Abstract
Knowledge of the 3D structure of glycoproteins and protein–carbohydrate complexes is indispensable to fully understand the biological processes they are involved in. Carbohydrate Structure Suite is an attempt to automatically analyse carbohydrate structures contained in the PDB and make the results publicly available on the internet. Characteristic torsion angles, glycoprotein sequences and carbohydrate–protein interactions are analysed. Furthermore, tools to crosslink the PDB and carbohydrate databases and to check the integrity of carbohydrate 3D structures are included. The service is available at (www.dkfz.de/spec/css/).
INTRODUCTION
Carbohydrate–protein interactions are implicated in a variety of cell–cell and cell–matrix recognition events, ranging from fertilization, cellular differentiation and development to pathological situations like inflammation, viral and bacterial infections, immune response and metastasis (1–3). These events require a specific recognition of different carbohydrate structures by carbohydrate-binding proteins, the lectins (4).
Most of the carbohydrates involved in these recognition processes are part of glycoproteins. Protein glycosylation is probably the most common and complex co- and post-translational modification. In many cases, the glycan chains are mandatory for the correct folding process (5). They also alter the properties of the proteins, e.g. by increasing solubility, protecting them from proteolysis or reducing backbone flexibility (2). Variations in the terminal residues of glycans attached to a protein enable a cell- and tissue-specific fine-tuning of protein properties without a change in the amino acid sequence (5).
To be able to understand these processes in detail, it is often indispensable to know the 3D structures of the glycoproteins or protein–carbohydrate complexes, respectively. The largest publicly available source of such 3D structures is the PDB (6). Unfortunately, there is no standard nomenclature for carbohydrate residues (2,7), which makes the analysis of these data difficult. Moreover, many of the structural characteristics of interest are secondary data, given only implicitly in the structures, e.g. linkage torsion angles. It is impossible to extract this information efficiently based on the nomenclature that is present in the PDB entries alone.
Carbohydrate Structure Suite (CSS) is an attempt to automatically detect and assign carbohydrate structures in the PDB in a nomenclature-independent manner, analyse them and make the results publicly available on the internet.
pdb2linucs: detection of carbohydrate structures in PDB entries
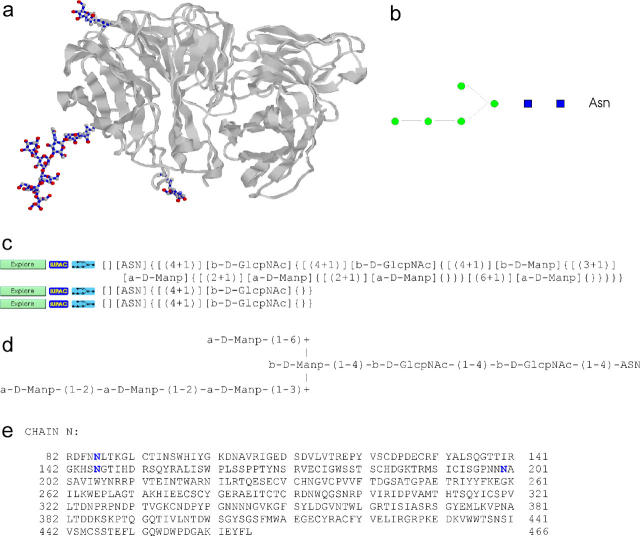
pdb2linucs (7) detects carbohydrate structures using an algorithm that is based on element types and atom coordinates only. The algorithm is independent of residue notation and thus bridges the gap of a missing standard nomenclature for carbohydrates in PDB entries. The program can be queried by PDB ID or by uploading a structure from a local computer. The results are displayed using the LINUCS nomenclature, a linear, unique notation for carbohydrate structures (8). This notation is also used to query GlycosciencesDB, the former SweetDB (9). In case there is a related entry in that database, a direct link is offered (Figure 1). Thereby, pdb2linucs establishes a cross-linking between a protein database and a carbohydrate database.
Figure 1.
pdb2linucs output. The 3D structure of the PDB entry 1a14 is displayed using Jmol or Chime, and the detected carbohydrate residues are highlighted (a). Below, the carbohydrate chains are listed in LINUCS nomenclature. Links to corresponding entries in the GlycosciencesDB are established (c). Furthermore, links to the IUPAC notation of the structures (d) and a graphical representation (b) are also provided. In case covalently bound glycans are present, the amino acid sequence is given as well. Glycosylated residues are highlighted (e).
Using an extended version of pdb2linucs, three data sets of carbohydrate- and glycoprotein-related data were generated from the PDB. These data sets, entitled GlyTorsionDB, GlySeqDB and GlyVicinityDB, are stored in extensible markup language (XML)-based text files and queried by specialized software, which can be accessed through web interfaces.
GlyTorsionDB: a database of carbohydrate torsion angles
3D protein structures are mainly characterized by their backbone torsion angles (10). Using a similar approach, carbohydrate structures can be described by the torsion angles of the glycosidic linkages connecting monosaccharide residues. GlyTorsionDB is a database containing the torsion angle values of all carbohydrate linkages present in the PDB. Furthermore, it includes the ring torsion angles of the single monosaccharides, omega torsion angles for exocyclic hydroymethyl-groups, side chain torsion angles of asparagine residues involved in N-linked glycosylation and the torsion angles of N-acetyl groups attached to carbohydrate rings. The database is queried by the software GlyTorsion.
In most cases, a statistic evaluation of the distribution of angles for a specific structural feature is the most informative way to look at the data. Therefore, GlyTorsion performs a graphical output of results by generating diagrams or plots (histograms). From these, the user can see at a glance, which angles are preferred by the residues of interest. In a subsequent step, the numerical values of the angles together with the information about the PDB entries they originate from can be displayed for the entire data, which have been used to generate the diagram. Additionally, the detailed data of user-defined ranges can be displayed.
carp: CArbohydrate Ramachandran Plot
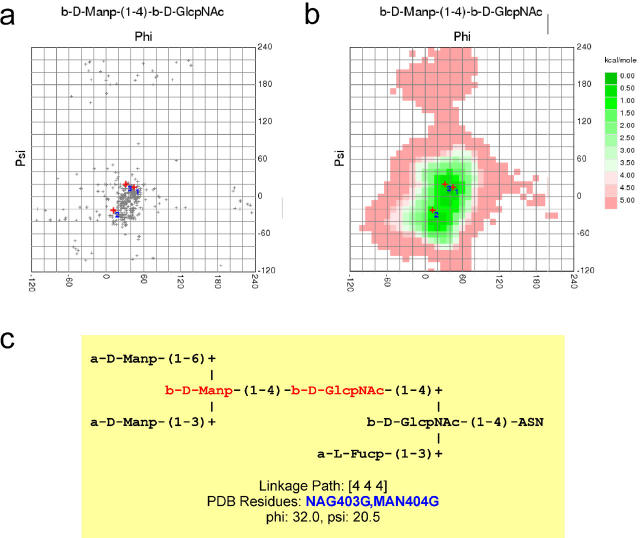
Another possibility to query GlyTorsionDB is carp, the CArbohydrate Ramachandran Plot. The ‘Ramachandran Plot’, where protein backbone torsion angles Φ and Ψ are plotted against each other, is a frequently used tool to evaluate the quality of a protein 3D structure (11). For carbohydrate structures, linkage torsions can be evaluated in a similar way. In contrast to proteins, however, the frequency of populated torsion angles depends on the monosaccharide residues involved in the linkage as well as the kind of linkage (1–3, 1–4, etc.). carp analyses carbohydrate data given in PDB files using the pdb2linucs algorithm and compares them to the data available in GlyTorsionDB (Figure 2a). For each different combination of monosaccharides and linkage type, a separate plot is generated.
Figure 2.
Carbohydrate Ramachandran plot. Carbohydrate linkage torsions can be analysed in an approach similar to the Ramachandran plot used for protein backbone torsions. For carbohydrates, a separate plot is needed for each combination of linkage type and monosaccharide residues. The observed torsions (PDB entry 1b5f ) are compared to other torsions of the same type found in the PDB (a) or to calculated energy plots (b). For each torsion, the corresponding monosaccharides are highlighted within the IUPAC representation of the carbohydrate chain (c).
For seldom occurring linkages, there is only few data available in the PDB. Therefore, carp offers the possibility to query another database, the GlycoMapsDB, available at www.dkfz.de/spec/glycomaps/. That database contains conformational maps derived from molecular dynamics simulations and is therefore independent of the number of entries found for a given linkage in the PDB (Figure 2b).
GlySeqDB: glycoprotein sequences database
About 70% of all proteins contain potential N-glycosylation sites (Asn-Xxx-Ser/Thr, Xxx not Pro). For reasons that are still unclear, not all of them are in fact glycosylated (12). Unfortunately, only very few experimental data are available, where it has been unambiguously shown, that a specific glycosylation site is occupied by a defined glycan. This lack in experimental data hampers to decipher the structural parameters, which are required for glycosylation. Therefore, glycoprotein sequences taken from the PDB and from carefully assigned SwissProt entries have been used to gain deeper insights into factors that regulate glycan attachment to a N-glycosylation site (13, 14). GlySeqDB makes these data now available, which had to be extracted manually until recently. GlySeqDB provides online access to glycoprotein sequences originating from PDB and SwissProt. The query software GlySeq analyses the statistics of amino acids in the sequential neighbourhood of N- and O-glycosylation sites. In addition to the results in text mode, GlySeq is able to display graphical results by generating diagrams.
Within the PDB, there are many redundant sequences. If, for example, a protein is a trimer consisting of three identical chains, and the structure of that protein was resolved at four different conditions, then there are 12 identical protein chains in the database. Therefore, GlySeqDB also contains datasets where redundant information is removed.
Besides the selection of the source dataset and the type of glycosylation site, GlySeq offers the use of additional filters, e.g. to analyse only those N-glycosylation sites where the residue at position +2 is Ser and not Thr or vice versa.
GlyVicinityDB: a database about carbohydrate–protein interactions
Besides the amino acids in sequential neighbourhood of glycosylation sites, those in the spatial vicinity of carbohydrate residues determine the characteristics of glycoproteins. The latter ones are of special interest for the examination of the interactions between carbohydrates and carbohydrate-binding proteins. Since carbohydrate ligands are not covalently bound, sequence analysis comparable to that for glycosylation sites is not applicable here.
GlyVicinityDB contains lists of distances about the amino acids in the spatial vicinity of carbohydrate residues present in the PDB. The database is queried by the software GlyVicinity. This program performs a statistical analysis of the frequency of amino acids within a user-definable distance (up to 10 Å) of carbohydrate residues. Like GlySeq, GlyVicinity is also able to display results as diagrams. In a subsequent step, examination of the atoms forming the closest contacts between carbohydrate and amino acid residues is also possible. Moreover, GlyVicinity enables the user to define subsets of the database by selecting the PDB entries to be considered, e.g. to investigate virus-related proteins only.
pdb-care: integrity check of carbohydrate structures
An analysis of the entire carbohydrate data in the PDB revealed that about 30% of all carbohydrate-containing PDB entries comprise one or several errors, most of which are due to inconsistencies in residue nomenclature or erroneous connection data (7). This results from a lack of check software for carbohydrate structures like it exists for the protein parts of PDB entries (11). pdb-care (PDB CArbohydrate REsidue check) (15) provides such a check by comparing the residue names derived by the pdb2linucs algorithm with those present in the file to be checked; and inconsistencies are reported. Furthermore, the program performs a check of the information given in the CONECT records of PDB files. Frequent use of this software will increase the quality of carbohydrate information within the PDB.
Updates
The PDB is updated weekly with new structures. The update process of the databases contained in CSS is mainly automatic; therefore the service always represents the actually available data in the PDB.
Implementation
CSS is running on a Linux PC with Apache web server software. Interaction with the user is realized by PHP interfaces. The data sets are stored in XML-formatted text files, and the programs to query these data are written in C.
Visualization of protein and carbohydrate structures ( pdb2lincus, GlyVicinity) or torsion angles (GlyTorsion, carp) is performed by the java applet Jmol (http://jmol.sourceforge.net) or the plugin Chime (www.mdlchime.com). Diagrams and plots are generated in scalable vector graphics (SVG) format. For browsers that cannot display SVG files, they are converted to graphics interchange format (GIF) files using ImageMagick (http://imagemagick.sourceforge.net).
The service is hosted at and maintained by the German Cancer Research Centre in Heidelberg, Germany. It can be accessed online at (www.dkfz.de/spec/css/). The databases and tools, which CSS consist of, are also integrated into the Glycosciences web portal (www.glycosciences.de).
Outlook
The download of retrieved data is currently only possible using copy/paste mechanisms. We are currently working to provide XML formats for all retrievable data, so that an efficient way of data exchange will be established. Additionally, we will provide access to structural parameters of the protein backbone in the vicinity of glycosylation sites. With the GlyProt application (available at the glycosciences web portal), an efficient tool is available to perform in silico glycosylation of a given 3D protein structure.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Thomas Götz for carefully extracting the glyco-related data contained in SwissProt. The development of CSS tools is funded by a grant from the German Research Council (Deutsche Forschungsgemeinschaft, DFG) within the digital library program.
REFERENCES
- 1.Yarema K.J. and Bertozzi,C.R. (2001) Characterizing glycosylation pathways. Genome Biol., 2, review S0004.0001–0004.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wormald M.R., Petrescu,A.J., Pao,Y.L., Glithero,A., Elliott,T. and Dwek,R.A. (2002) Conformational studies of oligosaccharides and glycopeptides: complementarity of NMR, X-ray crystallography, and molecular modelling. Chem. Rev., 102, 371–386. [DOI] [PubMed] [Google Scholar]
- 3.Imberty A. and Pérez,S. (1995) Stereochemistry of the N-glycosylation sites in glycoproteins. Protein Eng., 8, 699–709. [DOI] [PubMed] [Google Scholar]
- 4.Loris R. (2002) Principles of structures of animal and plant lectins. Biochim. Biophys. Acta, 1572, 198–208. [DOI] [PubMed] [Google Scholar]
- 5.Helenius A. and Aebi,M. (2001) Intracellular functions of N-linked glycans. Science, 291, 2364–2369. [DOI] [PubMed] [Google Scholar]
- 6.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lütteke T., Frank,M. and von der Lieth,C.-W. (2004) Data mining the protein data bank: automatic detection and assignment of carbohydrate structures. Carbohydr. Res., 339, 1015–1020. [DOI] [PubMed] [Google Scholar]
- 8.Bohne-Lang A., Lang,E., Forster,T. and von der Lieth,C.W. (2001) LINUCS: linear notation for unique description of carbohydrate sequences. Carbohydr. Res., 336, 1–11. [DOI] [PubMed] [Google Scholar]
- 9.Loss A., Bunsmann,P., Bohne,A., Schwarzer,E., Lang,E. and von der Lieth,C.W. (2002) SWEET-DB: an attempt to create annotated data collections for carbohydrates. Nucleic Acids Res., 30, 405–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith L.J., Bolin,K.A., Schwalbe,H., MacArthur,M.W., Thornton,J.M. and Dobson,C.M. (1996) Analysis of main chain torsion angles in proteins: prediction of NMR coupling constants for native and random coil conformations. J. Mol. Biol., 255, 494–506. [DOI] [PubMed] [Google Scholar]
- 11.Abola E.E., Bairoch,A., Barker,W.C., Beck,S., Benson,D.A., Berman,H.M., Cameron,G., Cantor,C., Doubet,S., Hubbard,J.P. et al. (2000) Quality control in databanks for molecular biology. Bioessays, 22, 1024–1034. [DOI] [PubMed] [Google Scholar]
- 12.Apweiler R., Hermjakob,H. and Sharon,N. (1999) On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta, 1473, 4–8. [DOI] [PubMed] [Google Scholar]
- 13.Petrescu A.J., Milac,A.L., Petrescu,S.M., Dwek,R.A. and Wormald,M.R. (2004) Statistical analysis of the protein environment of N-glycosylation sites: implications for occupancy, structure and folding. Glycobiology, 14, 103–114. [DOI] [PubMed] [Google Scholar]
- 14.Ben-Dor S., Esterman,N., Rubin,E. and Sharon,N. (2004) Biases and complex patterns in the residues flanking protein N-glycosylation sites. Glycobiology, 14, 95–101. [DOI] [PubMed] [Google Scholar]
- 15.Lütteke T. and von der Lieth,C.W. (2004) pdb-care (PDB carbohydrate residue check): a program to support annotation of complex carbohydrate structures in PDB files. BMC Bioinformatics, 5, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]