Abstract
Currently, clinical gene therapy is experiencing a renaissance, with new products for clinical use approved in Europe and clinical trials for multiple diseases reporting positive results, especially those using recombinant adeno-associated viral (rAAV) vectors. Amid this new success, it is prudent to recall that the field of gene therapy experienced tragic setbacks in 1999 and 2002 because of the serious adverse events related to retroviral and adenoviral gene delivery in two clinical trials that resulted in the death of two patients. In both cases, the toxicity observed in humans had been documented to occur in animal models. However, these toxicities were either undetected or underappreciated before they arose in humans. rAAVs have been tested extensively in animals and animal models of disease, largely without adverse events, except for transient elevation in liver enzymes in some patients. However, a small but growing number of murine studies have documented that adeno-associated viral gene delivery can result in insertional mutagenesis. Herein, the aggregate data are reviewed from multiple murine studies where genotoxicity associated with rAAV treatment has been observed. The data emphasize the need for a proactive position to evaluate the potential risks and possible solutions associated with AAV-mediated gene therapy.
Keywords: : AAV, gene therapy, insertional mutagenesis, genotoxicity, cancer, HCC
Introduction
The current level of enthusiasm for gene therapy as a viable therapeutic approach to treat a variety of human diseases is unprecedented. No longer is gene therapy a largely academic undertaking as biotechs, pharma companies, venture capitalists, and investors become increasingly more involved in the field.1–3 However, after two highly publicized deaths occurring in two separate gene therapy clinical trials in the late 1990s and early 2000s, the future of gene therapy appeared bleak.4,5 The toxicity observed in these clinical trials was caused by immunogenicity to an adenoviral vector and then to insertional mutagenesis by a retroviral vector. These types of toxicities had been observed in animals,6,7 but the risk in humans was deemed to be minimal. Current viral vectors are designed to be less mutagenetic, as in the case of SIN lentiviruses, or are intrinsically less immunogenic than the those that caused severe adverse events in earlier clinical trials.
One such vector, recombinant adeno-associated virus (rAAV), is inherently non-pathogenic and less likely to result in insertional mutagenesis because the majority of vector genomes remain episomal after transduction.8,9 This viral vector is suitable for both localized and systemic gene delivery and is capable of transducing a variety of cell types. To date, numerous naturally occurring AAV serotypes have been isolated from a number of different species, and novel capsids have been genetically engineered.10 A number of rAAVs have been shown to exhibit tropisms for hepatocytes, myocytes, and neuronal cells.
AAV has several advantages over other commonly used viral vectors that have been adapted for gene delivery, such as retroviral, lentiviral, and adenoviral vectors, including an ability to transduce dividing and nondividing cells, highly efficient in vivo cell transduction, sustained transgene expression, tropisms for specific tissues and cell types, low immunogenicity, and a history of clinical safety. The rAAVs have been successfully used as gene delivery vehicles in numerous animal models of human disease and have yielded long-term transgene expression without apparent vector-related toxicity. Furthermore, clinical trials using rAAV have involved hundreds of subjects, supporting the safety of these vectors for use in humans.11,12
The rAAVs used in gene delivery are physically different and exhibit different characteristics relative to the wild-type AAV precursor (reviewed by Daya and Berns13). The life cycle of wild-type AAV includes both latent and lytic phases. The AAV genome is 4.7 kb in size and consists of three promoters and multiple open reading frames coding for the replication proteins (Rep), the capsid proteins (Cap), and assembly-activating protein (AAP), which are flanked by inverted terminal repeats (ITR). The ITRs are DNA elements that are 145 bp in length and that are integral to AAV genome replication and proper DNA encapsulation.14–16 During the latent stage, the AAV2 provirus preferential integrates into a specific location on human chromosome 19.17–19
The wild-type AAV genome is substantially altered to create the rAAV genome used in gene therapy, and these alterations result in dramatic differences in physical characteristics between the wild-type AAV and rAAV. When rAAVs are genetically engineered to carry exogenous sequences, such as enhancers, promoters, transgenes, and polyA signals, most of the wild-type AAV genome is removed because AAVs have a packaging constraint of approximately 4.7 kb. While the Rep and Cap proteins are necessary for rAAV production, the AAV ITRs are the only sequences necessary for rAAV packaging of the regulatory and transgene sequences.20 Hence, rAAV integration into the host genome is predicted to be greatly reduced because the sequences coding for the native AAV proteins needed for integration are not present in the recombinant AAV. However, a low number of random integrations do occur following gene delivery using rAAV.21 The rAAV vectors' reduced ability to integrate also means that the therapeutic transgene exists predominantly in the host cell nucleus as non-replicating episomes. Because rAAV largely does not integrate, vector genomes and transgene expression are lost over time because of the inability to replicate with the host cell's DNA.22
Unlike adenoviral and gamma-retroviral vectors, rAAV has been used in numerous clinical trials with no vector- or viral-related severe adverse events, other than transient transaminitis.23 A gene therapy pilot safety study using an adenovirus to treat a patient with ornithine transcarbamylase deficiency resulted in a fatality thought to be the result of a systemic inflammatory response to the adenoviral capsid.24 This type of severe adverse immune response has not been observed in clinical trials that utilize rAAV gene therapy, presumably because AAV is less immunogenic than adenovirus.25,26 In fact, only mild elevations of liver enzymes have been reported after systemic rAAV delivery in patients.23,27 Clinical trials that utilized gamma-retroviral mediated ex vivo gene transfer to treat X-linked severe combined immunodeficiency and Wiskott–Aldrich syndrome have reported insertional mutagenesis by the gene therapy vector, which caused leukemia in patients.28,29 Because the rAAV transgene exists in a predominately episomal state and integrates at a very low frequency following rAAV transduction, the chances of insertional mutagenesis by a rAAV transgene are greatly reduced in comparison with retroviral and lentiviral vectors, which usually integrate into the host genome. However, an increased incidence in hepatic carcinoma (HCC) in mice after rAAV gene delivery has been reported, and some of these studies have shown that insertional mutagenesis by rAAV was the most likely cause of the HCC. Herein, some of the studies, listed in Table 1, that observed or interrogated HCC formation after rAAV gene delivery are discussed. The key observations from each study are highlighted, and unresolved issues are discussed.
Table 1.
Studies that observed or investigated HCC formation after rAAV gene delivery
| Publication | AAV serotype | Genome configuration | Enhancer/promoter (*promoters only) | Transgene | Route of delivery | Dose | Time of treatment | Loci with integration(s) associated with HCC |
|---|---|---|---|---|---|---|---|---|
| Donsante et al. Gene Ther (2001)39 | 2 | ss | CMV/CBA | GUSB | Superficial temporal vein | 1 × 1014 vg/kg | Neonatal | Rian |
| Donsante et al. Science (2007)40 | ||||||||
| Bell et al. Mol Ther (2005)45 | 1,2,5,7,8,9, others | ss | Alb, CBA, SV40, TBG, Z12* | A1AT, EGFP, Epo, FIX, LacZ, LDLR, TF1 | Portal vein | 3 × 109–1 × 1012 vg/mouse | 6–8 weeks | None |
| Tail vein | ||||||||
| Bell et al. Mol Ther (2006)43 | 2,7,8,9 | ss | AMPB/TBG | Oct, LacZ | Portal vein | 3 × 109–1 × 1012 vg/mouse | 9–11 weeks | Tax1bp1, Rian |
| Zhong et al. Hum Gene Ther (2013)44 | ||||||||
| Reiss et al. Hum Mutat (2011)57 | 1/2 | ss | CMV/CBA | MOCS1 | Intrahepatic | 1–3 × 109 vg/pup | Neonatal | None |
| Embury et al. Gene Ther Mol Biol (2008)42 | 2 | ss | CBA* (WPRE) | mPah | Portal vein | 3.7 × 109–7.3 × 1013 vg/mouse | 10–14 weeks | None |
| Li et al. Blood (2011)46 | 2 | ss | APOE/hAAT | FIX | Portal vein | 5 × 1012–1 × 1014 vg/kg | 4 weeks | None |
| Wang et al. Proc Natl Acad Sci U S A (2012)41 | 2 | ss | CMV/CBA | Homology Arms to Rian | Temporal vein | 3 × 109 vg/pup | Neonatal | Rian |
| Rosas et al. Mol Ther (2012)48 | rh74 | sc | CMV, CBA* | GFP, null | Tail vein | 5 × 1010 vg/mouse | 8–12 weeks | Hras1, Sos1, Fgf3 |
| Kao et al. Thromb Haemost (2013)47 | 8 | ss | APOE/hAAT | FIX-WT, FIX-Triple, FIX-Padua | Tail vein | 1.6 × 1012–2 × 1010 vg/mouse | Adult | None |
| Chandler et al. J Clin Invest (2015)50 | 2,8,9 | ss | AMPB/TBG CMV/CBA APOE/hAAT | hMUT, mMut, GFP | Intrahepatic | 7 × 1012–1 × 1014 vg/kg | Neonatal | Rian |
| Walia et al. Mol Ther (2015)49 | 9 | ss | CMV/CMV | HexB, LacZ | Superficial temporal vein | 2.5 × 1014 vg/kg | Neonatal | Rian |
HCC, hepatic carcinoma; rAAV, recombinant adeno-associated virus.
Aav and Hcc: An Overview of Published Studies
Lysosomal storage diseases (LSDs) are a relatively large class of inherited metabolic disease that are typically caused by a deficiency in a soluble lysosomal enzyme.30 LSDs are ideal candidates for systemic gene therapy.31 They are simple, monogenic diseases, and some proportion of lysosomal enzymes can be secreted from corrected cells and taken up by distant cells through a receptor-mediated process known as cross-correction.32 Mucopolysaccharidosis type VII (MPSVII) is an invariably fatal LSD caused by a deficiency in the lysosomal enzyme, β-glucuronidase (GUSB).33 β-Glucuronidase is ubiquitously expressed, and its deficiency leads to the accumulation of undegraded glycosaminoglycans in most tissues, including the liver spleen, heart, kidney, bones, eyes, and brain. A spontaneously arising mouse mutant was discovered at The Jackson Laboratory that has a single base pair deletion in the Gusb gene, is completely deficient in Gusb activity, and mimics most of the biochemical, histological, and clinical features of human MPSVII.34,35 The MPSVII mouse has been used extensively in preclinical studies to determine the efficacy of enzyme replacement therapy, stem cell–mediated therapy, and gene replacement therapy.
One of the first systemic AAV-mediated gene therapy experiments was performed using MPSVII mice.36,37 Intravenous injection of a first-generation AAV2 vector into newborn MPSVII mice resulted in widespread transduction, relatively high levels of human GUSB activity, and prevention of lysosomal storage material in most tissues of the body, including the brain. A follow-up study in the MPSVII mice showed that this approach led to persistent GUSB expression and resulted in dramatic improvements in bone development, retinal function, auditory function, body weight, and life-span.38 However, it was discovered that a number of long-lived (≥1 year) rAAV-treated MPSVII mice from this study developed HCC.39 It was virtually impossible to determine the cause of the tumors in that study, since the experiment was not designed as a toxicity study, and the number of animals with HCC was relatively small. In addition, the tumor samples were negative for GUSB activity, and the rAAV genome could not be detected by polymerase chain reaction (PCR) using primers specific for the GUSB cDNA.
A larger follow-up study in newborn MPSVII and normal mice was performed in an attempt to replicate the findings of Donsante et al. and to determine the cause of the HCC. Intravenous injection of the same AAV2 vector resulted in 30–60% of both normal and MPSVII mice developing HCC at ≥13 months of age.40 It was also shown that the incidence of HCC in long-lived MPSVII mice receiving bone-marrow transplantation and a transgenic mouse expressing ∼20-fold greater than normal levels of GUSB was nearly zero. The most striking and unexpected finding was that four of the six tumors evaluated had the recombinant AAV vector integrated within a 6,000 bp region of the Rian locus on mouse chromosome 12. This region contains several genes and is rich in micro- and snoRNAs. Integration of the rAAV vector dysregulated this locus. Another interesting finding was that all of the integrants were rearranged such that the GUSB cDNA was deleted. This explains the lack of a PCR product and absent GUSB activity in the tumors examined in the original report.39
Although rAAV integration into the Rian locus was associated with the development of HCC, a cause and effect relationship was suggested but not proven. However, a subsequent study showed that there was a high frequency of integration into the same locus following intravenous injection of an rAAV vector containing a promoter and enhancer combination (no transgene) flanked by sequences that are homologous to the Rian locus.41 Integration of the promoter/enhancer elements and subsequent dysregulation of this locus led to nearly 100% penetrance of the HCC phenotype. These data strongly suggest that disruption of this locus by integration of an rAAV vector containing a strong promoter/enhancer combination causes HCC in mice.
The development of HCC following intravenous injection of an AAV vector was still generally considered an isolated event, even though there were several additional reports of tumorigenesis following rAAV administration. Portal-vein injection of an rAAV vector containing the woodchuck hepatitis virus post-transcriptional regulatory element and the phenylalanine hydroxylase cDNA into the mouse model of phenylketonuria also resulted in a high frequency of HCC.42 The authors of this report concluded that the HCC was caused by expression of the hepatitis virus protein X-phenylalanine hydroxylase fusion protein. Although immunohistochemical analysis showed that the hepatitis virus protein X-phenylalanine hydroxylase fusion protein was expressed in the liver of experimental animals, the expression appeared to be limited to the normal liver tissue and not the tumor. In retrospect, these data suggest that the tumors were not caused by expression of the hepatitis virus protein X-phenylalanine hydroxylase fusion protein, rather a potential integration of a rearranged rAAV vector that did not express the transgene, but manifested genotoxicity similar to what was seen with the MPSVII mice and vectors.
A high incidence of HCC was also observed in ornithine transcarbamolase (OTC)-deficient mice following portal-vein injection of rAAV vectors encoding either OTC or LacZ.43 OTC-deficient mice are known to have a high incidence of spontaneous liver tumors. Statistical analyses of the various groups suggested that mice treated with the LacZ vector had an increased risk of HCC. A retrospective analysis of some of the samples from this study discovered numerous integration sites in the tumor tissue, including integrations into the Rian locus.44
There is also evidence suggesting that systemic delivery of rAAV vectors does not lead to the development of HCC. In 2005, a large retrospective study of mice treated with rAAV did not find an association between rAAV treatment and HCC in mice.45 A second, relatively large study, involving 132 mice, specifically designed to address the issue of HCC following intravenous administration of rAAV vectors, showed no statistically significant increase in HCC in mice injected with an rAAV vector encoding clotting factor IX.46 However, the small number of HCCs observed in this study, five in total, four of which occurred in the rAAV-treated group, may not have been sufficient to detect a significant association. Nevertheless, this study also used integration profiling and gene expression analysis to demonstrate that the HCCs appeared free of clonal integration events, or gene expression changes related to rAAV integration.46 A separate study showed that hepatic tumors were observed in young adult mice following intravenous injection of a high dose (2 × 1010 vp/mouse) of an rAAV vector expressing a factor IX cDNA containing the Padua mutation, but occurred at an insignificant rate of ∼15%. Genomic characterization was not performed on the HCC in this study. One significant difference between studies has been the developmental timing of the injections. Donsante et al. injected the mice on postnatal day (PND) 1–2; Bell et al. and Li et al. injected young adult animals.39,45,46 These data suggest that mice injected during the newborn period may be more susceptible to the development of rAAV-associated HCC and, furthermore, that the HCC association was not mouse model dependent.
In a complex study using varied scAAV vectors, applied with and without the agent camptothecin, and/or partial hepatectomy, Rosas et al. assayed HCC formation in cancer-prone and immunodeficient mice, with the intention that genotoxic effects of rAAV may be magnified.48 While the authors did observe HCC formation, and even integration events into cellular oncogenes, the very high background rate of HCC formation and relatively low number of recovered integration events failed to resolve whether rAAV was carcinogenic.
The next set of studies confirmed what was initially observed by Donsante et al., specifically that mice treated in the neonatal period with high doses of rAAV had a tendency to develop HCCs later in life. As with earlier studies that have observed HCC after rAAV administration, the most recent studies were designed to assess efficacy not toxicity in mouse models of either Sandhoff disease (SD) or methylmalonic acidemia (MMA). In the case of SD, neonatal mice were injected via the superficial temporal vein with a relatively high dose (2.5 × 1014 vg/kg) of an rAAV9 vector that used the CMV promoter to drive the expression of the HEXB cDNA, and followed for beneficial effects.49 At the experimental end point of 43 weeks, gross examination during necropsy revealed tumors in 8/10 neonatally rAAV9-HexB–injected control and SD mice; seven animals had liver tumors, and one had multiple lung tumors. Tumors were not observed in untreated mice, but it should be noted that untreated SD mice uniformly perished, typically by 15 weeks of age. In 2/7 of the liver tumors, integrations into the Rian locus were observed, while the single mouse that developed lung cancer was found to have an rAAV integration in fibroblast growth factor receptor 2 (Fgfr2), a gene known to have altered expression in cancer. Although this study lacked large numbers of control animals, the penetrance of HCC in the mice treated as neonates clearly implicates rAAV therapy as a predisposing factor to carcinogenesis.
The recent report by Chandler et al. is the largest mouse study to report rAAV genotoxicity to date. It used large numbers of mice that were treated with a variety of rAAV vectors and serotypes, or that were untreated.50 In this study, the colony-background incidence of HCC measured at <10% (n = 51 mice), whereas control (n = 24) and MMA mice (n = 24) treated in the neonatal period with a dose of 1 × 1011 vg/pup of therapeutic or reporter rAAVs developed HCC at a rate of 50–75% by 2 years of age. This study confirmed the findings of several previous reports that identified clonal rAAV integrations in the Rian locus, with upregulation of proximally located micro-RNAs and genes as the likely cause of the HCC. Identifying insertional mutagenesis of the Rian locus as the cause of HCC was not a surprising finding because previous studies to identify genes involved in carcinogenesis have identified mutations in the Rian locus as a cause of HCC in mice.51,52
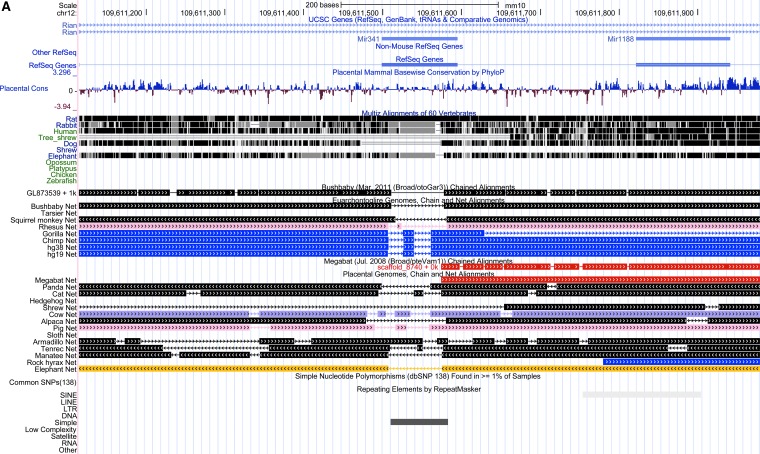
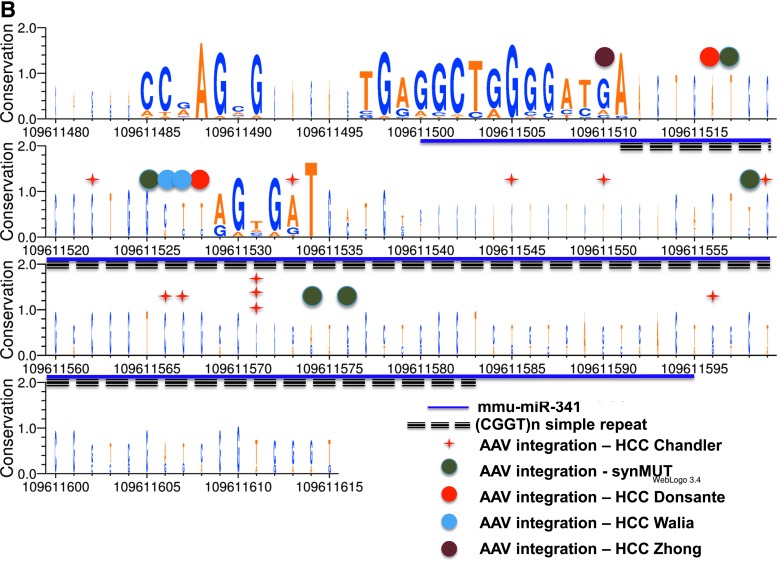
This study also reported several unique findings with regard to rAAV toxicity and integration patterns. First, the risk of the HCC was found to be rAAV dose-dependent. Second, a majority of the causative HCC-rAAV integrations were mapped to Mir341, a locus within Rian. As depicted in Fig. 1A, a genomic alignment centered on Mir341 revealed several unusual features. The locus appears only to be present in rodents, and contains an embedded repeat. Upon closer inspection, this repeat contains a unique array of two simple quadranucleotide repeats (CCGT and GGCT), some of which are internally repeated, as well as 11 CpG dinucleotides, yet is flanked by conserved sequences (Fig. 1B). Most rAAV HCC integrations mapped into this tiny microRNA, and perhaps most importantly, one rAAV, which utilized an hAAT promoter and synthetic MUT allele (designated synMUT in the Fig. 1B), did not cause upregulation of proximally located miRNAs and genes or HCC, suggesting that rAAV vector design could influence genotoxicity.
Figure 1.
(A) A genomic alignment showing an alignment of 31 syntenic vertebrate sequences around the miR341 locus of the mouse, and the position of a small simple repeat within the gene. (B) A base pair resolution map shows the CGGT repeat with mapped AAV integrations indicated as colored dots, with the corresponding study. The relative conservation of each base was measured and depicted with a logo plot where the height is proportional to conservation and boldness indicates more sequences.
The use of a sensitive integration-capture method that relied on high throughput sequencing (HTS) allowed for the detection of thousands of rAAV integration events from approximately 40 samples has afforded a limited assessment of integration preferences.50 To query integration preferences in a very direct fashion, the number of integrations in a locus was divided by the size of the locus as measured in base pairs in order to normalize for frequency. Approximately 149 of a total of 1,205 loci with an rAAV integration had multiple rAAV integrations (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/hum). Table 2 shows the loci with the highest rAAV integration frequency, which ranges from 2.19 × 10−1 to 4.40 × 10−7 integrations per base pair. Of importance, and in contrast to previous studies that have suggested the rAAV integration is largely random, it is noted that rAAV integration frequencies varied significantly between different loci, suggesting an integration preference by rAAV (Table 2) not yet observed in smaller rAAV data sets. This analysis also suggests that the characterization of many more integration events will be needed to further define sites preferred for integration by rAAV.
Table 2.
Genes that are frequent targets of rAAV integration from Chandler et al.50
| Gene name | Unique HCC integrations | Unique normal liver integrations | Total number of integrations in gene | Locus size (bp) | Integration frequency normalized to gene size |
|---|---|---|---|---|---|
| Mir341 (Rian) | 18 | 3 | 21 | 96 | 2.19 × 10−1 |
| AC152063.1 (Rian) | 1 | 0 | 1 | 87 | 1.15 × 10−2 |
| Mir212 | 0 | 1 | 1 | 91 | 1.10 × 10−2 |
| Alb | 68 | 75 | 143 | 15,715 | 9.10 × 10−3 |
| Igfbp1 | 14 | 9 | 23 | 4,765 | 4.83 × 10−3 |
| Gm14719 | 0 | 1 | 1 | 231 | 4.33 × 10−3 |
| 7SK | 1 | 0 | 1 | 284 | 3.52 × 10−3 |
| Afp | 23 | 28 | 51 | 18,194 | 2.80 × 10−3 |
| Gm16326 | 1 | 0 | 1 | 414 | 2.42 × 10−3 |
| AY036118 | 0 | 1 | 1 | 725 | 1.38 × 10−3 |
| Gm10800 | 1 | 0 | 1 | 755 | 1.32 × 10−3 |
| Gm11620 | 2 | 0 | 2 | 1,845 | 1.08 × 10−3 |
| Olfr1113 | 0 | 1 | 1 | 981 | 1.02 × 10−3 |
| Dnajb8 | 0 | 1 | 1 | 989 | 1.01 × 10−3 |
Concluding Remarks and Future Directions
Given the recent observation that AAV integrations appear to be present in some human HCCs and, in subset, may be pathologically relevant, a careful reconsideration of possible genotoxicity mediated by rAAV in preclinical models is warranted as vectors are studied before entering the clinic.53 What is clear from a number of reports is that mice treated in the neonatal period with high doses of rAAV that contains strong promoter-enhancer elements, or vectors that target integration into Rian, have a very high chance of developing HCC, and in the HCC, remnants of the vector will be likely be present in Mir341. However, it needs to be emphasized that distinct, non-Mir341 rAAV integrations within Rian do occur and appear pathogenic. The results of the limited reanalysis of available integration data presented in Table 2 confirms that site preferences for rAAV do exist, especially when one considers that some view a common integration site (CIS) as a region of ≤30 kb that harbors two or more integrations (the equivalent of ≥6.7 × 10−5 integration per base pair).54 Many more rAAV integrations must be captured and analyzed in order to define rAAV CISs and to determine whether sequence motifs, chromatin states/marks, and vector characteristics influence integration preferences. A recent publication describing a hybrid AAV-piggyBac transposase delivery method might also add important information about whether AAV integration events can cause HCC.55
Another area of future experimental need surrounds the precise characterization of rAAV integration events. A particularly relevant observation is that both sides of the integration junction have never been captured in any rAAV HCC study. Thus, it remains possible that rAAV integration might be associated with a complex rearrangement or even translocation, in addition to the possibility of vector concatamerization.56 Classical genomic cloning, tiling, and/or new methods may need to be applied to elucidate fully the genomic features of rAAV integration and whether they might contribute to carcinogenesis.
A set of parallel studies should also be undertaken, allowing the growing genomic resources associated with human HCCs to be queried to ascertain whether there is a recurrent set of genes, RNAs, and/or regulatory elements commonly dysregulated in HCC. By integrating HTS-based analyses of integrations in liver tissue from rodents (and other species) treated with rAAV, an assessment of genotoxic risk might be approximated by simply mapping rAAV integrations, especially if they fall into a CIS, onto a framework of genetic elements that are associated with human HCC. A pattern of vector integration into loci associated with HCC, if seen in numerous samples from different animals, might indicate a potential hepatic toxicity, and suggest further monitoring, such as the assessment of circulating alpha-fetoprotein and/or abdominal imaging, in clinical applications.
While the current safety profile of rAAV in human clinical applications remains impeccable, the preclinical animal model data, coupled with an incomplete understanding of rAAV integration, should continue to motivate vector safety studies in preclinical models using modern genomic methods.
Supplementary Material
Acknowledgments
R.J.C. and C.V. were supported by the Intramural Research Program of the National Human Genome Research Institute, and M.S.S. was partially supported by an NIH grant R01 NS043205.
Author Disclosure
No competing financial interests exist.
References
- 1.Naldini L. Gene therapy returns to centre stage. Nature 2015;526:351–360 [DOI] [PubMed] [Google Scholar]
- 2.Bender E. Gene therapy: industrial strength. Nature 2016;537:S57–59 [DOI] [PubMed] [Google Scholar]
- 3.Schimmer J, Breazzano S. Investor outlook: gene therapy picking up steam; at a crossroads. Hum Gene Ther Clin Dev 2016;27:87–92 [DOI] [PubMed] [Google Scholar]
- 4.Hacein-Bey-Abina S, von Kalle C, Schmidt M, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med 2003;348:255–256 [DOI] [PubMed] [Google Scholar]
- 5.Raper SE, Chirmule N, Lee FS, et al. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol Genet Metab 2003;80:148–158 [DOI] [PubMed] [Google Scholar]
- 6.Li Z, Dullmann J, Schiedlmeier B, et al. Murine leukemia induced by retroviral gene marking. Science 2002;296:497. [DOI] [PubMed] [Google Scholar]
- 7.Bruder JT, Kovesdi I. Adenovirus infection stimulates the Raf/MAPK signaling pathway and induces interleukin-8 expression. J Virol 1997;71:398–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schnepp BC, Clark KR, Klemanski DL, et al. Genetic fate of recombinant adeno-associated virus vector genomes in muscle. J Virol 2003;77:3495–3504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schnepp BC, Jensen RL, Chen CL, et al. Characterization of adeno-associated virus genomes isolated from human tissues. J Virol 2005;79:14793–14803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asokan A, Schaffer DV, Samulski RJ. The AAV vector toolkit: poised at the clinical crossroads. Mol Ther 2012;20:699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mueller C, Flotte TR. Clinical gene therapy using recombinant adeno-associated virus vectors. Gene Ther 2008;15:858–863 [DOI] [PubMed] [Google Scholar]
- 12.Mingozzi F, High KA. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat Rev Genet 2011;12:341–355 [DOI] [PubMed] [Google Scholar]
- 13.Daya S, Berns KI. Gene therapy using adeno-associated virus vectors. Clin Microbiol Rev 2008;21:583–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LeFebvre RB, Berns KI. Unique events in parvovirus replication. Microbiol Sci 1984;1:163–167 [PubMed] [Google Scholar]
- 15.Bohenzky RA, LeFebvre RB, Berns KI. Sequence and symmetry requirements within the internal palindromic sequences of the adeno-associated virus terminal repeat. Virology 1988;166:316–327 [DOI] [PubMed] [Google Scholar]
- 16.Samulski RJ, Chang LS, Shenk T. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J Virol 1989;63:3822–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samulski RJ, Zhu X, Xiao X, et al. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. Embo J 1991;10:3941–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotin RM, Siniscalco M, Samulski RJ, et al. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci U S A 1990;87:2211–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotin RM, Menninger JC, Ward DC, et al. Mapping and direct visualization of a region-specific viral DNA integration site on chromosome 19q13-qter. Genomics 1991;10:831–834 [DOI] [PubMed] [Google Scholar]
- 20.Grimm D, Kern A, Rittner K, et al. Novel tools for production and purification of recombinant adenoassociated virus vectors. Hum Gene Ther 1998;9:2745–2760 [DOI] [PubMed] [Google Scholar]
- 21.Nakai H, Montini E, Fuess S, et al. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat Genet 2003;34:297–302 [DOI] [PubMed] [Google Scholar]
- 22.Duan D, Sharma P, Yang J, et al. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle tissue. J Virol 1998;72:8568–8577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manno CS, Pierce GF, Arruda VR, et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat Med 2006;12:342–347 [DOI] [PubMed] [Google Scholar]
- 24.Raper SE, Chirmule N, Lee FS, et al. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol Genet Metab 2003;80:148–158 [DOI] [PubMed] [Google Scholar]
- 25.Jooss K, Yang Y, Fisher KJ, et al. Transduction of dendritic cells by DNA viral vectors directs the immune response to transgene products in muscle fibers. J Virol 1998;72:4212–4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Somanathan S, Breous E, Bell P, et al. AAV vectors avoid inflammatory signals necessary to render transduced hepatocyte targets for destructive T cells. Mol Ther 2010;18:977–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nathwani AC, Reiss UM, Tuddenham EG, et al. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N Engl J Med 2014;371:1994–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 2003;302:415–419 [DOI] [PubMed] [Google Scholar]
- 29.Braun CJ, Boztug K, Paruzynski A, et al. Gene therapy for Wiskott–Aldrich syndrome—long-term efficacy and genotoxicity. Sci Transl Med 2014;6:227ra233. [DOI] [PubMed] [Google Scholar]
- 30.Part 16: Lysosomal Disorders. In: Valle D, Beaudet AL, Vogelstein B, eds. The Online Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill Global Education Holdings, LLC, 2017 [Google Scholar]
- 31.Sands MS, Davidson BL. Gene therapy for lysosomal storage diseases. Mol Ther 2006;13:839–849 [DOI] [PubMed] [Google Scholar]
- 32.Neufeld EF, Fratantoni JC. Inborn errors of mucopolysaccharide metabolism. Science 1970;169:141–146 [DOI] [PubMed] [Google Scholar]
- 33.Sly WS, Quinton BA, McAlister WH, et al. Beta glucuronidase deficiency: report of clinical, radiologic, and biochemical features of a new mucopolysaccharidosis. J Pediatr 1973;82:249–257 [DOI] [PubMed] [Google Scholar]
- 34.Birkenmeier EH, Davisson MT, Beamer WG, et al. Murine mucopolysaccharidosis type VII. Characterization of a mouse with beta-glucuronidase deficiency. J Clin Invest 1989;83:1258–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sands MS, Birkenmeier EH. A single-base-pair deletion in the beta-glucuronidase gene accounts for the phenotype of murine mucopolysaccharidosis type VII. Proc Natl Acad Sci U S A 1993;90:6567–6571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daly TM, Vogler C, Levy B, et al. Neonatal gene transfer leads to widespread correction of pathology in a murine model of lysosomal storage disease. Proc Natl Acad Sci U S A 1999;96:2296–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daly TM, Okuyama T, Vogler C, et al. Neonatal intramuscular injection with recombinant adeno-associated virus results in prolonged beta-glucuronidase expression in situ and correction of liver pathology in mucopolysaccharidosis type VII mice. Hum Gene Ther 1999;10:85–94 [DOI] [PubMed] [Google Scholar]
- 38.Daly TM, Ohlemiller KK, Roberts MS, et al. Prevention of systemic clinical disease in MPS VII mice following AAV-mediated neonatal gene transfer. Gene Ther 2001;8:1291–1298 [DOI] [PubMed] [Google Scholar]
- 39.Donsante A, Vogler C, Muzyczka N, et al. Observed incidence of tumorigenesis in long-term rodent studies of rAAV vectors. Gene Ther 2001;8:1343–1346 [DOI] [PubMed] [Google Scholar]
- 40.Donsante A, Miller DG, Li Y, et al. AAV vector integration sites in mouse hepatocellular carcinoma. Science 2007;317:477. [DOI] [PubMed] [Google Scholar]
- 41.Wang PR, Xu M, Toffanin S, et al. Induction of hepatocellular carcinoma by in vivo gene targeting. Proc Natl Acad Sci U S A 2012;109:11264–11269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Embury JE, Frost S, Charron CE, et al. Hepatitis virus protein X-phenylalanine hydroxylase fusion proteins identified in PKU mice treated with AAV-WPRE vectors. Gene Ther Mol Biol 2008;12:69–76 [Google Scholar]
- 43.Bell P, Moscioni AD, McCarter RJ, et al. Analysis of tumors arising in male B6C3F1 mice with and without AAV vector delivery to liver. Mol Ther 2006;14:34–44 [DOI] [PubMed] [Google Scholar]
- 44.Zhong L, Malani N, Li M, et al. Recombinant adeno-associated virus integration sites in murine liver after ornithine transcarbamylase gene correction. Hum Gene Ther 2013;24:520–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bell P, Wang L, Lebherz C, et al. No evidence for tumorigenesis of AAV vectors in a large-scale study in mice. Mol Ther 2005;12:299–306 [DOI] [PubMed] [Google Scholar]
- 46.Li H, Malani N, Hamilton SR, et al. Assessing the potential for AAV vector genotoxicity in a murine model. Blood 2011;117:3311–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kao CY, Yang SJ, Tao MH, et al. Incorporation of the factor IX Padua mutation into FIX-Triple improves clotting activity in vitro and in vivo. Thromb Haemost 2013;110:244–256 [DOI] [PubMed] [Google Scholar]
- 48.Rosas LE, Grieves JL, Zaraspe K, et al. Patterns of scAAV vector insertion associated with oncogenic events in a mouse model for genotoxicity. Mol Ther 2012;20:2098–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walia JS, Altaleb N, Bello A, et al. Long-term correction of Sandhoff disease following intravenous delivery of rAAV9 to mouse neonates. Mol Ther 2015;23:414–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chandler RJ, LaFave MC, Varshney GK, et al. Vector design influences hepatic genotoxicity after adeno-associated virus gene therapy. J Clin Invest 2015;125:870–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ranzani M, Cesana D, Bartholomae CC, et al. Lentiviral vector-based insertional mutagenesis identifies genes associated with liver cancer. Nat Methods 2013;10:155–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dupuy AJ, Rogers LM, Kim J, et al. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res 2009;69:8150–8156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nault JC, Datta S, Imbeaud S, et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat Genet 2015;47:1187–1193 [DOI] [PubMed] [Google Scholar]
- 54.Paneda A, Lopez-Franco E, Kaeppel C, et al. Safety and liver transduction efficacy of rAAV5-cohPBGD in nonhuman primates: a potential therapy for acute intermittent porphyria. Hum Gene Ther 2013;24:1007–1017 [DOI] [PubMed] [Google Scholar]
- 55.Cunningham SC, Siew SM, Hallwirth CV, et al. Modeling correction of severe urea cycle defects in the growing murine liver using a hybrid recombinant adeno-associated virus/piggyBac transposase gene delivery system. Hepatology 2015;62:417–428 [DOI] [PubMed] [Google Scholar]
- 56.Gil-Farina I, Fronza R, Kaeppel C, et al. Recombinant AAV integration is not associated with hepatic genotoxicity in nonhuman primates and patients. Mol Ther 2016;24:1100–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reiss J, Hahnewald R. Molybdenum cofactor deficiency: mutations in GPHN, MOCS1, and MOCS2. Hum Mutat 2011;32:10–18 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.