cytosolic Ca2+ that potentiates diverse signaling outcomes, depending on the cellular context.
Loss of IP3R (Subramanian et al., 2013b) and STIM (Baumbach et al., 2014) leads to obesity in adult Drosophila. Importantly, adults of a hypomorphic IP3R mutant heteroallelic combination, itprka1091/ug3 (hereafter: itprku) exhibit obesity, starvation resistance and hyperphagia, which are all rescued by overexpression of IP3R in NE cells (Subramanian et al., 2013a). This adult metabolic phenotype prompted us to investigate the role of IP3R and InR in NE cells during larval development.
RESULTS AND DISCUSSION
Downregulation of InR- or IP3R-mediated intracellular Ca2+ signaling in NE cells reduces pupariation under starvation
In Drosophila, a large subset of NE cells express the transcription factor DIMM (Park et al., 2008). We downregulated InR, TOR and intracellular Ca2+ signaling pathways in dimm+ NE cells using the UAS-GAL4 system (Brand and Perrimon, 1993), and monitored pupariation of larvae on a sucrose-only diet from 88 h after egg laying (AEL; Fig. 1A), a time point used previously (Cheng et al., 2011).
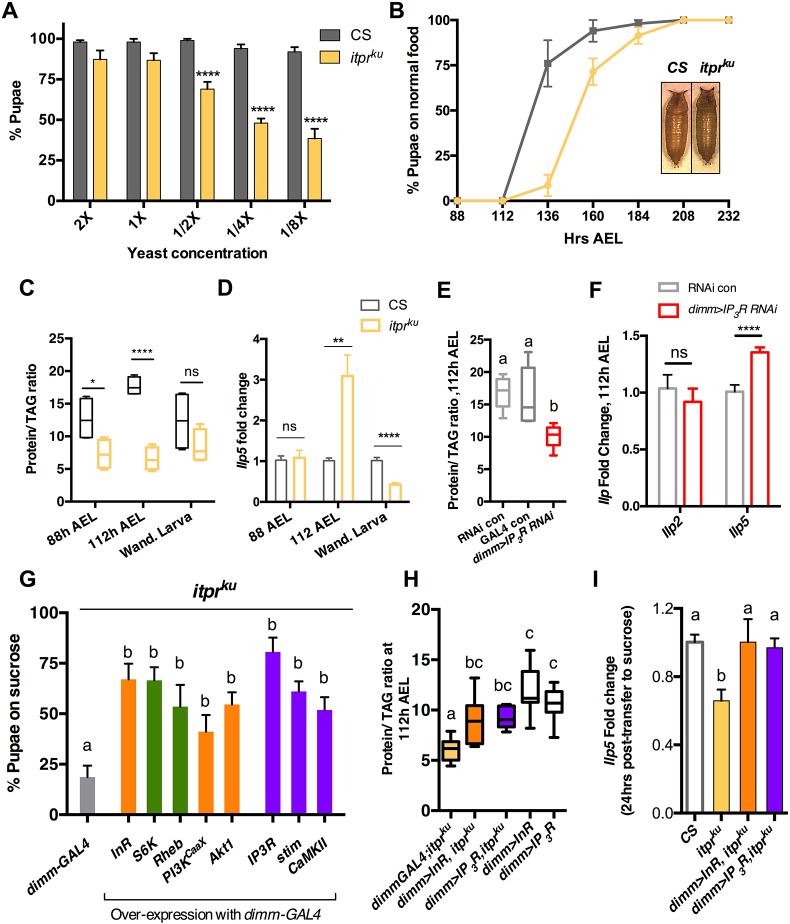
Fig. 1.
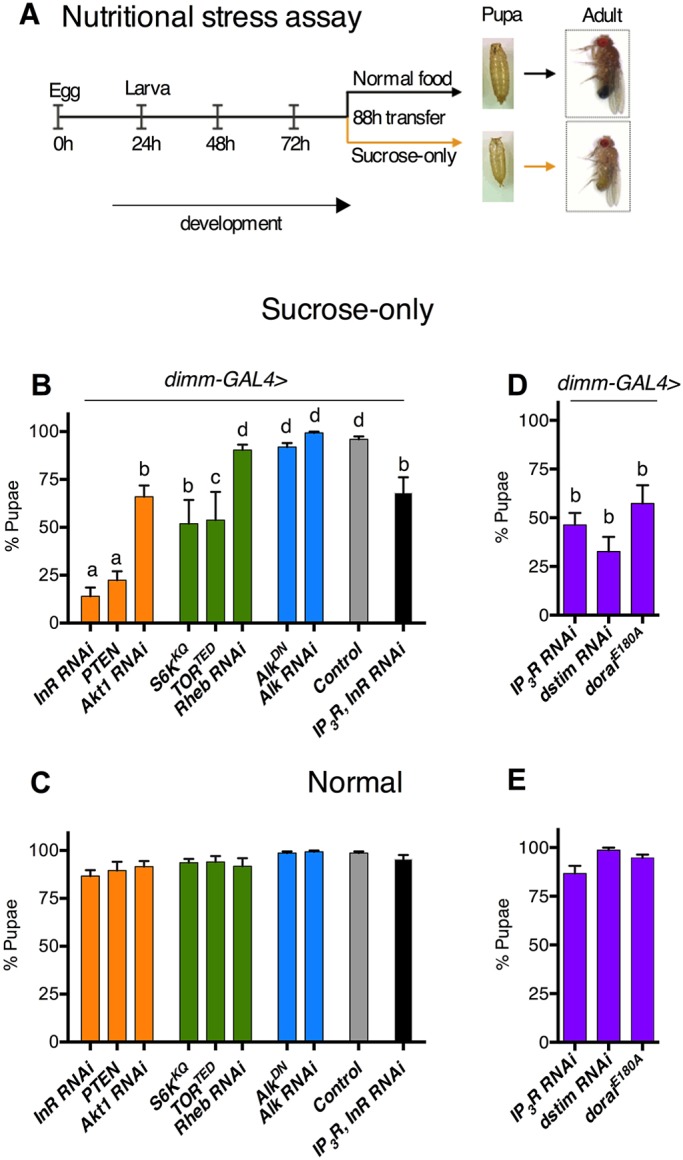
Downregulation of InR, TOR and intracellular Ca2+ signaling pathways in NE cells impairs larval development on sucrose. (A) Schematic of assay protocol with representative examples of pupa and adult appearance. (B,C) Pupariation upon reduction of InR/TOR signaling in dimm-GAL4 cells on sucrose (B) or normal diet (C). (D,E) Pupariation upon reduction of intracellular Ca2+ signaling on sucrose (D) or normal diet (E). Regulators of the InR (orange), TOR (green), Alk (blue) or intracellular Ca2+ (purple) signaling. UAS controls are shown in Fig. S1. Bars with the same letter represent statistically indistinguishable groups (one-way ANOVA with post-hoc Tukey's test, P<0.05). n=6 batches of 25 larvae each. Data represent mean±s.e.m.
Less than 25% of larvae pupariated on sucrose either after InR knockdown or after overexpression of a negative regulator of InR signaling, Pten (Fig. 1B). Manipulation of InR/TOR signaling components by overexpression of dominant-negative versions (TORTED, S6KKQ) or RNAi (Akt1, Rheb) affected pupariation mildly or not at all (Fig. 1C). Unlike NSCs (Cheng et al., 2011), neither overexpression of dominant-negative Alk (AlkDN) nor reduction via Alk RNAi, in NE cells, affected larval development, regardless of diet (Fig. 1B,C). Perhaps because NE cells are differentiated, they employ another mechanism to maintain insulin signaling during starvation. InR/TOR signaling affects NE cell size (Luo et al., 2013) and, interestingly, the pupariation rate that we observed in larvae on a sucrose-only diet correlates with the observations of Luo et al. For example, InR knockdown resulted in a NE cell size reduction of ∼18% (Luo et al., 2013) and gave a strong phenotype in our assay, whereas reduction of Rheb or Alk, which does not change NE cell size, gave no phenotype in our assay (Fig. 1B). Robust pupariation on normal food for the above genetic manipulations (Fig. 1C) suggested that dietary nutrients compensate for reduced InR/TOR signaling in NE cells. Together, these observations underscore the importance of NE cell function in overcoming nutrient stress.
Reducing intracellular Ca2+ signaling in NE cells by knockdown of either IP3R or dSTIM, or overexpression of a dominant-negative form of Orai (OraiE180A) (Pathak et al., 2015), reduced pupariation on the sucrose-only diet (Fig. 1D) but not on normal food (Fig. 1E). The similarities in outcome upon downregulation of either InR/TOR signaling or intracellular Ca2+ signaling prompted us to test the genetic interactions among components of the two pathways. Overexpression of IP3R in NE cells with InR knockdown led to increased pupariation on sucrose, compared with InR reduction alone (Fig. 1B), suggesting that under nutrient stress, IP3R can compensate for InR. Next, we investigated the IP3R mutant, itprku.
IP3R mutant larvae are deficient in protein metabolism
Although itprku exhibited robust pupariation on normal food (Fig. S2A), its pupariation was sensitive to reduction of yeast, the major source of dietary protein in ‘normal food’ (Fig. 2A). Pupariation was also reduced on sucrose (Fig. S2A), and rescued by supplementation with amino acids (Jayakumar et al., 2016) or amino acids and vitamins, but not lipids or vitamins alone (Fig. S2A).
Fig. 2.
Dysregulated protein metabolism in the IP3R hypomorph itprku can be rescued by overexpression of either InR or IP3R in NE cells. (A) Pupariation of 65 h larvae transferred into media with varying amounts of yeast. Two-way ANOVA, ****P<0.0001. (B) Pupariation over time after transfer to normal food at 88 h. Inset shows representative pupa. Relative pupal volume is shown in Fig. S2C. (C,E) Temporal changes in protein/TAG ratio, normalized to weight, for different genotypes. n≥5. See also Fig. S2D-F. (D,F) Ilp5 transcript levels in larval CNS normalized to rp49. n=6. (G) Pupariation of itprku on sucrose diet upon overexpression of positive regulators of InR and TOR signaling (orange and green) or intracellular Ca2+ signaling (purple) in NE cells. See also Fig. S3D. (H) Protein/TAG ratios normalized to weight. See also Fig. S2D-F. n≥8. (I) Ilp5 transcript levels in larval CNS normalized to rp49. n=4. Statistics: C,D,F, unpaired t-test, *P<0.05, **P<0.01, ****P<0.0001; E,G,H,I, one-way ANOVA. Bars with the same letter represent statistically indistinguishable groups (one-way ANOVA with a post-hoc Tukey's test, P<0.05). Data represent mean±s.e.m. CS, Canton S; ns, not significant; Wand., wandering.
At 88 h, an equal proportion of second (2L) and third (3L) instar itprku larvae co-exist (Fig. S2B), suggesting pleiotropic development delay. Additionally, when the pupariation rate of 88 h 3Ls was monitored, itprku displayed a lag of ∼24 h (Fig. 2B). Surprisingly, longer development time did not result in greater pupal volume (Fig. 2B; Fig. S2C), typically seen when larvae spend more time feeding (McBrayer et al., 2007). Although the weight of 3L itprku at 88 h, 112 h and as wandering larvae were not different from control (Fig. S2D), protein and triacylglyceride (TAG) levels were different (Fig. S2E,F). At 88 h and 112 h, itprku had higher TAG levels and lower protein levels. In wandering 3L, these levels were near normal (Fig. S2E,F). When plotted as protein/TAG ratio (Fig. 2C), it appeared that itprku had a slower rate of protein assimilation. Increased developmental time on normal food by itprku might therefore be a strategy to accumulate sufficient protein, and also explain why it does not result in increased body size.
Abnormal protein/TAG ratios suggested perturbed insulin signaling in itprku. We therefore measured transcript levels of Drosophila insulin-like peptides (dILPs) 2, 3, 5 and 6 from larval brains on normal food (Fig. 2D; Fig. S3A). Except Ilp5, which varied temporally to a significant degree (Fig. 2D), the trend for other dILPs (Fig. S2F) was similar to control. Although produced in the same set of NE cells (insulin-producing cells, IPCs), Ilp2, Ilp3 and Ilp5 transcripts are independently regulated. Ilp2 transcription is considered to be a systemic response, whereas Ilp3 is regulated by sugar (Kim and Neufeld, 2015), and Ilp5 by protein concentration (Geminard et al., 2009; Okamoto and Nishimura, 2015). Selective variation of Ilp5 thus indicated dysfunctional protein sensing in itprku. Overexpression of Ilp2 results in larger adults (Sato-Miyata et al., 2014); in contrast, the size of itprku pupae (Fig. S2C) is similar to that of controls, suggesting that the small increase of Ilp2 at 112 h (Fig. S2F) could be a response to Ilp5 upregulation.
We next downregulated IP3R in various cells/organs known to coordinate metabolism and development (Fig. S3B). As expected, IP3R knockdown in the prothoracic gland (PG) decreased pupariation on sucrose (Fig. S3B) because IP3R is required for ecdysone release from the PG (Venkatesh and Hasan, 1997; Yamanaka et al., 2015). However, unlike itprku or larvae with reduced IP3R in either NE cells or all neurons (Fig. S3B), supplementation with amino acids did not improve viability. This suggested that IP3R functions differently in PG cells and neurons. It is also likely that PG function in itprku is not as compromised as it is in the PG-IP3R-knockdown condition, as Ca2+ release in itprku neurons is reduced but not abolished (Joshi et al., 2004; Srikanth et al., 2004; Venkiteswaran and Hasan, 2009). Notably, reduction of IP3R in the fat body, or oenocytes (Fig. S3B), other sites of metabolic regulation in Drosophila, had no effect on larval development on sucrose.
IP3R reduction in NE cells also resulted in larvae with a lower protein/TAG ratio (Fig. 2E) and elevated Ilp5 expression (Fig. 2F). These features were similar (although reduced in magnitude) to itprku (Fig. 2C,D), suggesting the contribution of non-NE cells to itprku phenotype. Indeed, a set of glutamatergic neurons have been identified in which overexpression of IP3R is sufficient to rescue lethality of itprku on sucrose (Jayakumar et al., 2016).
Because itprku displayed abnormal transcript levels of Ilp5 and Ilp2, loss of IP3R specifically in the IPCs was tested, and was found not to affect pupariation on sucrose (Fig. S3B). This is consistent with previous observations that IP3R knockdown in IPCs does not phenocopy IP3R mutant phenotypes (Agrawal et al., 2009; Subramanian et al., 2013a). Together, these data suggest that IP3R in the IPCs does not affect dILPs directly. Increases in Ilp5 and Ilp2 transcripts in itprku (Fig. 2D; Fig. S3A) might instead be diet-dependent compensatory systemic responses. This is supported by the modest rescue of itprku pupariation on sucrose, with overexpression of either Ilp2 in IPCs (Jayakumar et al., 2016) or Ilp5 in NE cells (Fig. S3C).
Increasing InR/TOR or intracellular Ca2+ signaling in NE cells restores protein synthesis levels in the IP3R mutant
Next, we tested the effect of upregulation of InR and intracellular Ca2+ signaling components in NE cells of itprku. Overexpression of positive regulators of either the InR (InR, PI3KCaaX, Akt1) or the TOR (S6K, Rheb) pathway rescued itprku development under nutritional stress (Fig. 2G; Fig. S3D). These manipulations increase growth by promoting ribosomal biogenesis (Grewal, 2009), and in NE cells by increasing their size (Luo et al., 2013). Restoring intracellular Ca2+ signaling by overexpression of wild-type IP3R or STIM, as well as overexpression of CaMKII, a kinase that propagates Ca2+ signaling, in NE cells of itprku, also rescued larval lethality on sucrose (Fig. 2G; Fig. S3D).
At the systemic level, overexpression of either InR or IP3R in NE cells (dimm>InR/IP3R, itprku) was sufficient to increase protein/TAG ratios of itprku (Fig. 2H) at 112 h to levels similar to wandering stage itprku on normal food (Fig. 2C), suggesting that both pathways ultimately affected systemic protein metabolism. Protein/TAG ratios of rescues (dimm>InR/IP3R, itprku) are compared with dimm>InR/IP3R because non-linear increases in weight, protein and TAG levels were observed when either InR or IP3R alone were overexpressed in NE cells (Fig. S2D-F). Of note are TAG levels in dimmGAL4;itprku control and dimm>InR/IP3R itprku rescues (Fig. S2E). In both rescue conditions, protein levels increase (Fig. S2F), whereas TAG levels remain high (like dimmGAL4;itprku). Thus, insufficient protein, and not higher TAGs, correlates with the pupariation defect of itprku on sucrose.
As overexpression of Ilp5 rescued itprku partially and itprku displayed upregulated Ilp5 at 112 h on normal food, we investigated whether dimm>InR/IP3R rescues involved Ilp5. Nutrient withdrawal typically reduces Ilp2, Ilp3 and Ilp5 transcript levels significantly (Ikeya et al., 2002) and 88 h control larvae tested for levels of these dILPs after 24 h on sucrose showed expected reductions (Fig. S3E). Interestingly, itprku displayed greater reduction in Ilp5 (Fig. 2I) but not Ilp2 (Fig. S3F), when tested 24 h after transfer to sucrose. This reduction in Ilp5 probably affects itprku because at this time point (112 h) on normal food, it requires a ∼3-fold upregulation of Ilp5 (Fig. 2D). On sucrose, overexpression of InR or IP3R in NE cells increases Ilp5 levels in itprku (Fig. 2I) to control levels, without affecting Ilp2 levels (Fig. S3F). This suggests that dimm>InR/IP3R rescues itprku in part by systemically upregulating Ilp5.
IP3R positively regulates protein translation in NE cells
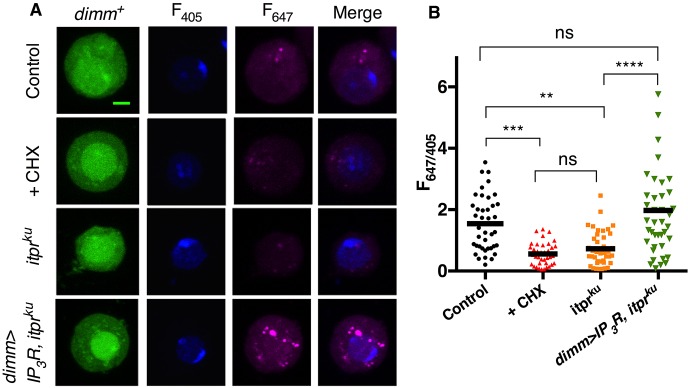
As systemic protein levels in itprku were rescued by overexpression of IP3R in NE cells, a cellular role for IP3R in protein translation was investigated. In itprku NE cells, obtained by culturing larval CNS, protein translation was reduced by ∼50%, similar to NE cells treated with the protein synthesis inhibitor cycloheximide (Fig. 3A,B). This reduction was rescued by overexpression of IP3R (Fig. 3A,B), strengthening the idea that IP3R, like InR, has a positive effect on protein synthesis.
Fig. 3.
NE cells from itprku display reduced protein synthesis. (A) Representative confocal images of NE cells (dimm+, GFP positive) in culture from the indicated genotypes and conditions. Newly synthesized peptides (F647) and nuclear volume (F405) were measured. Cells were treated with 10 µM cycloheximide (CHX) for 30 min. Scale bar: 2 µm. (B) Quantification of F647 and F405 from confocal images. n≥40. One-way ANOVA with post-hoc Holms–Sidak. **P<0.01, *** P<0.001, ****P<0.0001. Data represent mean±s.e.m. ns, not significant.
This observation is opposite to that reported for mammalian cell cultures (Brostrom and Brostrom, 2003), suggesting novel regulation of protein synthesis in neuropeptidergic cells. Reduced protein synthesis observed in mammalian cells treated with vasopression, angiotensin II and cholecystokinin (Brostrom et al., 1986; Kimball and Jefferson, 1990), agents that mobilize IP3R-mediated ER Ca2+ stores, can be rescued by addition of extracellular 2 mM Ca2+ during stimulation (Brostrom et al., 1986; Kimball and Jefferson, 1990; Sans et al., 2002). This suggests that extracellular Ca2+ entry counteracts ER-store Ca2+ effects on protein synthesis. Interestingly, when IP3R function is compromised in neurons, extracellular Ca2+ entry via SOCE is diminished (Venkiteswaran and Hasan, 2009). Thus, it is possible that a signaling cascade connects SOCE to protein translation, via IP3R.
Unlike NE cells, the rate of protein translation in itprku neurons was found to be no different from control neurons (Fig. S4A,B), suggesting that IP3R compensation of InR signaling is cell specific. Consistent with this, InR overexpression in cholinergic neurons of itprku did not rescue its viability on sucrose (Fig. S4C).
Peptide release from a subset of NE cells is regulated by IP3R-mediated Ca2+ transients from a subset of glutamatergic neurons (Jayakumar et al., 2016). Our results show that IP3R-mediated Ca2+ release also regulates protein translation in NE cells. Together, these observations illustrate the plurality of cellular processes controlled by IP3/Ca2+ signaling in the context of nutrient stress.
In summary, IP3R-mediated Ca2+ signaling helps maintain normal protein translation levels in NE cells, and this activity promotes systemic protein metabolism during larval development. On a nutrient-rich diet, loss of IP3R signaling is not detrimental, because dietary cues maintain insulin/TOR signaling, and thereby keep protein levels normal for completing development. Under starvation, dietary cues are lost. IP3/Ca2+ signaling possibly provides a nutrient-independent mechanism in order to maintain protein synthesis in cells essential to surviving nutrient stress, such as NE cells in which increased levels of cell surface receptors or neuropeptides might be required for modulating relevant neural circuits. As yet, there are no receptors or neuropeptides reported to be upregulated upon starvation in dimm+ NE cells, but there is precedence to suggest that they might exist. For example, in starved Drosophila, the receptor for short Neuropeptide F is upregulated in the antenna (Root et al., 2011), and in starved mammals levels of agouti-related peptide, which affects appetite and feeding, are increased (Henry et al., 2015). A recent screen identified IP3/Ca2+-coupled neuropeptide receptors, on glutamatergic neurons, that are required for larval adaptation to nutrient stress (Jayakumar et al., 2016). Neuropeptides from NE cells that couple to such receptors might function during starvation in our model (Fig. S5).
By focusing on animal development, this study integrates cellular observations and organismal phenotype. Therefore, it sets the framework for the discovery of mechanistic details of how stimulus-coupled increases in cytosolic Ca2+ can regulate protein synthesis in a cell-specific manner, and how that consequently regulates protein metabolism in the whole animal.
MATERIALS AND METHODS
Fly husbandry and stocks
Flies were reared on ‘normal’ laboratory food (1 L recipe: 80 g corn flour, 20 g glucose, 40 g sugar, 15 g yeast extract, 4 ml propionic acid, 5 ml p-hydroxybenzoic acid methyl ester in ethanol, 5 ml ortho butyric acid) in an incubator at 25°C under 12 h/12 h light/dark conditions. Fly strains are listed in supplementary Materials and Methods.
Larval nutritional stress assay
Eggs were collected on normal laboratory food for 6-8 h (depending on cross fecundity; ∼100 eggs per bottle) and allowed to mature for 88 h. For each genotype, six batches of 25 third instar larvae of similar size were transferred to a fresh vial of normal food or 100 mM sucrose in 1% agar. Pupae were scored 10 days after transfer. For development time, pupariation was scored every 24 h.
Pupal volume measurement
Pupal volume was approximated by measuring the width and height from pupal pictures, and applying the volumetric formula for cylinders, πr2h.
Weight, protein and TAG measurements on whole larvae
Thirty larvae were weighed on a microbalance (Shimadzu, Libror AEG 220). From this, ten larvae were homogenized in 1 ml of 0.2% Tween 20, followed by heating at 70°C for 10 min. Lysates were spun for 4 min at 4000 rpm (1500 g). For the BCA assay, 5 µl of the supernatant was withdrawn and protein estimated following the manufacturer's protocol (Pierce BCA Protein Assay Kit). To 50 µl of supernatant, 150 µl of enzyme-substrate mix (BeneSphera GPS TAG Kit, Avantor Performance Materials) was added to measure TAG levels.
RT-PCR
RNA was isolated from 10-12 larval brains at specified time points. cDNA synthesis was carried out as described (Pathak et al., 2015). All mRNA levels are reported as fold change normalized to rp49 (RpL32 – FlyBase). Primers are listed in supplementary Materials and Methods.
In vivo protein translation assay
Neuronal cultures from late third instar larval brains were prepared as described (Deb et al., 2016). After 16-18 h, cultures were processed for in vivo protein synthesis labeling using the manufacturer's protocol provided with the Click-iT Plus OPP Protein Synthesis Assay Kit (C10458). Confocal fluorescence images were collected using an Olympus FV1000 at 60× with 0.5 µm z-stacks. Between 10 and 15 cells were imaged per dish and at least three independent dishes were cultured for each genotype. Identical confocal settings were used for all imaging. Total fluorescence in each channel for the entire stack was measured using ImageJ and the background in each channel for each individual cell was subtracted for the measured region of interest.
Acknowledgements
We thank Paul Taghert, Michael O'Connor, Manfred Frasch, Ernst Hafen, Bloomington Drosophila Stock Center and Vienna Drosophila Resource Center for fly strains. The NCBS Central Imaging and Flow Facility helped with imaging.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
M. performed the experiments. M. and G.H. designed experiments, analyzed results and wrote the manuscript.
Funding
This work was supported by the Wellcome Trust/DBT India Alliance (Early Career Award IA/E/12/1/500742 to M.) and National Centre for Biological Sciences core funding (G.H.). Deposited in PMC for immediate release.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.145235.supplemental
References
- Agrawal N., Padmanabhan N. and Hasan G. (2009). Inositol 1,4,5- trisphosphate receptor function in Drosophila insulin producing cells. PLoS ONE 4, e6652 10.1371/journal.pone.0006652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal N., Venkiteswaran G., Sadaf S., Padmanabhan N., Banerjee S. and Hasan G. (2010). Inositol 1,4,5-trisphosphate receptor and dSTIM function in Drosophila insulin-producing neurons regulates systemic intracellular calcium homeostasis and flight. J. Neurosci. 30, 1301-1313. 10.1523/JNEUROSCI.3668-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbach J., Hummel P., Bickmeyer I., Kowalczyk K. M. M., Frank M., Knorr K., Hildebrandt A., Riedel D., Jäckle H., Kühnlein R. P. et al. (2014). A Drosophila in vivo screen identifies store-operated calcium entry as a key regulator of adiposity. Cell Metab. 19, 331-343. 10.1016/j.cmet.2013.12.004 [DOI] [PubMed] [Google Scholar]
- Brand A. H. and Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401-415. [DOI] [PubMed] [Google Scholar]
- Brostrom M. A. and Brostrom C. O. (2003). Calcium dynamics and endoplasmic reticular function in the regulation of protein synthesis: implications for cell growth and adaptability. Cell Calcium 34, 345-363. 10.1016/S0143-4160(03)00127-1 [DOI] [PubMed] [Google Scholar]
- Brostrom C. O., Bocckino S. B., Brostrom M. A. and Galuska E. M. (1986). Regulation of protein synthesis in isolated hepatocytes by calcium-mobilizing hormones. Mol. Pharmacol. 29, 104-111. [PubMed] [Google Scholar]
- Cheng L. Y., Bailey A. P., Leevers S. J., Ragan T. J., Driscoll P. C. and Gould A. P. (2011). Anaplastic lymphoma kinase spares organ growth during nutrient restriction in Drosophila. Cell 146, 435-447. 10.1016/j.cell.2011.06.040 [DOI] [PubMed] [Google Scholar]
- Deb B. K., Pathak T. and Hasan G. (2016). Store-independent modulation of Ca2+ entry through Orai by Septin 7. Nat. Commun. 7 10.1038/ncomms11751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbing J. and Sands J. (1971). Vulnerability of developing brain. IX. The effect of nutritional growth retardation on the timing of the brain growth-spurt. Biol. Neonate 19, 363-378. [DOI] [PubMed] [Google Scholar]
- Geminard C., Rulifson E. J. and Léopold P. (2009). Remote control of insulin secretion by fat cells in Drosophila. Cell Metab. 10, 199-207. 10.1016/j.cmet.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Grewal S. S. (2009). Insulin/TOR signaling in growth and homeostasis: a view from the fly world. Int. J. Biochem. Cell Biol. 41, 1006-1010. 10.1016/j.biocel.2008.10.010 [DOI] [PubMed] [Google Scholar]
- Henry F. E., Sugino K., Tozer A., Branco T. and Sternson S. M. (2015). Cell type-specific transcriptomics of hypothalamic energy-sensing neuron responses to weight-loss. Elife 4, e09800 10.7554/eLife.09800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeya T., Galic M., Belawat P., Nairz K. and Hafen E. (2002). Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr. Biol. 12, 1293-1300. 10.1016/S0960-9822(02)01043-6 [DOI] [PubMed] [Google Scholar]
- Jayakumar S., Richhariya S., Reddy O. V., Texada M. J. M., Hasan G., Richariya S., Reddy V. O., Texada M. J. M. and Hasan G. (2016). Drosophila larval to pupal switch under nutrient stress requires IP3R/Ca2+ signalling in glutamatergic interneurons. Elife 2016, e17 10.7554/eLife.17495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R., Venkatesh K., Srinivas R., Nair S. and Hasan G. (2004). Genetic dissection of itpr gene function reveals a vital requirement in aminergic cells of Drosophila larvae. Genetics 166, 225-236. 10.1534/genetics.166.1.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. and Neufeld T. P. (2015). Dietary sugar promotes systemic TOR activation in Drosophila through AKH-dependent selective secretion of Dilp3. Nat. Commun. 6, 6846 10.1038/ncomms7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball S. R. and Jefferson L. S. (1990). Mechanism of the inhibition of protein synthesis by vasopressin in rat liver. J. Biol. Chem. 265, 16794-16798. [PubMed] [Google Scholar]
- Luo J., Liu Y. and Nässel D. R. (2013). Insulin/IGF-regulated size scaling of neuroendocrine cells expressing the bHLH transcription factor Dimmed in Drosophila. PLoS Genet. 9, e1004052 10.1371/journal.pgen.1004052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBrayer Z., Ono H., Shimell M., Parvy J.-P., Beckstead R. B., Warren J. T., Thummel C. S., Dauphin-Villemant C., Gilbert L. I. and O'Connor M. B. (2007). Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Dev. Cell 13, 857-871. 10.1016/j.devcel.2007.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nässel D. R. and Winther A. M. E. (2010). Drosophila neuropeptides in regulation of physiology and behavior. Prog. Neurobiol. 92, 42-104. 10.1016/j.pneurobio.2010.04.010 [DOI] [PubMed] [Google Scholar]
- Okamoto N. and Nishimura T. (2015). Signaling from glia and cholinergic neurons controls nutrient-dependent production of an insulin-like peptide for drosophila body growth. Dev. Cell 35, 295-310. 10.1016/j.devcel.2015.10.003 [DOI] [PubMed] [Google Scholar]
- Padmanabha D. and Baker K. D. (2014). Drosophila gains traction as a repurposed tool to investigate metabolism. Trends Endocrinol. Metab. 25, 518-527. 10.1016/j.tem.2014.03.011 [DOI] [PubMed] [Google Scholar]
- Park D., Veenstra J. A., Park J. H. and Taghert P. H. (2008). Mapping peptidergic cells in Drosophila: where DIMM fits in. PLoS ONE 3, e1896 10.1371/journal.pone.0001896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak T., Agrawal T., Richhariya S., Sadaf S. and Hasan G. (2015). Store-operated calcium entry through orai is required for transcriptional maturation of the flight circuit in Drosophila. J. Neurosci. 35, 13784-13799. 10.1523/JNEUROSCI.1680-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakriya M. and Lewis R. S. (2015). Store-operated calcium channels. Physiol. Rev. 95, 1383-1436. 10.1152/physrev.00020.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root C. M., Ko K. I., Jafari A. and Wang J. W. (2011). Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell 145, 133-144. 10.1016/j.cell.2011.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sans M. D., Kimball S. R. and Williams J. A. (2002). Effect of CCK and intracellular calcium to regulate eIF2B and protein synthesis in rat pancreatic acinar cells. Am. J. Physiol. Gastrointest. Liver Physiol. 282, G267-G276. 10.1152/ajpgi.00274.2001 [DOI] [PubMed] [Google Scholar]
- Sato-Miyata Y., Muramatsu K., Funakoshi M., Tsuda M. and Aigaki T. (2014). Overexpression of dilp2 causes nutrient-dependent semi-lethality in Drosophila. Front. Physiol. 5, 147 10.3389/fphys.2014.00147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth S., Wang Z., Tu H., Nair S., Mathew M. K., Hasan G. and Bezprozvanny I. (2004). Functional properties of the Drosophila melanogaster inositol 1,4,5-trisphosphate receptor mutants. Biophys. J. 86, 3634-3646. 10.1529/biophysj.104.040121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian M., Jayakumar S., Richhariya S. and Hasan G. (2013a). Loss of IP3 receptor function in neuropeptide secreting neurons leads to obesity in adult Drosophila. BMC Neurosci. 14, 157 10.1186/1471-2202-14-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian M., Metya S. K., Sadaf S., Kumar S., Schwudke D. and Hasan G. (2013b). Altered lipid homeostasis in Drosophila InsP3 receptor mutants leads to obesity and hyperphagia. Dis. Model. Mech. 6, 734-744. 10.1242/dmm.010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghert P. H. and Nitabach M. N. (2012). Peptide neuromodulation in invertebrate model systems. Neuron 76, 82-97. 10.1016/j.neuron.2012.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh K. and Hasan G. (1997). Disruption of the IP3 receptor gene of Drosophila affects larval metamorphosis and ecdysone release. Curr. Biol. 7, 500-509. 10.1016/S0960-9822(06)00221-1 [DOI] [PubMed] [Google Scholar]
- Venkiteswaran G. and Hasan G. (2009). Intracellular Ca2+ signaling and store-operated Ca2+ entry are required in Drosophila neurons for flight. Proc. Natl. Acad. Sci. USA 106, 10326-10331. 10.1073/pnas.0902982106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka N., Marqués G. and O'Connor M. B. (2015). Vesicle-mediated steroid hormone secretion in Drosophila melanogaster. Cell 163, 907-919. 10.1016/j.cell.2015.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]