Abstract
The use of recombinant adeno-associated viruses (rAAVs) ushered in a new millennium of gene transfer for therapeutic treatment of a number of conditions, including congenital blindness, hemophilia, and spinal muscular atrophy. rAAV vectors have remarkable staying power from a therapeutic standpoint, withstanding several ebbs and flows. As new technologies such as clustered regularly interspaced short palindromic repeat genome editing emerge, it is now the delivery tool—the AAV vector—that is the stalwart. The long-standing safety of this vector in a multitude of clinical settings makes rAAV a selling point in the advancement of approaches for gene replacement, gene knockdown, gene editing, and genome modification/engineering. The research community is building on these advances to develop more tailored delivery approaches and to tweak the genome in new and unique ways. Intertwining these approaches with newly engineered rAAV vectors is greatly expanding the available tools to manipulate gene expression with a therapeutic intent.
Keywords: : AAV vectors, genome editing, non-coding RNA, miRNA, RNAi
Gene Replacement is the Standard Bearer of Gene Therapeutics
The promise of rAAV gene therapy has most notably been realized from a gene replacement standpoint. The road to the clinic for rAAV vectors expressing transgenes replacing mutant or absent genes has been met with increasing success and safety in clinical trials. Notable examples include replacement of the factor IX gene in hemophilia,1,2 the α1-antitrypsin gene in α1-antitrypsin deficiency,3 the retinal pigment epithelium-specific protein 65 kDa (RPE65) gene in Leber Congenital Amaurosis,4,5 and survival motor neuron (SMN) transgene delivery in spinal muscular atrophy.6 Furthermore, rAAV delivery for lipoprotein lipase deficiency7 is approved in Europe, paving the way for additional conditions to be deemed safe and efficacious. Optimizing AAV capsids for refined properties will expand the range of tissues and conditions that can be considered, ultimately enhancing the number of disease targets. The continued identification of mutations causative for Mendelian conditions means novel gene replacement strategies will be attempted, leading to continued clinical success stories.8 However, this review will focus on non-traditional uses for rAAV vectors (Fig. 1) and what these hold in store for therapeutic strategies.
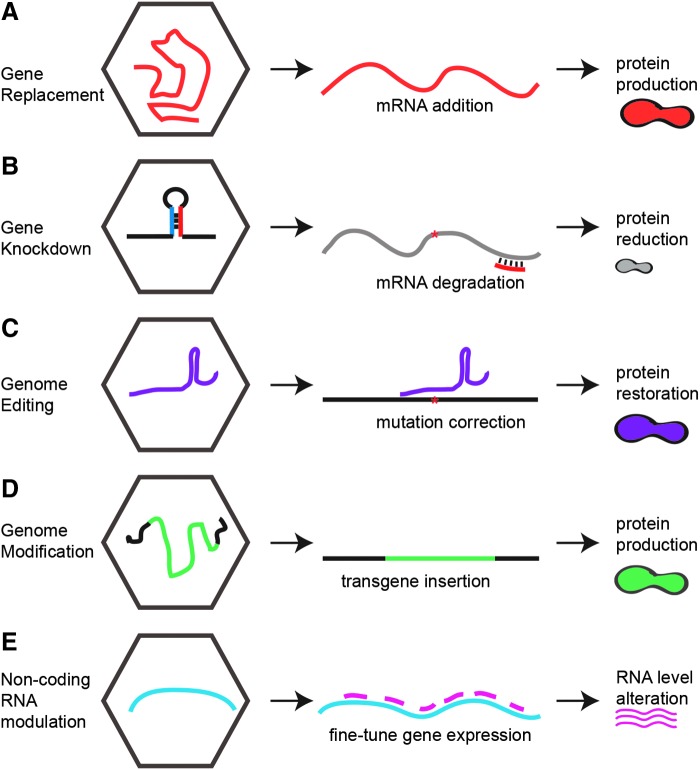
Figure 1.
Broad categories of recombinant adeno-associated virus therapeutic strategies. (A) Gene delivery, often of a cDNA that replaces a missing or defective gene, driven by an exogenous promoter. (B) Gene knockdown through delivery of sequences that generate small interfering RNAs that can degrade a mutant gene or infectious virus. (C) Genome editing by zinc finger nucleases, transcription activator-like effector nucleases, or the clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 system to introduce or correct mutations in the host or virus genome or introduce transcripts at double-stranded genome breaks. (D) Genome modulation through introduction of DNA sequences with homology to the host genome to correct a mutation or provide a therapeutic gene. (E) Non-coding RNA modulation or sequestration to alter transcript and protein levels, for example through introduction of sequences with engineered binding sites.
RNA Interference Remains a Powerful Approach for Gene Knockdown
The capability of eliciting specific reduction of a mutant gene or virus has considerable appeal for gene therapy. RNA interference (RNAi) is an evolutionarily conserved mechanism to suppress endogenous and exogenous genes through complementarity between a small RNA and the target sequence.9 Typically, primary microRNA transcripts are processed in two steps by Drosha10 and Dicer, associate with Argonaute (Ago) proteins, and enter the RNA-induced silencing complex (RISC) where the resulting 21–24nt single stranded RNA is recruited to suppress target genes.11–13 Regions with complete complementary between the microRNA and mRNA target lead to most efficient mRNA knockdown through Ago2-mediated cleavage and degradation.14 RNAi can be co-opted from a gene therapy perspective to suppress target genes of interest through use of synthetic double-stranded oligonucleotides or as genetic DNA templates from which hairpin RNAs are transcribed in the target cells.15 Indeed, the discovery that genes could be targeted using the RNAi pathway was rapidly validated in mammals by the use of oligonucleotides16 or small hairpin RNAs,17 underscoring the potential therapeutic application of this technology. The ability to utilize rAAV vectors as delivery tools for RNAi-based gene knockdown has profound consequences for the treatment of disease.
Several gene knockdown strategies have been particularly effective in using rAAVs to deliver microRNAs (miRNAs) or short hairpin RNAs (shRNAs), especially in the preclinical evaluation of disease in rodents.18 RNAi-based approaches are best suited for targeting mutations that confer gain-of-function properties, including expanded repeat disorders or to eliminate transcripts from pathogenic organisms. In the liver, delivery of shRNAs that target the hepatitis C virus (HCV) have been validated in nonhuman primate models of HCV infection19 and attempted in clinical trials. However, because of the recent success of small molecule HCV inhibitors, interest in pursuing shRNA knockdown approaches has instead shifted to treating chronic hepatitis B.17,20 The success of this approach, though, likely requires AAVs that can transduce the majority of hepatocytes, something that has yet to be achieved in humans.
Targeting the mutant SERPINA1 gene is a viable option for the treatment of the most common form of α-1 antitrypsin deficiency.21,22 These patients can suffer from liver disease and/or early onset pulmonary emphysema. The former is the result of accumulated mutant protein in hepatocytes, while the latter is the consequence of a deficiency in the neutrophil elastase inhibitor. Liver disease can present at any time but commonly occurs in childhood and is the leading genetic cause for liver transplantation in the Western world. A combinatorial approach has thus been attempted whereby the administered vector can supply both the wild-type SERPINA1 coding sequence and an shRNA that specifically targets the mutant allele, reducing the mutant protein.
Delivery of shRNAs for gene knockdown has had success in other tissues, including the muscle for facioscapulohumeral muscular dystrophy23–25 and the eye for age-related macular degeneration and retinitis pigmentosa.26,27 The brain represents a complex organ where shRNA-mediated removal of mutant RNAs and protein aggregates would have a profound impact on several neuronal disorders. For example, shRNAs have been administered to knock down Ataxin1 (Atxn1) harboring expanded CAG trinucleotide repeats that recapitulate Spinocerebellar ataxia in mice, ultimately reducing intranuclear inclusions and restoring cerebellar function.28 Likewise, delivery of rAAVs bearing shRNAs that target CAG expansions in Huntingtin (Htt) have improved phenotypic outcomes in mouse models of Huntington's disease,29,30 and rAAV-shRNAs have been used successfully to target superoxide dismutase 1 (Sod1) in rodent31–33 and monkey34 models of amyotrophic lateral sclerosis and α-synuclein (Scna) in Parkinson's disease.35 Together, the functionality of these approaches has been validated and can be translated to other related mutant genes and disorders.
Despite the success of rAAV-based RNAi, fewer clinical trials are in the pipeline than, for instance, antisense oligonucleotides (ASOs) or other nucleotide delivery mechanisms. The advantage of an AAV system is that a single administration can provide sustained expression of shRNAs, with levels of therapeutic oligonucleotide potentially increasing rather than decreasing longitudinally. Some of these discrepancies have been minimized through the development of optimized formulations of oligonucleotides and delivery conjugates that extend their potency and bioavailability and reduce cytotoxic responses.36 Mammals are more amenable to repeated infusions of oligonucleotides at present, enabling a dosing regimen that can be titered and halted if necessary.
Several barriers still need to be overcome before the use of rAAV-shRNAs is broadly accepted. Complications including toxicity arising from high doses of shRNA delivery need to be recognized and avoided in order to ensure safe and sustained gene knockdown. This toxicity is associated with too high a dose of delivered shRNAs, irrespective of their sequence or the presence of an endogenous target.20 In the context of the liver, the effect of endogenous miR-122 levels are an important indicator of sustained gene knockdown and unintended toxicity.37 Several approaches have been designed to circumvent the severe effects of this toxicity. The most effective appears to be to design a sequence that first requires Drosha processing38 in contrast to shRNAs that form a stem-loop structure that enters the RNA interference pathway at the Dicer cleavage step, bypassing Drosha processing but facilitating gene silencing.16,39–42 While the efficiency of Drosha processed miRNA-like structures may not approach shRNA expression, the level of target knockdown is sufficient to ameliorate many conditions.38,43 Alternatively, a shorter pre-miRNA scaffold can be generated that bypasses Dicer processing and instead is generated via Ago2 cleavage.44 Ago2 can also be co-delivered along with an shRNA to permit long-term gene suppression,45 or RNA decoys can be co-administered to sequester sense strand by-products of shRNA processing.46 shRNAs are continually being optimized with regards to expression in order to design a more robust processing site with fewer unintended small RNAs that are generated, reducing off-target effects.47 Together, these refinements will permit the generation of safer and more effective vectors.
In the future, host genomes could be modified to express shRNAs that target a mutant gene or viral sequence continually. For instance, hepatocytes could be proliferated in the presence of shRNAs, targeting various viral sequences and preventing their replication. rAAVs could in theory be used to deliver the appropriate gene set to reprogram progenitor cells such as induced pluripotent stem cells (iPSCs), as well as expressing antiviral shRNAs prior to their transplantation to combat a badly damaged liver resulting from a chronic viral infection.
Gene Editing: Transcription Activator-Like Effector Nucleases and Zinc Finger Nucleases Lay the Groundwork for Targeted Genome Modifications
Classical rAAV provides an episomal genome in transduced cells. For quiescent tissues, this can provide long-term transgene expression, although the true length of persistence for each individual tissue is not yet known. In regenerative tissues or in infants, long-term expression is unlikely.48 However, using any of the genome modification approaches would in most tissues provide lifelong expression. Hence, another advantage of the approaches to follow includes the possibility of treating neonates with a single vector dose administration. This is important because repeat administration of rAAVs may prove to be difficult due to the induction of humoral immune responses.
The zinc finger nucleases (ZFNs),49 transcription activator-like effector nucleases (TALENs),50 and engineered meganucleases51 have provided an exciting starting point for the modification of host genomes. Broadly, they introduce a double-stranded break in the genomic DNA to be modified, followed by replacement of the desired sequence with a DNA donor template. Although TALENs and ZFNs may be considerably more time-consuming and laborious to construct than clustered regularly interspaced short palindromic repeat (CRISPR) gene editing (see below), this pales in comparison to the length and cost of a clinical trial, and should not necessarily be considered an impediment. In addition, there is greater difficulty in identifying a sequence that can be effectively targeted, but when found, it is possible that this affords a greater on versus off targeting rate. These approaches have translated into clinical trials, for instance by Sangamo Biosciences. Electroporation of ZFN mRNA targeting a safe harbor site such as the AAVS1 site on chromosome 19 and administration of a rAAV expressing a transgene of interest. One example is the ex vivo introduction of gp91phox in hematopoietic stem and progenitor cells for intervention in X-linked chronic granulomatous disease.52 Likewise, the factor VIII or factor IX transgene can be inserted at the highly expressed albumin locus for amelioration of hemophilia A and B.53,54 The open clinical trial on this indication is certainly of great interest, though concerns remain about the continued expression of a nuclease and the long-term consequences of potential off-targeting and/or immunogenicity associated with expression of a foreign protein. Provided the safety profile is acceptable and preclinical studies demonstrate few off-targeting events (and most importantly none that are harmful), this technology may be what ends up with the greatest translational potential.
CRISPR/Cas9 Opens a Pandora's Box of Gene Editing Prospects
The adaption of the CRISPR adaptive immune defense system in bacteria and archaea to mammalian systems has revolutionized molecular biology.55–57 CRISPR and the CRISPR-associated endonuclease Cas9 sample segments of phage or plasmid DNA to incorporate into CRISPR loci,58–60 which are then transcribed and processed into CRISPR RNAs (crRNAs).61 These crRNAs guide Cas9 to homologous regions in invading sequences, facilitating their recognition and elimination.62 The CRISPR system can be adopted and engineered to mediate double-stranded genomic breaks in any genomic sequence adjacent to a 5′-NGG protospacer-adjacent motif (PAM) through the association of a chimeric single-guide RNAs (sgRNA) with the Cas9 nuclease.57 These breaks are typically rejoined through non-homologous end joining (NHEJ), and as this process is imperfect, short insertions and deletions (indels) at the targeted region are often generated.63 In the event of an indel, the target region is no longer a perfect complement to the sgRNA, and the region becomes largely impervious to further editing by the sgRNA. Ideally, when employed correctly, this approach introduces frameshift mutations that lead to loss of expression of the target gene. Co-delivery of an oligonucleotide or rAAV bearing sequence homology to the target region can act as a template for homologous recombination (HR) mediated repair of the break to alter the resulting genetic sequence, often for the purpose of correcting a disease-causing mutation.55,56 Together, while much of the groundwork for understanding the CRISPR/Cas9 complex in a natural and therapeutic setting has only recently been established, the work clearly reveals the remarkable potential of this system.
Just like with RNAi, the discovery that CRISPR/Cas9 approaches can be used to modify mammalian genomes has piqued interest into how this new technology can be extended from a powerful molecular biology approach to a therapeutic context. This is a nascent field, meaning that it will be intriguing to determine which low-hanging fruits (such as disrupting the HIV CCR5 co-receptor64) are available for study. Researchers have already reported success in utilizing the CRISPR/Cas9 system and rAAV vectors for a multitude of expanding approaches. This includes correcting ornithine transcarbamylase (OTC) deficiency in newborn mice,65 eliminating fumarylacetoacetate hydrolase (Fah)-positive hepatocytes by eliminating Fah splicing mutations,66 reducing Pcsk9 levels for reduction of serum cholesterol,67 introducing and correcting PRKAG2 mutations in mice for therapy of the cardiac Wolff–Parkinson–White syndrome,68 ex vivo correction of the β-globin gene in sickle cell disease,69 and skipping mutant exons in a mouse model of Duchenne muscular dystrophy.70–72 Proof-of-principle approaches have also shown the ability to disrupt multiple genes in the adult mouse brain.73 Without question, CRISPR/Cas9 technology has already facilitated the development of animal models for diseases that previously were impractical or inconceivable to generate, for instance generation of several combined mutations causative for lung cancer.74 Other technological developments include targeting non-genic regions to modulate gene expression. Potential allele-specific knockdown can be achieved when one allele is heavily methylated at CpG islands, for instance targeting the mutant, unmethylated, and expressed allele of p16INK4a in the HCT116 colorectal cancer cell line.75 Allele-specific removal of an expanded CAG repeat has also recently been demonstrated in the HTT gene using adjacent common single nucleotide polymorphisms (SNPs) present within a patient that alter a PAM motif.76 The use of common SNPs for allele-specific silencing is not new; this has been attempted previously for si/shRNA knockdown of several mutant genes, including HTT itself.77 However, often the wild-type allele was simultaneously reduced. The definitive nucleotide sequence requirement of the PAM motif affords the allele specificity that was previously challenging. Several biotech companies have raced to develop therapeutic strategies employing CRISPR-mediated gene editing. While the operational success of these companies may hinge on the outcome of ongoing patent disputes, several strategies are already in the works. The results of these discoveries are eagerly anticipated and add an exciting new dimension to the American Society of Gene and Cell Therapy annual meeting.
Countless optimizations are still in the works to improve this system from a therapeutic standpoint (Fig. 2). Drawing parallels with oligonucleotide therapeutics, chemical modifications such as 2′-O-methyl and related analogs of the sgRNA can improve stability and bioavailability.78 From a development standpoint, humanizing Cas9 to dampen immune recognition, as has been employed for antibody development,79 and biasing to HR instead of NHEJ could present fewer complications. Mutating one of the HNH and RuvC nuclease domains to generate a Cas9 nickase enzyme57,80 that cuts one DNA strand can facilitate specificity.81 Delivering two guide RNAs that recognize adjacent genomic regions can vastly reduce the number of off-target cuts. Furthermore, the generation of a completely catalytically inert (dead) dCas9 with Asp10Ala and His840Ala mutations57 can be used to tether proteins that modulate the genome. This takes advantage of the target specificity of the Cas9-gRNA complex. Theoretically, the consequence of targeting other genomic regions is also vastly minimized. For instance, combining dCas9 with a cytodine deaminase enzyme can introduce C-to-T or G-to-A edits without introducing DNA breaks, albeit with a more narrow range of targetable regions in the context of a PAM motif.82 Activation or inhibition (CRISPRa or CRISPRi) of genomic loci can be accomplished by fusing dCas9 to transcriptional activators or repressors as a fascinating approach to fine-tune gene regulation.83,84 Expanding the repertoire of approaches, the CRISPR/Cas9 complex has also been engineered to target single-stranded RNA.85 Many of these combinatorial delivery approaches likely exceed the rAAV vector capacity and may require co-delivery or other clever tricks. Not all of these approaches will translate to the clinic either but serve as examples of modifications to the CRISPR/Cas9 system that can drive more exquisite modification of a target gene or genome and curb concerns of off-target genome editing.
Figure 2.
Broad categories of CRISPR/Cas9 genome editing strategies. (A) The use of the CRISPR/Cas9 with a single guide RNA (sgRNA) targeting a specific genomic region introduces double-stranded breaks that lead to insertions and deletions (indels) through the non-homologous end joining pathway. A donor template can be co-delivered to introduce a specific DNA segment at the double-stranded break by homology directed repair. (B) Cas9 nickase enzymes have a mutation that directs single-stranded DNA breaks. When two sgRNAs are engineered to target regions in a reasonable proximity along with Cas9 nickase enzymes, off-targeting is reduced because the chance that similar off-targets are close by is low. This also facilitates larger genomic deletions. (C) The use of catalytically inactive Cas9 (dCas9) can leverage the recruitment of the enzyme tethered to an additional transcription factor to modulate the genome or epigenome without introducing breaks. In all situations, the use of patient-specific or common single nucleotide polymorphisms can direct editing to the mutant allele.
The AAV research community should be heartened that the development of CRISPR/Cas9 approaches has embraced AAV vectors for delivery. The size limitation of rAAV vectors, however, has necessitated some optimization (as is usually the case) to ensure appropriate expression and gene editing. One approach to leave sufficient space for therapeutic transgenes is to use the Staphylococcus aureus (saCas9) system, reducing the length of Cas9 by about 1 kb from the 4.2 kb version derived from the widely used Staphylococcus pyogenes. This more manageably fits the 4.7 kb size constraints of AAV without compromising editing, initially requiring a modestly less abundant 5′-NNGRRT PAM motif,67 which has been further optimized to confer additional targets using a NNNRRT PAM.86 Importantly, using saCas9 permits delivery of Cas9 and one or more promoter-driven sgRNAs in the same vector. Another approach involves dual-rAAV delivery, including an intein for trans-splicing, again leaving sufficient space for sgRNAs.87 The optimal approach likely will be to devise a mechanism to express Cas9 transiently, enabling hit-and-run edits at the intended target with next to no edits at unintended regions. Importantly, the stability (and clearly the functionality) of the sgRNA appears to be dependent on the presence Cas9.78
For CRISPR/Cas9 gene editing technology to really take hold in a clinical context, several critical considerations need to be taken into account. This includes the consequences of off-target genome edits and immune responses to components of the CRISPR/Cas9 machinery, along with an acceptable delivery regimen, promoter choice, and sgRNA expression levels. It is impossible to overstate how dangerous it might be to have an endonuclease such as Cas9 continually present within the cell potentially editing more and more off-targets over time, leading to potentially drastic and dire consequences on a cell and therefore also in humans. Presently, this represents a formidable obstacle. Furthermore, it is exceedingly difficult to ensure that only the target genome region is edited, although optimized Cas9 nucleases have drastically reduced or undetectable off-targets based on current technology.88,89 Even by high-resolution sequencing approaches, the consequences of potential off-target editing on each individual cell is difficult to ascertain. This is further complicated by cell-specific chromatin architecture and Cas9 accessibility precluding the direct translation of findings in cell culture to safety and success in animals and ultimately in humans. It should also be noted that most drugs have some level of off-targeting from their intended target, and ultimately how rare nuclease mediated off-targeting translate into truly detrimental events is not known.
Other concerns of the CRISPR/Cas9 system include the widespread use of the U6 Pol-III promoter to drive sgRNAs; this promoter can lead to toxicity when used to express shRNAs at high levels.20,37 Situations that preclude the efficacy of this approach should be reported to ensure that the field is aware of scenarios that are prohibitive. A case in point is when attempting to edit and then correct using oligonucleotide-templated repair a hypomorphic sparse fur ash (spfash) allele causing OTC in adult mice. The efficiency of repair was incomplete, resulting in complete loss of the transcript and a more adverse consequence than the initial relatively subtle mutation.65 This underscores some of the difficulties in attempting multifaceted therapies such as combining editing and repair. Even if safe concentrations of CRISPR/Cas9 in vivo are achievable in humans, reaching effective levels to treat various disorders will need to be demonstrated. Finally, from an ethical standpoint, the field has monitored and attempted to discourage the potential for human embryo editing and other ramifications of the CRISPR/Cas9 system.90 Over time, society will have to make the decisions on how far to move in this direction.
These challenges notwithstanding, the potential of CRISPR/Cas9 technology is remarkable, and contemplating the various uses is extremely exciting. For instance, one could envision a scenario whereby methods could be employed to modify somatic variants in a tumor back to the original host genomic sequence or correct a disease-causing mutation in carriers of a particular condition prior to symptom onset. These and other possibilities make for a promising future.
RNAi Knockdown and Nuclease-Mediated Knockout Comparisons
RNAi and nucleases such as CRISPR/Cas9 approaches differ in their consequence for gene targets, efficiency, and off-target effects. Genome editing has the potential to effectively knock out the resulting mRNA transcript, in contrast to gene knockdown with shRNAs that reduces and in some cases essentially eliminates existing transcripts. With shRNAs, the dose can be titrated to reduce but not necessarily eliminate a mutant transcript (or mutant and wild type, where it is problematic to affect only one allele exclusively). The convergence of hits or pathways from both complementary approaches can reveal scientifically meaningful insights. For instance, whole-transcriptome libraries can be generated using shRNAs or CRISPR-mediated edits of each gene.91,92 These studies establish that results from one approach rarely if ever will perfectly match that of the other. Together they can cover more ground and reveal which genes prevent or accelerate phenotype when suppressed or completely eliminated. Both approaches could conceivably be combined to target a gene and knock down any residual or mutant transcripts. In the long term, it is likely that both RNAi and nuclease-mediated genome editing will find their specific therapeutic niches.
Advances in Modulating the Genome
Rather than gene knockdown or editing, a more traditional strategy involves the delivery of sequences complementary to a mammalian genome and the use of homologous recombination to integrate portions of the delivered viral sequence in vivo.93 This can be used to model disease such as the generation of a porcine model of hereditary tyrosinemia type 1.94 This also has several therapeutic applications, including the targeting of a disease gene to correct a mutant allele back to wild-type state, for instance the in vitro correction of a LAMA3 mutation in epidermolysis bullosa with the idea that this can then be reintroduced into patients.95 Alternatively, rAAV-bearing homology arms flanking a coding and/or non-coding therapeutic sequences can be precisely integrated into a locus, allowing the expression of a completely new transgene of interest such as factor IX for the treatment of hemophilia.96 In this example, the factor IX coding sequence was inserted near the 3′ end of the albumin locus but just upstream of the stop codon. Using a ribosomal skipping sequence, just 5′ of the factor IX coding results in a chimeric mRNA transcript that produces both albumin and factor IX proteins. Because the albumin promoter is so robust, up to 20% of the normal factor IX level was stably expressed from mouse liver, even though <1% of the albumin alleles were targeted. This “generide” approach is advantageous over classical AAV and nuclease-mediated gene therapy in that off-target integrations should have reduced capacity for expression given the avoidance of a promoter sequence. However, the low level of homologous recombination-mediated integration events97,98 means the few successful integrants have to drive considerable transgene expression and mostly in conditions where incomplete gene replacement is sufficient to alleviate phenotypic consequences. However, improvements in gene targeting efficiency or utilizing a selection scheme can enhance the potential of this approach.99 Recently, including an additional intron that encodes an shRNA into the factor IX sequence enabled a metabolic selection scheme when a short course of drug therapy was provided. This resulted in a stable 50-fold expansion of the gene-modified cells.100 If the efficiency of this technology can be expanded considerably, the idea of directing rAAV to modulate genomic sequence can have a lasting effect on the cell, permitting selection of hepatocytes that have been corrected of a particular genetic mutation. In general, these approaches enable stable genomic modification and sustained intracellular effects.
The rare but non-negligible level of rAAV integration events, especially at unintended genomic intervals, needs to be monitored and addressed when contemplating the safety profile of rAAV therapy. Much has been discussed as to whether there is cause for concern over rAAV integration, especially at the mouse chr12qF1 Dlk1-Dio3 locus leading to hepatocellular carcinoma, including in this issue of Human Gene Therapy. The similarity in sequence conservation of all protein coding genes, noncoding RNAs, and most microRNAs along with parent-of-origin methylation status between the mouse chr12qF1 locus and the syntenic region on human chr14q32.2 (as is the case in all eutherian mammals)101 means that findings in the mouse likely could have analogous complications in humans. The absence of a promoter in the delivered transgene can at least minimize complications and aberrant activation of genes neighboring these integrants.
Targeting Non-Coding RNAs
The expanding world of non-coding RNAs affords additional means of therapeutic intervention through the fine-tuning of their gene expression, and thus represents a new frontier of sequences to target or express. Several classes of RNAs from long non-coding RNAs (lncRNAs) to microRNAs are characterized. Other categories include a large complement of small nucleolar RNAs (snoRNAs) and small RNAs derived from tRNAs. The cellular roles of these RNAs are still being elucidated, and represent new facets of human biology and disease. rAAVs can be utilized to supplement microRNAs that are depleted in certain conditions such as miR-26a in liver cancer102 or enhance decay of a target such as miR-196a in spinal and bulbar muscular atrophy.103 rAAVs can also be exploited to administer a genomic sequence capable of sequestering microRNAs or RNA binding proteins, perhaps through the generation of a stable circular RNA such as ones that have been identified to be a sponge for miR-7.104,105 Intricate interactions similarly exist between pseudogenes and their coding counterparts such as Pten.106,107 These approaches can be contemplated for the removal of microRNAs or other non-coding RNAs that are elevated in various conditions, particularly in various forms of cancer.108 Ultimately, as the non-coding RNA field matures and mechanisms of disease are established, the relationships between coding and non-coding elements of the genome can be exploited in a therapeutic context.
AAV Optimization for Enhanced Tropism and Delivery
While highly effective as a starting point, just like any drug, optimization of rAAV broadens the applicability and specificity of this technology. The differences in mice and humans in both transduction efficiencies of various rAAV serotypes109 and immune responses mean a considerable amount of research is still warranted to ensure optimal safety and potency. At present, certain tissues are much more prone to rAAV transduction, meaning the study of associated diseases such as those affecting the liver can be studied with confidence. In other tissues, the ability for efficient targeting and transduction still presents a challenge. Consequently, the field of rAAV biology has graduated from a discovery-based approach for the identification of AAV serotypes110 to the generation of shuffled capsid and peptide display libraries that can confer novel targeting properties111–114 to the rational design and engineering of novel serotypes. This is most notable in nervous system transduction where the ability of AAV to cross the blood–brain barrier and transduce neurons reveals tremendous potential for neuronal disease. Building on this work, groups have identified serotypes with peptide insertions that have been rigorously evaluated for enhanced neuronal transduction properties115 or for retrograde transport to projection neurons.116 Other approaches involve enhanced neuronal tropism via grafting the AAV9 galactose binding motif onto AAV2117 through addition of a poly-alanine peptide at the N-terminus of the VP2 capsid gene118 or through an AAV-capsid shuffling strategy.119 rAAV serotypes have been generated that have optimized transduction in tissue culture111 or that selectively infect human hepatocytes.109 Additional strategies have been employed to enhance in vivo or ex vivo tropism through, for instance, incorporation of designed ankyrin repeat proteins in the AAV capsid protein VP2.120 Work evaluating the crystal structure of AAV is imperative for comprehending how these modifications are tolerated and alter physiochemical properties of AAV. An understanding of some of the cellular receptors and machinery that permit cellular AAV entry can be modulated to enhance tropism of a vector or to de-target a particular organ (especially the liver), permitting more circulating AAV that can access an organ of interest. Notable examples include the identification of the KIAA0319L type I transmembrane (AAVR) receptor for AAV entry,121 the heparan sulfate proteoglycan identified for AAV2 attachment,122 and the N-linked terminal galactosyl residue binding specificity for AAV9.123 Together, the combination of approaches will undoubtedly lead to a more diverse toolkit of rAAV vectors for selective transduction of one or more organs previously refractory or suboptimal to rAAV entry and transgene expression.
Conclusions
Just like the CRISPR/Cas9 field that has adopted rAAV vectors for delivery, future technologies will benefit from a gene delivery tool that has been studied for decades with established safety profiles. The field of gene therapy has benefited from a strong long-standing community. It is imperative that this collaboration continues to raise and adequately address issues associated with gene therapy approaches and AAV vectors. With these responsible approaches, the AAV community can enjoy continued successes and can be married to new technologies as they are developed and implemented.
Acknowledgments
We are grateful to members of the Kay lab for their critiques and suggestions. This work was supported by grants NIH 1R01 DK 078424, 1R01 AI11698, R01 HL064274 (M.A.K.).
Author Disclosure
No competing financial interests exist for Paul Valdmanis. Mark Kay is a scientific founder of Voyager Therapeutics, and founder of LogicBio Therapeutics.
References
- 1.Snyder RO, Miao CH, Patijn GA, et al. Persistent and therapeutic concentrations of human factor IX in mice after hepatic gene transfer of recombinant AAV vectors. Nat Genet 1997;16:270–276 [DOI] [PubMed] [Google Scholar]
- 2.Nathwani AC, Reiss UM, Tuddenham EG, et al. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N Engl J Med 2014;371:1994–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song S, Morgan M, Ellis T, et al. Sustained secretion of human alpha-1-antitrypsin from murine muscle transduced with adeno-associated virus vectors. Proc Natl Acad Sci U S A 1998;95:14384–14388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maguire AM, High KA, Auricchio A, et al. Age-dependent effects of RPE65 gene therapy for Leber's congenital amaurosis: a Phase 1 dose-escalation trial. Lancet 2009;374:1597–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cideciyan AV, Hauswirth WW, Aleman TS, et al. Vision 1 year after gene therapy for Leber's congenital amaurosis. N Engl J Med 2009;361:725–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valori CF, Ning K, Wyles M, et al. Systemic delivery of scAAV9 expressing SMN prolongs survival in a model of spinal muscular atrophy. Sci Transl Med 2010;2:35ra42. [DOI] [PubMed] [Google Scholar]
- 7.Gaudet D, Methot J, Dery S, et al. Efficacy and long-term safety of alipogene tiparvovec (AAV1-LPLS447X) gene therapy for lipoprotein lipase deficiency: an open-label trial. Gene Ther 2013;20:361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mingozzi F, High KA. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat Rev Genet 2011;12:341–355 [DOI] [PubMed] [Google Scholar]
- 9.Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998;391:806–811 [DOI] [PubMed] [Google Scholar]
- 10.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003;425:415–419 [DOI] [PubMed] [Google Scholar]
- 11.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell 2009;136:642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gregory RI, Chendrimada TP, Cooch N, et al. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005;123:631–640 [DOI] [PubMed] [Google Scholar]
- 13.Matranga C, Tomari Y, Shin C, et al. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 2005;123:607–620 [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Carmell MA, Rivas FV, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004;305:1437–1441 [DOI] [PubMed] [Google Scholar]
- 15.Haussecker D, Kay MA. RNA interference. Drugging RNAi. Science 2015;347:1069–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCaffrey AP, Meuse L, Pham TT, et al. RNA interference in adult mice. Nature 2002;418:38–39 [DOI] [PubMed] [Google Scholar]
- 17.McCaffrey AP, Nakai H, Pandey K, et al. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol 2003;21:639–644 [DOI] [PubMed] [Google Scholar]
- 18.Kay MA. State-of-the-art gene-based therapies: the road ahead. Nat Rev Genet 2011;12:316–328 [DOI] [PubMed] [Google Scholar]
- 19.Suhy DA, Kao SC, Mao T, et al. Safe, long-term hepatic expression of anti-HCV shRNA in a nonhuman primate model. Mol Ther 2012;20:1737–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimm D, Streetz KL, Jopling CL, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 2006;441:537–541 [DOI] [PubMed] [Google Scholar]
- 21.Mueller C, Tang Q, Gruntman A, et al. Sustained miRNA-mediated knockdown of mutant AAT with simultaneous augmentation of wild-type AAT has minimal effect on global liver miRNA profiles. Mol Ther 2012;20:590–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cruz PE, Mueller C, Cossette TL, et al. In vivo post-transcriptional gene silencing of alpha-1 antitrypsin by adeno-associated virus vectors expressing siRNA. Lab Invest 2007;87:893–902 [DOI] [PubMed] [Google Scholar]
- 23.Wallace LM, Garwick-Coppens SE, Tupler R, et al. RNA interference improves myopathic phenotypes in mice over-expressing FSHD region gene 1 (FRG1). Mol Ther 2011;19:2048–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallace LM, Liu J, Domire JS, et al. RNA interference inhibits DUX4-induced muscle toxicity in vivo: implications for a targeted FSHD therapy. Mol Ther 2012;20:1417–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bortolanza S, Nonis A, Sanvito F, et al. AAV6-mediated systemic shRNA delivery reverses disease in a mouse model of facioscapulohumeral muscular dystrophy. Mol Ther 2011;19:2055–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Reilly M, Palfi A, Chadderton N, et al. RNA interference-mediated suppression and replacement of human rhodopsin in vivo. Am J Hum Genet 2007;81:127–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tam LC, Kiang AS, Kennan A, et al. Therapeutic benefit derived from RNAi-mediated ablation of IMPDH1 transcripts in a murine model of autosomal dominant retinitis pigmentosa (RP10). Hum Mol Genet 2008;17:2084–2100 [DOI] [PubMed] [Google Scholar]
- 28.Xia H, Mao Q, Eliason SL, et al. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat Med 2004;10:816–820 [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez-Lebron E, Denovan-Wright EM, Nash K, et al. Intrastriatal rAAV-mediated delivery of anti-Huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington's disease transgenic mice. Mol Ther 2005;12:618–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harper SQ, Staber PD, He X, et al. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc Natl Acad Sci U S A 2005;102:5820–5825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foust KD, Salazar DL, Likhite S, et al. Therapeutic AAV9-mediated suppression of mutant SOD1 slows disease progression and extends survival in models of inherited ALS. Mol Ther 2013;21:2148–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller TM, Kaspar BK, Kops GJ, et al. Virus-delivered small RNA silencing sustains strength in amyotrophic lateral sclerosis. Ann Neurol 2005;57:773–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomsen GM, Gowing G, Latter J, et al. Delayed disease onset and extended survival in the SOD1G93A rat model of amyotrophic lateral sclerosis after suppression of mutant SOD1 in the motor cortex. J Neurosci 2014;34:15587–15600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H, Yang B, Qiu L, et al. Widespread spinal cord transduction by intrathecal injection of rAAV delivers efficacious RNAi therapy for amyotrophic lateral sclerosis. Hum Mol Genet 2014;23:668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khodr CE, Sapru MK, Pedapati J, et al. An alpha-synuclein AAV gene silencing vector ameliorates a behavioral deficit in a rat model of Parkinson's disease, but displays toxicity in dopamine neurons. Brain Res 2011;1395:94–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Satterlee A, Huang L. In vivo gene delivery by nonviral vectors: overcoming hurdles? Mol Ther 2012;20:1298–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valdmanis PN, Gu S, Chu K, et al. RNA interference-induced hepatotoxicity results from loss of the first synthesized isoform of microRNA-122 in mice. Nat Med 2016;22:557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boudreau RL, Martins I, Davidson BL. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol Ther 2009;17:169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science 2002;296:550–553 [DOI] [PubMed] [Google Scholar]
- 40.McManus MT, Petersen CP, Haines BB, et al. Gene silencing using micro-RNA designed hairpins. RNA 2002;8:842–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paddison PJ, Caudy AA, Bernstein E, et al. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev 2002;16:948–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell 2002;9:1327–1333 [DOI] [PubMed] [Google Scholar]
- 43.McBride JL, Boudreau RL, Harper SQ, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci U S A 2008;105:5868–5873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu YP, Schopman NC, Berkhout B. Dicer-independent processing of short hairpin RNAs. Nucleic Acids Res 2013;41:3723–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borner K, Niopek D, Cotugno G, et al. Robust RNAi enhancement via human Argonaute-2 overexpression from plasmids, viral vectors and cell lines. Nucleic Acids Res 2013;41:e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mockenhaupt S, Grosse S, Rupp D, et al. Alleviation of off-target effects from vector-encoded shRNAs via codelivered RNA decoys. Proc Natl Acad Sci U S A 2015;112:E4007–4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu S, Jin L, Zhang Y, et al. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell 2012;151:900–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, Wang H, Bell P, et al. Hepatic gene transfer in neonatal mice by adeno-associated virus serotype 8 vector. Hum Gene Ther 2012;23:533–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urnov FD, Miller JC, Lee YL, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 2005;435:646–651 [DOI] [PubMed] [Google Scholar]
- 50.Christian M, Cermak T, Doyle EL, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 2010;186:757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith J, Grizot S, Arnould S, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res 2006;34:e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Ravin SS, Reik A, Liu PQ, et al. Targeted gene addition in human CD34(+) hematopoietic cells for correction of X-linked chronic granulomatous disease. Nat Biotechnol 2016;34:424–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J, Exline CM, DeClercq JJ, et al. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat Biotechnol 2015;33:1256–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, Haurigot V, Doyon Y, et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 2011;475:217–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science 2013;339:823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012;337:816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolotin A, Quinquis B, Sorokin A, et al. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005;151(Pt 8):2551–2561 [DOI] [PubMed] [Google Scholar]
- 59.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, et al. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 2005;60:174–182 [DOI] [PubMed] [Google Scholar]
- 60.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 2005;151(Pt 3):653–663 [DOI] [PubMed] [Google Scholar]
- 61.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature 2012;482:331–338 [DOI] [PubMed] [Google Scholar]
- 62.Brouns SJ, Jore MM, Lundgren M, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008;321:960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bibikova M, Golic M, Golic KG, et al. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 2002;161:1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cho SW, Kim S, Kim JM, et al. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 2013;31:230–232 [DOI] [PubMed] [Google Scholar]
- 65.Yang Y, Wang L, Bell P, et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol 2016;34:334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yin H, Song CQ, Dorkin JR, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol 2016;34:328–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ran FA, Cong L, Yan WX, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015;520:186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xie C, Zhang YP, Song L, et al. Genome editing with CRISPR/Cas9 in postnatal mice corrects PRKAG2 cardiac syndrome. Cell Res 2016; 26:1099–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dever DP, Bak RO, Reinisch A, et al. CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells. Nature 2016;539:384–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nelson CE, Hakim CH, Ousterout DG, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 2016;351:403–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tabebordbar M, Zhu K, Cheng JK, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 2016;351:407–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Long C, Amoasii L, Mireault AA, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 2016;351:400–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Swiech L, Heidenreich M, Banerjee A, et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol 2015;33:102–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Platt RJ, Chen S, Zhou Y, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 2014;159:440–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fujita T, Yuno M, Fujii H. Allele-specific locus binding and genome editing by CRISPR at the p16INK4a locus. Sci Rep 2016;6:30485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shin JW, Kim KH, Chao MJ, et al. Permanent inactivation of Huntington's disease mutation by personalized allele-specific CRISPR/Cas9. Hum Mol Genet 2016. September 15 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boudreau RL, McBride JL, Martins I, et al. Nonallele-specific silencing of mutant and wild-type Huntingtin demonstrates therapeutic efficacy in Huntington's disease mice. Mol Ther 2009;17:1053–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hendel A, Bak RO, Clark JT, et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol 2015;33:985–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Riechmann L, Clark M, Waldmann H, et al. Reshaping human antibodies for therapy. Nature 1988;332:323–327 [DOI] [PubMed] [Google Scholar]
- 80.Gasiunas G, Barrangou R, Horvath P, et al. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A 2012;109:E2579–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ran FA, Hsu PD, Lin CY, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013;154:1380–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Komor AC, Kim YB, Packer MS, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016;533:420–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gilbert LA, Horlbeck MA, Adamson B, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 2014;159:647–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maeder ML, Linder SJ, Cascio VM, et al. CRISPR RNA-guided activation of endogenous human genes. Nat Methods 2013;10:977–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.O'Connell MR, Oakes BL, Sternberg SH, et al. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 2014;516:263–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kleinstiver BP, Prew MS, Tsai SQ, et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol 2015;33:1293–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chew WL, Tabebordbar M, Cheng JK, et al. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat Methods 2016;13:868–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kleinstiver BP, Pattanayak V, Prew MS, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016;529:490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Slaymaker IM, Gao L, Zetsche B, et al. Rationally engineered Cas9 nucleases with improved specificity. Science 2016;351:84–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Baltimore D, Berg P, Botchan M, et al. Biotechnology. A prudent path forward for genomic engineering and germline gene modification. Science 2015;348:36–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Morgens DW, Deans RM, Li A, et al. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat Biotechnol 2016;34:634–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Deans RM, Morgens DW, Okesli A, et al. Parallel shRNA and CRISPR-Cas9 screens enable antiviral drug target identification. Nat Chem Biol 2016;12:361–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Miller DG, Wang PR, Petek LM, et al. Gene targeting in vivo by adeno-associated virus vectors. Nat Biotechnol 2006;24:1022–1026 [DOI] [PubMed] [Google Scholar]
- 94.Hickey RD, Lillegard JB, Fisher JE, et al. Efficient production of Fah-null heterozygote pigs by chimeric adeno-associated virus-mediated gene knockout and somatic cell nuclear transfer. Hepatology 2011;54:1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Melo SP, Lisowski L, Bashkirova E, et al. Somatic correction of junctional epidermolysis bullosa by a highly recombinogenic AAV variant. Mol Ther 2014;22:725–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Barzel A, Paulk NK, Shi Y, et al. Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature 2015;517:360–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chamberlain JR, Schwarze U, Wang PR, et al. Gene targeting in stem cells from individuals with osteogenesis imperfecta. Science 2004;303:1198–1201 [DOI] [PubMed] [Google Scholar]
- 98.Russell DW, Hirata RK. Human gene targeting by viral vectors. Nat Genet 1998;18:325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Paulk NK, Wursthorn K, Wang Z, et al. Adeno-associated virus gene repair corrects a mouse model of hereditary tyrosinemia in vivo. Hepatology 2010;51:1200–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nygaard S, Barzel A, Haft A, et al. A universal system to select gene-modified hepatocytes in vivo. Sci Transl Med 2016;8:342ra379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Edwards CA, Mungall AJ, Matthews L, et al. The evolution of the DLK1-DIO3 imprinted domain in mammals. PLoS Biol 2008;6:e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kota J, Chivukula RR, O'Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 2009;137:1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Miyazaki Y, Adachi H, Katsuno M, et al. Viral delivery of miR-196a ameliorates the SBMA phenotype via the silencing of CELF2. Nat Med 2012;18:1136–1141 [DOI] [PubMed] [Google Scholar]
- 104.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384–388 [DOI] [PubMed] [Google Scholar]
- 105.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333–338 [DOI] [PubMed] [Google Scholar]
- 106.Johnsson P, Ackley A, Vidarsdottir L, et al. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol 2013;20:440–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010;465:1033–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov 2014;13:622–638 [DOI] [PubMed] [Google Scholar]
- 109.Lisowski L, Dane AP, Chu K, et al. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature 2014;506:382–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gao G, Vandenberghe LH, Wilson JM. New recombinant serotypes of AAV vectors. Curr Gene Ther 2005;5:285–297 [DOI] [PubMed] [Google Scholar]
- 111.Grimm D, Lee JS, Wang L, et al. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J Virol 2008;82:5887–5911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li W, Asokan A, Wu Z, et al. Engineering and selection of shuffled AAV genomes: a new strategy for producing targeted biological nanoparticles. Mol Ther 2008;16:1252–1260 [DOI] [PubMed] [Google Scholar]
- 113.Maheshri N, Koerber JT, Kaspar BK, et al. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat Biotechnol 2006;24:198–204 [DOI] [PubMed] [Google Scholar]
- 114.Dalkara D, Byrne LC, Klimczak RR, et al. In vivo-directed evolution of a new adeno-associated virus for therapeutic outer retinal gene delivery from the vitreous. Sci Transl Med 2013;5:189ra176. [DOI] [PubMed] [Google Scholar]
- 115.Deverman BE, Pravdo PL, Simpson BP, et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat Biotechnol 2016;34:204–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tervo DG, Hwang BY, Viswanathan S, et al. A designer AAV variant permits efficient retrograde access to projection neurons. Neuron 2016;92:372–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Murlidharan G, Sakamoto K, Rao L, et al. CNS-restricted transduction and CRISPR/Cas9-mediated gene deletion with an engineered AAV vector. Mol Ther Nucleic Acids 2016;5:e338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Choudhury SR, Harris AF, Cabral DJ, et al. Widespread central nervous system gene transfer and silencing after systemic delivery of novel AAV-AS vector. Mol Ther 2016;24:726–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Choudhury SR, Fitzpatrick Z, Harris AF, et al. In vivo selection yields AAV-B1 capsid for central nervous system and muscle gene therapy. Mol Ther 2016;24:1247–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Munch RC, Muth A, Muik A, et al. Off-target-free gene delivery by affinity-purified receptor-targeted viral vectors. Nature Commun 2015;6:6246. [DOI] [PubMed] [Google Scholar]
- 121.Pillay S, Meyer NL, Puschnik AS, et al. An essential receptor for adeno-associated virus infection. Nature 2016;530:108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Summerford C, Samulski RJ. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol 1998;72:1438–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shen S, Bryant KD, Brown SM, et al. Terminal N-linked galactose is the primary receptor for adeno-associated virus 9. J Biol Chem 2011;286:13532–13540 [DOI] [PMC free article] [PubMed] [Google Scholar]