Abstract
The Human Ageing Genomic Resources (HAGR) is a collection of online resources for studying the biology of human ageing. HAGR features two main databases: GenAge and AnAge. GenAge is a curated database of genes related to human ageing. Entries were primarily selected based on genetic perturbations in animal models and human diseases as well as an extensive literature review. Each entry includes a variety of automated and manually curated information, including, where available, protein–protein interactions, the relevant literature, and a description of the gene and how it relates to human ageing. The goal of GenAge is to provide the most complete and comprehensive database of genes related to human ageing on the Internet as well as render an overview of the genetics of human ageing. AnAge is an integrative database describing the ageing process in several organisms and featuring, if available, maximum life span, taxonomy, developmental schedules and metabolic rate, making AnAge a unique resource for the comparative biology of ageing. Associated with the databases are data-mining tools and software designed to investigate the role of genes and proteins in the human ageing process as well as analyse ageing across different taxa. HAGR is freely available to the academic community at http://genomics.senescence.info.
INTRODUCTION
Human ageing is a major but poorly understood biological problem (1). Even though ageing is universal amongst humans, there is little information on the genetics of human ageing and few online resources. Herein, we present the Human Ageing Genomic Resources (HAGR), a novel collection of resources for studying the biology of human ageing. HAGR is composed of two core databases associated with a variety of tools to analyse and interpret the datasets. First, we developed GenAge, a curated database of genes related to human ageing. Given that identifying the genes that determine the different rates of ageing amongst organisms is a major endeavour (2), HAGR also includes AnAge, an integrative database of the ageing phenotype in several species.
In the wake of the human genome, high-throughput projects in functional genomics are generating an immense amount of data. The next challenge is to digest and analyse these different types of data to enhance our understanding of biological processes (3). In other biomedical sciences, such as oncology, several databases already exist to integrate information about human genes in health and disease. Our aim in building HAGR is to provide researchers with a new set of online resources to study the genetic basis of human ageing.
GenAge DATA SOURCES, SELECTION AND ANNOTATION PROCESS
Great care was taken to ensure that the selection of GenAge entries was as unbiased as possible. Since we wanted to focus on genes that may influence ageing, rather than being a consequence of ageing, the selection process was largely dependent on genetic perturbations. For example, mutations that either delay or accelerate ageing in mice provide some of the few clear hints on the genetics of ageing. Consequently, genes that may modulate ageing in humans (4) or mammals (3) served as a first selection to GenAge (http://genomics.senescence.info/genes/clues.html). As advocated by many others (1), care was taken to determine whether a given gene affects the ageing process or simply preserves health. For example, many murine genes are lethal at early ages and certainly decrease longevity but do not affect ageing; as such, only genes that may affect the ageing process were considered. The selection is, of course, subjective and so a gene had to accelerate or delay multiple age-related changes or change the rate of ageing for it to be considered.
GenAge entries were largely selected based on findings from model organisms and many entries represent human homologues of genes shown to affect ageing in model organisms. Since it is difficult to extrapolate results from model organisms to human biology (5), great care was used to assess whether a human homologue of a gene discovered in a model organism may be related to human ageing. If applicable, the following criteria were employed: (i) the influence of the gene in the model organism's ageing process; (ii) literature suggesting the human homologue has a similar function; (iii) information on the phenotype of human variants or mutations of the gene; and (iv) effects on ageing of the genetic manipulation—e.g. overexpression or knock-out—of the gene's product(s) in mammals. Consequently, evolutionary distant models such as invertebrates had a much smaller impact on GenAge than mammalian models such as mice.
In addition to genes directly shown to affect ageing in humans or model organisms, entries were also selected by association. Following the criteria set above and always depending on the available literature, further entries were selected due to their interaction with genes or pathways shown to affect ageing in humans or model organisms. For instance, proteins found highly associated with other proteins or pathways previously linked with ageing were selected. A functional clustering of pathways involved was also derived to identify the pathways of interest (http://genomics.senescence.info/genes/function.html), in line with previous such works (2). Together, this resulted in a number of genes being selected as ‘guilt-by-association’ due to their high connectivity to previously selected entries.
GenAge uses data from a number of other databases. Promoter sequences were derived from the Eukaryotic Promoter Database (EPD) (6), while open reading frame (ORF) and protein sequences were obtained from RefSeq (7). Homologues were retrieved from HomoloGene (7) and cytogenetic information was taken from the UCSC Genome Browser (8). Protein–protein interactions were obtained from HPRD (9), itself a curated database and thus less prone to the high number of false positives that affect many protein interaction databases. Additional information was obtained from GeneCards (10), HPRD (9), OMIM, LocusLink, PubMed and UniGene (7). GenAge employs the nomenclature set by the HUGO Gene Nomenclature Committee (11), though commonly used aliases are also included.
Each entry consists of a mix of automatically extracted annotation and manually curated information. Subsequent updates will profit from the automated methods implemented, and we expect to release new versions of GenAge on a regular basis. Manually curated information, such as the description of the gene's relevance to human ageing, was largely based on a careful review of the literature. A database design plan is available online (http://genomics.senescence.info/genes/schema.gif).
AnAge DATA SOURCES, CURATION AND ANNOTATION PROCESS
Rates of ageing were taken from a variety of sources for inclusion in AnAge and expressed as mortality rate doubling time. Alternatively, the mortality rate doubling time was calculated according to published mortality data, as described (12). If available, a description of the ageing process and age-related decline was included based on the literature. The ageing process for a given species was described under near-optimal environmental and typical genetic backgrounds. In other words, animals in captivity were preferred but genetic manipulations that affected ageing were not included in AnAge. The rationale being that humans live in protected environments and since AnAge was designed for studying human ageing through a comparative biology approach, it is logical that animal species are also represented in near-optimal conditions.
Metabolic rates were taken from various sources as well as additional data supplied by Van Savage et al. (13). Life history traits, such as gestation time and age at sexual maturity were taken from numerous sources and references are provided in each entry. Longevity records and taxonomic classifications were initially derived from the work of James Carey and Debra Judge (14), and further refined with data from a variety of sources as well as communications with zoos and parks. While longevity records were used to estimate maximum life span, they are a function of population size and so more error-prone than rates of ageing.
As with GenAge, although data was automatically entered into the database, several entries were manually verified. The automated procedures implemented will be useful in subsequent updates. AnAge's schema is also available online (http://genomics.senescence.info/species/schema.gif).
THE GenAge AND AnAge DATABASES
The primary resource in HAGR is GenAge, a manually curated database of genes related to human ageing. Each GenAge entry includes, where available, protein, ORF and promoter sequences, chromosomal location, protein–protein interactions, function, tissue expression, protein functional domains, and a description of the gene's relevance to human ageing (Figure 1). To permit a rough quantification of the impact of a given gene on human ageing, each entry includes a brief explanation of why the gene was chosen for the database. Although only human entries are included, homologues in other organisms are mentioned so researchers working in model organisms may relate their findings to human ageing. At present, there are over 200 manually curated entries in GenAge with over 1000 associated homologues (Supplementary Table 1).
Figure 1.

GenAge entry for CKN1, Cockayne syndrome type 1. This entry includes basic information regarding the CKN1 gene and protein, a description of CKN1's relevance to ageing research, CKN1's protein–protein interactions, homologues in model organisms, a selection of references regarding CKN1 and its possible involvement in human ageing, and an extensive list of hyperlinks to external resources.
Although GenAge is not a literature database, it contains a selection of literature references for a total of over 1000 references—each including, if applicable, a hyperlink to PubMed. In addition, hyperlinks in each entry point the user to a variety of additional sources of information; excluding hyperlinks to PubMed, roughly 2000 hyperlinks are featured in GenAge. Our goal is not only to provide the most relevant information about each entry in the context of human ageing, but also make it easy for researchers to find additional online information. At the time of writing, GenAge's latest version is build 8 (30/09/2004), available at http://genomics.senescence.info/genes/.
Another important database in HAGR is AnAge, which provides a description of the ageing process in animals. The aim of AnAge is to allow comparisons between human ageing and that of animals as well as help understand the evolutionary forces shaping ageing and longevity. Each entry features, if available, the organism's taxonomic classification, its maximum life span, mortality rate doubling time, initial mortality rate, metabolic rate and a description of its ageing phenotype. Species with negligible senescence are highlighted. Data on life history traits, such as developmental schedules, typical body weight, gestation period and age of sexual maturation, are also included in AnAge (Figure 2).
Figure 2.

AnAge entry for the tiger salamander, Ambystoma tigrinum. This entry includes the taxonomy of the tiger salamander, its maximum life span, life history traits and a description of its ageing phenotype. Information on metabolism is also displayed as well as a selection of references and hyperlinks to external resources.
The taxonomic classification includes kingdom, phylum, class, order, family, genus and species. Over 400 literature references are also included. Of course, AnAge does not aim to include a rigorous description of ageing in each entry for that would be impossible in the case of well-studied species, such as humans and mice. Instead, our aim is to provide the most relevant features of ageing for a given animal as well as include works using a comparative biology approach to study ageing and life history events. Since our focus is on human ageing, we paid particular attention to evolutionary close species: mammals, reptiles, birds, amphibians and fishes, though other commonly used models in biology are also included. At present AnAge includes over 2000 entries, of which roughly a third are mammals (Table 1). As of writing, AnAge's latest version is build 5 (15/09/2004), available at http://genomics.senescence.info/species/.
Table 1. Overview of chordates present in AnAge.
| Chordates (class) | AnAge entries |
|---|---|
| Amphibia | 83 |
| Aves | 860 |
| Mammalia (order) | 801 |
| Artiodactyla | 123 |
| Carnivora | 147 |
| Cetacea | 38 |
| Chiroptera | 59 |
| Insectivora | 35 |
| Marsupialia | 77 |
| Primates | 137 |
| Rodentia | 131 |
| Others | 54 |
| Reptilia | 304 |
| Others (Piscis, etc.) | 233 |
DATA-ANALYSIS TOOLS
GenAge is a relational database that aims not only to provide information about genes in the context of ageing and human biology, but also allow an overview of the current knowledge on the genetics of human ageing. Consequently, GenAge features methods to search and analyse genes or clusters of genes, construct networks, and study pathways. For instance, it is possible to search genes related to a certain function or present in a given cellular organelle, allowing users to analyse the genetic network of their choice and find novel relations between the genes involved. It is also possible to analyse entries based on the selection criteria. GenAge's browser is available at http://genomics.senescence.info/genes/browser.php.
Using GenAge, it is possible to seek genes related to common pathways through, for example, protein–protein interactions and a number of visualization tools are available. One example is the Interactions Graphical Display (IGD) script, used to display protein–protein interactions. More advanced users may download protein–protein interactions for use with biological pathway analysis software (Figure 3). In addition, it is possible to find links between GenAge entries by mining the bibliography. The links can then be displayed through IGD.
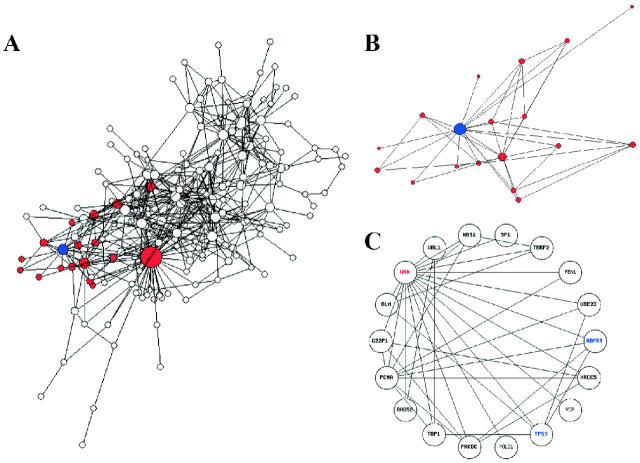
Figure 3.
(A) Global protein–protein interaction map of GenAge. A total of 502 protein–protein interactions are displayed. The WRN protein is highlighted in blue and connecting nodes, with one degree of freedom, are highlighted in red. Layout computed and displayed using InterViewer 3.7 (Inha University, WI Lab, South Korea). (B) Magnification of the WRN network taken from the global protein–protein interaction map using InterViewer 3.7 (Inha University, WI Lab, South Korea). (C) Protein–protein interactions of WRN with one degree of freedom as generated by IGD. Entries identified through their relevance to human (red) or mammalian ageing (blue) are displayed in different colours. Genes only indirectly associated with ageing are displayed using a non-bold font.
AnAge's design allows users to browse through AnAge from a phylogenetic perspective (Supplementary Figure 1). Once a taxum of interest is located, it is possible to find longevity information about that taxum or compare it with other taxa. It is also possible to browse through the phylogenetic tree until a specific entry is found. AnAge's browser is available at http://genomics.senescence.info/species/browser.php.
In order to understand the evolution of ageing and longevity, a number of visualization tools are associated with AnAge. One example is the Phylogentic Tree Plotter (PTP), which is used to display phylogenetic relationships of species in the database (Supplementary Figure 2). PTP is available at http://genomics.senescence.info/species/ptp.php.
SOFTWARE AND PLATFORM
In addition to PTP and IGD, several tools are available in HAGR. It is possible to analyse a query sequence to locate, for instance, CpG islands. Hyperlinks also link to other online resources and tools. For example, associated with GenAge are visual tools to identify the chromosomal location of genes, which can be used, for instance, in gene expression studies to locate chromosomal clustering. All tools in HAGR are available for download. Included in HAGR are the Ageing Research Computational Tools (ARCT), a Perl toolkit designed to investigate the function and relevance of genes in the ageing process. ARCT includes several tools of general use in ageing research, such as tools for searching putative transcription factor binding sites associated or not with ageing, multiple alignment and phylogenetic footprinting tools, a script to automate PubMed searches and perform text-mining of PubMed records, and a variety of data-mining algorithms. Although ARCT was tailored to study the human ageing process, other researchers working on comparative genomics may profit from it. The latest version of the ARCT toolkit is 0.8 (31/01/2004), which includes more detailed examples and is freely available online for non-commercial purposes at http://genomics.senescence.info/software/.
All programs were developed in Perl 5.8.1 often making use and integrating with the Bioperl toolkit (15). The databases are implemented using the MySQL 4.0.18 relational database management system running on an Intel Pentium III platform with Linux. The query form Web pages are created dynamically in PHP 4.3.7 or CGI scripts written in the Perl programming language.
ACCESS AND USER INTERACTION
HAGR is accessed over the Internet using a web browser. Searching HAGR is simple and intuitive. Users may retrieve an entry through its HAGRID, HAGR's unique identifier, or use HUGO's nomenclature in GenAge. AnAge is searchable by having either the organism's species or its common name. Individual families, orders or classes may also be selected and analysed. In addition to key words, it is also possible to choose a subset of GenAge entries, as described above. Lastly, bibliographic references may be searched by either knowing the reference's PubMed ID, if any, or by using key words from the reference's title. The HAGR is freely available to the academic community at http://genomics.senescence.info.
We chose to display GenAge using an intuitive graphical display to make it accessible to researchers with various degrees of computer proficiency. For bioinformaticians who prefer automated methods, GenAge is available for download and is easy to integrate with other resources. For example, sequences may be downloaded as FASTA, and protein–protein interactions may be downloaded for visualization with other programs such as InterViewer (Inha University, WI Lab, South Korea) or PathwayAssist (Stratagene, La Jolla, CA).
Since the aim of GenAge is to provide the most complete and comprehensive database of genes related to human ageing, the interaction with the gerontological community is of critical importance. The opinion of biologists specialized in the organisms we describe in AnAge is also crucial. Therefore, we expect a response from the community in providing feedback for our project. In each HAGR page, we provide a hyperlink to an easy-to-use feedback form and will continue to update HAGR on a regular basis.
DISCUSSION AND FUTURE DIRECTIONS
GenAge aims to become a major resource in understanding the genetics of human ageing. Not only is GenAge useful as a reference for researchers, but it may serve as a basis for experimental work: through GenAge, researchers may focus on specific pathways and find novel links between the players involved or even construct new hypothesis that can be experimentally tested. Moreover, GenAge may be useful to develop DNA microarrays specific for studying ageing in mammals. Having as goal the understanding of the genetics of human ageing, GenAge is an important step and provides a framework upon which a systems biology understanding of ageing can be developed.
There are other databases of genes involved in ageing such as AgeingDB (16) and, most notably, AGEID (17). GenAge is unique in its improved data selection (see above), fully automated processes that can be downloaded by users, and by focusing on genes and proteins in the context of human biology. Moreover, GenAge focuses on the ageing process, the deleterious process affecting all human beings that exponentially increases our chances of dying with age, and not merely health-promoting genes.
Change is essential in a systems biology approach to any biological problem (3). Intra- and inter-species differences are observed in ageing, most notably between species (12). That is why comparative biology is such an important tool to understand ageing and that is why we created AnAge. With the advent of having several fully sequenced animal genomes, AnAge is a unique and valuable resource with phylogenetic and ageing phenotype information for a large number of species. While AnAge features over 2000 animals, since ageing has only been detailed in a proportion of these, maximum life span should be seen as an approximation and used carefully. Where available, rate of ageing should be preferred. Though our goal was to include species that share a high degree of similarity, both at a genomic and phenotypic level, with humans, researchers working in the ageing process of model organisms may use GenAge and AnAge to place their research in the perspective of human ageing. In addition, AnAge may provide clues about the evolutionary forces shaping ageing and longevity.
With so many discoveries yet to be made in the biology of ageing, our main goal is to keep the databases up-to-date and continue to enhance the HAGR system, but a few new features are in our plans. As described above, GenAge is mostly derived from the phenotypes witnessed following genetic perturbations either in humans or model organisms. A complementary approach involves the identification of genes and proteins differently expressed with age. Indeed the next major enhancement to GenAge is the inclusion and analysis of genes differently expressed with age, which may be useful, for instance, to determine biomarkers of ageing. Since it is not our goal to describe age-related changes or include entries that are likely to be solely effects of ageing, this will require a new methodology (3). Moreover, the inclusion of promoters in GenAge and transcription factor analysis in ARCT provides the perfect starting point for investigating transcriptional regulation during ageing. Our ultimate aim is to use GenAge to understand the regulatory basis of age-related gene expression changes.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
Thanks to Jamie Gillooly, Van Savage and Andrew McKechnie for supplying us with data prior to publication. Many thanks to Matt Kaeberlain, Fabian Bastin, Jason Stajich, George Church, Domingos Magalhães, and everyone of the Linux and Perl/Bioperl communities for their invaluable assistance. Further thanks to the many visitors of senescence.info for their feedback, and to the biologists and researchers who have contributed with their knowledge to establish AnAge and GenAge. J.P.M. is supported by NIH-NHGRI CEGS grant to George Church. He also wishes to thank the FCT, Portugal, for research support. O.T. is a Research Associate from the FNRS, Belgium.
REFERENCES
- 1.Hayflick L. (2000) The future of ageing. Nature, 408, 267–269. [DOI] [PubMed] [Google Scholar]
- 2.Butler R.N., Austad,S.N., Barzilai,N., Braun,A., Helfand,S., Larsen,P.L., McCormick,A.M., Perls,T.T., Shuldiner,A.R., Sprott,R.L. et al. (2003) Longevity genes: from primitive organisms to humans. J. Gerontol. A Biol. Sci. Med. Sci., 58, 581–584. [DOI] [PubMed] [Google Scholar]
- 3.de Magalhaes J.P. and Toussaint,O. (2004) How bioinformatics can help reverse engineer human aging. Ageing Res. Rev., 3, 125–141. [DOI] [PubMed] [Google Scholar]
- 4.Martin G.M. and Oshima,J. (2000) Lessons from human progeroid syndromes. Nature, 408, 263–266. [DOI] [PubMed] [Google Scholar]
- 5.Pound P., Ebrahim,S., Sandercock,P., Bracken,M.B. and Roberts,I. (2004) Where is the evidence that animal research benefits humans? Br. Med. J., 328, 514–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmid C.D., Praz,V., Delorenzi,M., Perier,R. and Bucher,P. (2004) The Eukaryotic Promoter Database EPD: the impact of in silico primer extension. Nucleic Acids Res., 32, D82–D85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wheeler D.L., Church,D.M., Edgar,R., Federhen,S., Helmberg,W., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Sequeira,E. et al. (2004) Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res., 32, D35–D40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karolchik D., Hinrichs,A.S., Furey,T.S., Roskin,K.M., Sugnet,C.W., Haussler,D. and Kent,W.J. (2004) The UCSC table Browser data retrieval tool. Nucleic Acids Res., 32, D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peri S., Navarro,J.D., Amanchy,R., Kristiansen,T.Z., Jonnalagadda,C.K., Surendranath,V., Niranjan,V., Muthusamy,B., Gandhi,T.K., Gronborg,M. et al. (2003) Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res., 13, 2363–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Safran M., Solomon,I., Shmueli,O., Lapidot,M., Shen-Orr,S., Adato,A., Ben-Dor,U., Esterman,N., Rosen,N., Peter,I. et al. (2002) GeneCards 2002: towards a complete, object-oriented, human gene compendium. Bioinformatics, 18, 1542–1543. [DOI] [PubMed] [Google Scholar]
- 11.Povey S., Lovering,R., Bruford,E., Wright,M., Lush,M. and Wain,H. (2001) The HUGO Gene Nomenclature Committee (HGNC). Hum. Genet., 109, 678–680. [DOI] [PubMed] [Google Scholar]
- 12.Finch C.E. (1990) Longevity, Senescence, and the Genome. The University of Chicago Press, Chicago and London. [Google Scholar]
- 13.Savage V.M., Gillooly,J.F., Woodruff,W.H., West,G.B., Allen,A.P., Enquist,B.J. and Brown,J.H. (2004) The predominance of quarter-power scaling in biology. Funct. Ecol., 18, 257–282. [Google Scholar]
- 14.Carey J.R. and Judge,D.S. (2000) Longevity Records: Life Spans of Mammals, Birds, Amphibians, Reptiles, and Fish. Odense University Press, Odense. [Google Scholar]
- 15.Stajich J.E., Block,D., Boulez,K., Brenner,S.E., Chervitz,S.A., Dagdigian,C., Fuellen,G., Gilbert,J.G., Korf,I., Lapp,H. et al. (2002) The bioperl toolkit: perl modules for the life sciences. Genome Res., 12, 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park D.U., Kim,C.H., Hong,S.E., Yu,B.P., Chung,H.Y. (2002) AgingDB: aging-related signal pathway imagemap and protein interaction visualization. Genome Informatics, 13, 537–538. [Google Scholar]
- 17.Kaeberlein M., Jegalian,B. and McVey,M. (2002) AGEID: a database of aging genes and interventions. Mech. Ageing Dev., 123, 1115–1119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.