Abstract
We describe here a one pot RNA production, packaging and delivery system based on bacteriophage Qβ. We demonstrate a method for production of a novel RNAi scaffold, packaged within Qβ virus-like particles (VLPs). The RNAi scaffold is a general utility chimera that contains a functional RNA duplex with paired silencing and carrier sequences stabilized by a miR-30 stem-loop. The Qβ hairpin on the 5΄ end confers affinity for the Qβ coat protein (CP). Silencing sequences can include mature miRNAs and siRNAs, and can target essentially any desired mRNA. The VLP–RNAi assembles upon co-expression of CP and the RNAi scaffold in E. coli. The annealing of the scaffold to form functional RNAs is intramolecular and is therefore robust and concentration independent. We demonstrate dose- and time-dependent inhibition of GFP expression in human cells with VLP–RNAi. In addition, we target the 3΄UTR of oncogenic Ras mRNA and suppress Pan-Ras expression, which attenuates cell proliferation and promotes mortality of brain tumor cells. This combination of RNAi scaffold design with Qβ VLP packaging is demonstrated to be target-specific and efficient.
INTRODUCTION
Functional RNAs such as small interfering RNAs, microRNAs (miRNAs) and short hairpin RNAs have therapeutic (1) and research applications (2,3). Here, we produce and assemble functional nanoparticles in which precursors of active RNAs are surrounded by a protective coat protein (CP). These ‘one pot’ particles form spontaneously when RNA and CP are co-expressed in Esherichia coli. The particles are stable and readily absorbed by human cells, where the contents are released and processed to active RNAs.
The RNA production, packaging and delivery system described here uses a virus-like particle (VLP) that is derived from bacteriophage Qβ. Qβ is a T = 3 icosahedral positive-sense single-strand RNA bacteriophage with a small mRNA genome. A Qβ VLP is a nanoparticle with a diameter of 30 nm formed by 180 replicas of the Qβ CP (4,5). Qβ VLPs lack components of the phage genome and are replication-incompetent (6). A region of Qβ genomic RNA with high affinity for CP has been isolated as a 29-nucleotide RNA hairpin, known as the Qβ hairpin (7,8). The Qβ hairpin provides a hook for preferential encapsidation of target RNA by Qβ CP during in vitro or in vivo capsid assembly (8–12). Qβ VLPs are stable over a broad range of temperatures (9) and contain pores (4), by which water, ions and small molecules can enter and exit (9).
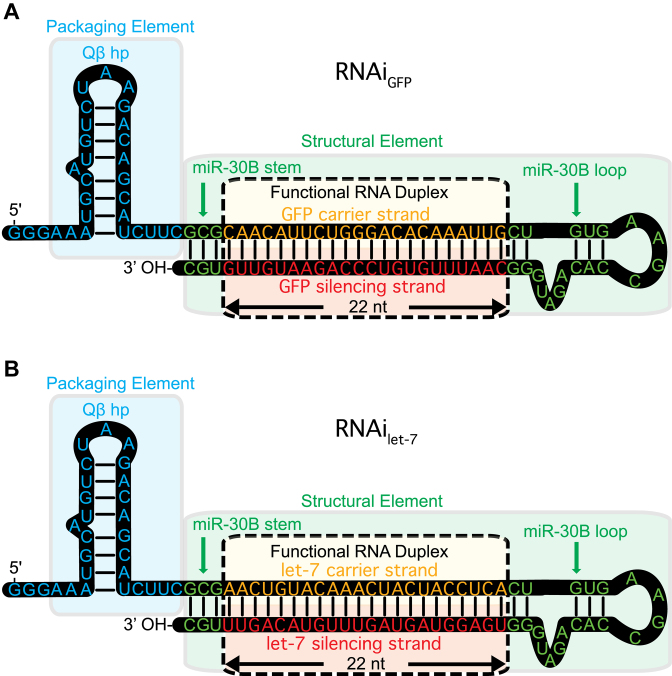
Here, we demonstrate a one pot in vivo method for production of functional RNA and CP, leading to spontaneous packaging of the RNA within VLPs. The method employs a novel RNAi scaffold (Figure 1), which we designed specifically for in vivo assembly. The RNAi scaffold folds by an intramolecular process and associates tightly with CP, leading to assembly of VLP–RNAi particles within E. coli. The RNAi scaffold is a general utility chimera that contains a functional sequence paired to a carrier sequence, combined with stability, packaging and processing elements. The scaffold contains the Qβ hairpin (7,8) on the 5΄ end to confer affinity for the CP, and a miR-30 stem-loop (13,14). The RNAi scaffold, once processed, releases the functional sequence, which can specifically inhibit gene expression (13,15).
Figure 1.
The Qβ RNAi scaffold is designed to fold into two hairpins and co-assemble with CP in vivo. A single RNA molecule forms the Qβ hairpin (blue) linked to a miR-30B stem loop (green), which contains the functional RNA duplex (yellow/red). The functional RNA duplex can encode miRNA or siRNA. The scaffold can incorporate any 22 base pair sequence, depending on the desired target. (A) An RNAi scaffold that targets GFP mRNA. (B) An RNAi scaffold that targets the Pan-Ras mRNA 3΄ UTR.
With our scaffold, functional duplex RNAs form by an intramolecular process, which is concentration independent and efficient in vivo. The silencing RNA component remains in the duplex state within the VLP and when delivered to the cytoplasm (16,17). The RNA is released from the VLP in cells, where Dicer protein liberates the functional ∼22 nucleotide element from the scaffold (18,19). The functional RNA enters the RNA-induced silencing complex (RISC complex) to induce RNA interference (13–15).
MATERIALS AND METHODS
Construction of expression vectors for Qβ–VLP, VLP–RNAi and GFP-containing VLPs
CP gene construction
DNA encoding Qβ CP monomer (NC_001890.1) flanked by Nco I and Avr II sites (Supplementary Table S1) was constructed by recursive-PCR (R-PCR) (20), cloned into pCDF-1b (Novagen) to generate plasmid pCDF-CP, and transformed into E. coli. All constructs described here were commercially sequenced after transformants were selected by antibiotic resistance and screened by colony PCR.
RNAi scaffold construction
The sequence of human miR-30B pre-miRNA, fused to the Qβ hairpin, was was added to 22-mer sequences from genes including GFP (AB038602.1, 5΄-CAA CAT TCT GGG ACA CAA ATT G-3΄) and Let-7 (NR_029660.1, 5΄-TGA GGT AGT AGT TTG TAC AGT T-3΄). Template DNAs, including the T7 promoter (T7p) and T7 terminator (T7t), were constructed by R-PCR. The DNA sequences encoding T7p-RNAi scaffold-T7t (RNAiGFP or RNAiLet-7, Supplementary Table S2) were cloned into pET28-b(+) (Novagen) to generate Qβ VLP–RNAi scaffold expression vectors.
Constructs for GFP expression
DNA encoding GFP was amplified by PCR from gWizGFP (Genlantis), appended to BglII and BlpI sites (Supplementary Table S1) and cloned into pET28-b(+) (Novagen) to generate plasmid pET28-b(+)-GFP.
Expression of Qβ VLPs and GFP-containing VLPs
E. coli containing pCDF–CP were inoculated in NZY medium with streptomycin, incubated overnight and diluted 1:100 in ZYM-5052 auto-induction medium (21). Cells were centrifuged at 4°C and 6500 ×g for 30 min, resuspended in an equal volume of Qβ buffer (10 mM MgCl2 and 20 mM Tris-HCl, pH 7.5) and lysed by sonication. The lysate was centrifuged for 30 min at 23 000 ×g. Ammonium sulfate precipitation and centrifugation yielded a pellet of crude VLPs. VLPs were resuspended in 1 ml Qβ buffer and extracted with 1:1 n-butanol:chloroform. The VLPs, from the aqueous layer, were purified by step sucrose gradient ultracentrifugation (10–40% w:v). VLPs were precipitated from the sucrose fraction with 20% w:v PEG8000, resuspended in Qβ buffer and dialyzed exhaustively against 50 mM NaCl, 20 mM Tris, pH 7.5. Final VLP concentration was assessed by the Pierce BCA Protein Assay kit.
Conjugation of Dylight 633 NHS to Qβ–VLPs
Conjugation of DyLight 633 NHS (N-hydroxysuccinimide) to the surface of VLPs was performed according the manufacturer protocols. VLPs were dialyzed against labeling buffer (0.05 M sodium borate, pH 8.5) and were incubated (100 μl of 1 mg ml−1) with 200 μl DyLight 633 NHS reagent (1 mg ml−1) at room temperature for 1 h. Excess DyLight 633 NHS was removed by centrifugal filtration.
In vitro encapsidation of GFP within Qβ–VLPs
DyLight 633-labeled VLPs (10 mg ml−1) were incubated in disassembly buffer (20 mM Tris-HCl, 50 mM NaCl, 6M urea, 10 mM dithiothreitol) at 4°C for 1 h and dialyzed against 10 mM acetic acid and 50 mM NaCl. Labeled CP was purified on a Sephadex G75 column and concentrated. CP was combined with a 5-fold molar excess of GFP and dialyzed against reassembling buffer (50 mM NaCl, 20 mM Tris-HCl, pH 7.5). Excess GFP and unassembled CP dimers were removed by centrifugation (100 kDa MWCO). The DyLight 633-labeled VLPs that encapsidated GFP were stored at −80°C.
Confocal microscopy
HeLa cells were seeded into glass bottom microwell dishes (MatTek, 2 × 105 cells/well) and incubated for 24 h in Dulbecco's modified Eagle's medium (DMEM) with 2.2 mg ml−1 sodium carbonate, 10% fetal bovine serum (FBS), 50 μg ml−1 gentamicin, 50 μg ml−1 penicillin and 50 μg ml−1 streptomycin at 37°C. All cells were grown and maintained at 37°C in 5% CO2. VLP633-GFPs were added to the HeLa cells to a final concentration of 45 nM. Treated cells were incubated for 0, 24 or 48 h, washed with phosphate buffered saline (PBS), stained with Hoechst for 20 min and washed again. Images were acquired on a Zeiss LSM 700 Confocal Microscope with 63x oil immersion objective and processed with Zeiss Zen software.
Fluorescence microscopy
In vivo assembled GFP-containing VLPs at 360 nM were incubated with PC3 cells for 48 h. After incubation, the cells were washed three times with PBS to remove external VLPs. Fluorescence images were captured using an Olympus U-LH100HG fluorescence microscope. Additional information on imaging is available in the Supplementary Data.
In vitro transcription of RNAiLet-7
DNA containing T7p–RNAiLet-7-T7t was amplified by PCR with primers RNAi-Fwd and RNAi–Rev (Supplementary Table S1) and used as a template for in vitro transcription. Approximately 1 μg of DNA was transcribed for 4 h at 37°C with the MEGAscript High Yield Transcription Kit (Applied Biosystems) followed by incubation with TURBO DNase for 15 min at 37°C. RNA was pelleted by ammonium acetate precipitation, washed with 80% ethanol and lyophilized. Pellets were resuspended in nuclease-free water and purified on illustra™ NAP-5™ columns.
In vitro Dicer assay
Dicer cleavage followed the manufacturer's protocol (Genlantis). Ten microliter reactions containing recombinant human Dicer enzyme (1 U) and in vitro transcribed RNAi scaffold (RNAiLet-7, 1 μg) were incubated at 37°C for 16 h. Products were analyzed by denaturing gel electrophoresis. RNA was stained with SYBR Green. Images were acquired using a Typhoon™ FLA 9500 biomolecular imager (GE Healthcare).
Northern blotting of processed RNAi scaffold
To test for in vivo Dicer processing, U87 cells were seeded in 12-well plates (∼2 × 106 cells per well) and incubated for 24 h with PBS, 1 μM VLPWT (containing endogenous E. coli RNAs) or 1 μM of VLP–RNAiLet7. Enriched small RNAs were extracted with the mirVana PARIS kit (Life Technologies), resolved by denaturing gel electrophoresis, transferred to a Hybond™-N+ membrane (GE Healthcare) by the capillary method and immobilized by UV transillumination (365 nm). Northern blotting was performed according to the manufacturer's protocols (North2South Chemiluminescent Hybridization and Detection Kit, Thermo Scientific). The membrane was probed with a biotin-labeled DNA oligonucleotide (5΄-GCA AAC TGT ACA AAC TAC TAC CTC ACC [BioTEG]-3΄, MWG Operon), which is complementary to the Let-7 siRNA.
Expression and inhibition of GFP
To express GFP in HeLa and U87 cells, 12-well plates were seeded with ∼2 × 105 cells per well and incubated for 24 h. Cells were transfected with GFP-expressing plasmid, pET28-b(+)-GFP, by addition of 1 μg of plasmid and lipofectamine. After 18 h of incubation, the cells, which were observed to express GFP, were washed three times with deionized water and given fresh media. To test for inhibition of GFP expression, VLPWT or VLP–RNAiGFP was added to the GFP-expressing cells. Cells were washed with PBS to remove residual VLPs and GFP expression was assayed by fluorescence microscopy, Western blotting and flow cytometry.
Flow cytometry
pET28-b(+)-GFP transfected HeLa cells were incubated with various concentrations of VLP-RNAiGFP (45, 90, 180, 270, 360, 540, 720 nM) or VLPWT (360, 720 nM, as negative control) for 24 h or 48 h. After washing three times with PBS, cells were detached, pelleted and resuspended in PBS. Inhibition of GFP expression was quantitated with a BD™ LSR II flow cytometer (BD Biosciences), and analyzed using FACSDiva™ software.
Pan-Ras and GFP expression by Western blotting
For Western blot analysis, U87 cells were added to 12-well plates to a density of ∼2 × 105 cells per well, 24 h before addition of 750 nM VLPWT or VLP–RNAiLet-7. After incubation for 48 h, cells were washed three times with serum-free DMEM and incubated for 24 h in DMEM, 10% FBS. U87 cells were incubated with 150 or 300 nM VLP-RNAiGFP or 300 nM VLPWT for 48 h. Inhibition efficiency of expression of Pan-Ras and GFP was quantitated by Western blotting. Chemically synthesized Let-7 siRNA without transfection agent was used as a negative control.
U87 cell cytotoxicity by VLP–RNAiLet-7
U87 cells were cultured in DMEM supplemented with 2.2 mg ml−1 sodium carbonate, 10% FBS, 50 μg ml−1 gentamicin, 50 μg ml−1 penicillin and 50 μg ml−1 streptomycin. Around 10 000 cells per well were cultured in 96-well plates for 24 h. Variable concentrations of VLP–RNAiLet-7 (62.5, 125, 250, 500, 1000 nM) were added followed by incubation for 24 h or 48 h. The culture medium was removed, and the cells were incubated in 120 μl of XTT solution for 2 h. Subsequently, 100 μl of XTT solution from each well was transferred to a 96-well counting plate. The extent of survival of U87 cells at different time points was evaluated by OD at 490 nm.
RESULTS
CP and RNAi scaffold co-express and co-assemble to form functional VLP–RNAi
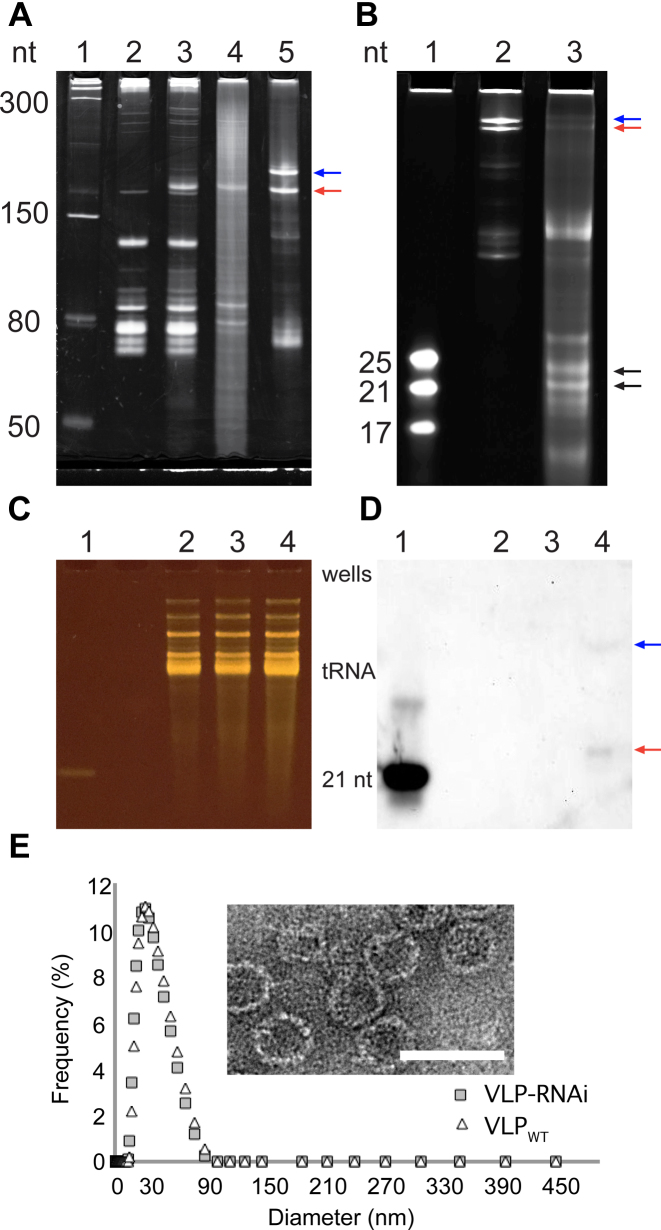
We have co-expressed Qβ CP and the RNAi scaffold in E. coli. Purified VLP–RNAi assemblies were characterized by gel electrophoresis, transmission electron microscopy and dynamic light scattering. The RNAi scaffold is overexpressed, accumulates in E. coli (Figure 2A, lane 3) and is successfully packaged inside VLPs (Figure 2A). VLPs also package endogenous RNA from E. coli during the in vivo self-assembly process. We observe some RNAi scaffold read-through product from in vitro transcription but not from in vivo expression in E. coli.
Figure 2.
Characterization of the RNAi scaffold and VLP–RNAi by electrophoresis, microscopy, Northern blotting and dynamic light scattering. (A) The in vivo transcribed RNAi scaffold was produced in E. coli. Lane 1: Size ladder. Lane 2: Total E. coli RNA before induction of expression of the RNAi scaffold and CP. Lane 3: Total E. coli RNA after induction. Lane 4: RNA extracted from purified VLP–RNAi. Lane 5: RNAi scaffold prepared by in vitro transcription. The red arrow indicates the RNAi scaffold. The blue arrow indicates RNAi scaffold extended by read-through of the terminator. (B) RNAi scaffold prepared by in vitro transcription and then processed by human Dicer. Lane 1: Size ladder. Lane 2: RNAi scaffold. Lane 3: RNAi scaffold treated with human Dicer. The black arrows indicate products of Dicer processing of the RNAi scaffold. The Dicer products are 21- to 25-mers. Panels C and D show an assay for products of in vivo processing of VLP–RNAiLet-7 in U87 cells. (C) All small RNAs visualized on a denaturing gel. (D) The same gel with the processed RNAiLet-7 scaffold visualized by Northern blotting with an antisense DNA probe. Lane 1: The 21 nt Let-7 siRNA is a positive control. Lane 2: RNA from cells incubated with PBS are a negative control. Lane 3: RNA from cells incubated with VLPWT are a negative control. Lane 4: RNA from cells incubated with VLP–RNAiLet-7. The red arrow indicates product of in vivo Dicer processing. The blue arrow indicates incomplete cleavage by Dicer. Six hundred nanograms of enriched small RNA were loaded in each lane, except for the positive control, which is 50 ng of RNA. (E) Characterization of Qβ-VLP with RNAi scaffold (gray rectangle) and wild-type Qβ-VLP (white triangle) by dynamic light scattering. Inset: Transmission electron microscope images of Qβ VLP containing the RNAi scaffold. Scale bar = 50 nm. The gels are denaturing.
Our RNAi scaffold is a substrate for human Dicer in vitro and in vivo. The scaffold is cleaved in vitro by Dicer to produce oligomers ranging from 21 to 25 nucleotides in length (Figure 2B). Treatment with VLP–RNAiLet-7 particles for 24 h causes U87 cells to produce a single small RNA product (Figure 2C and D) that contains a small RNA derived from VLP–RNAiLet-7. Control experiments show that U87 cells treated with VLPWT particles or with PBS buffer do not express the Let-7 small RNA. In contrast to production of multiple Dicer products in vitro, processing in vivo appears to produce a single Let-7 small RNA product. The results indicate that Dicer cleaves RNAiLet-7 scaffold in vivo generating a small RNA.
Packaging of the RNAi scaffold in the capsid does not change the morphology of the VLP (Figure 2E). By light scattering and transmission microscopy, VLP–RNAiLet-7 particles are indistinguishable from previous Qβ VLPs (9,22).
To further confirm that the RNAi scaffold assembles with Qβ CP in vivo to form functional VLPs, we compared the activities of scaffold-containing and scaffold-deficient VLPs. RNA sequences inserted into the scaffold were directed against several well-characterized targets whose expression levels can be readily monitored. The targets included GFP and oncogenic Pan-Ras proteins. VLP–RNAiGFP (Figure 1A) is designed to cause degradation of the mRNA of GFP. VLP–RNAiLet-7 (Figure 1B) is designed to decrease Pan-Ras expression at the level of translation (23). Both of these VLP–RNAi are active against their targets. Control experiments confirm that neither VLPWT (containing endogenous E. coli RNA) nor VLP–RNAi containing off-target RNAi are active. The results are consistent with VLP–RNAi activity against specified RNAi targets.
VLPs are taken up by human cells
Finn and coworkers have previously shown that Qβ VLPs targeted by attachment of epidermal growth factor are rapidly internalized in epidermoid carcinoma cell line A431 by receptor-mediated endocytosis (24). During those relatively short incubations, undecorated Qβ VLPs associate weakly with A431 cells but are not internalized. Here, we extend incubation times to promote internalization of untargeted Qβ VLPs.
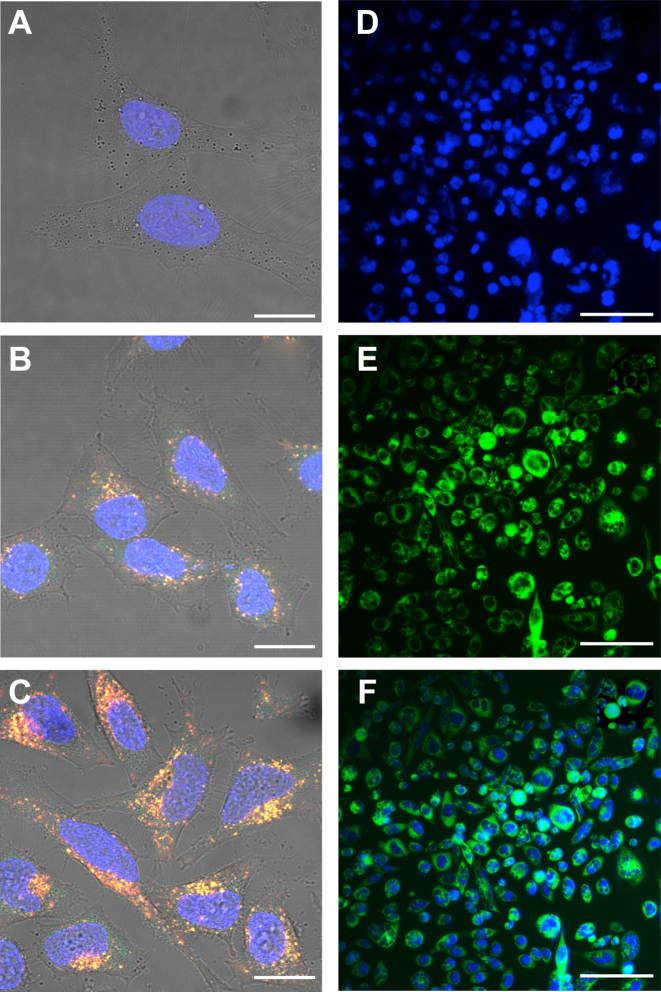
The results show that untargeted Qβ VLPs, like untargeted MS2 VLPs (25), are internalized by HeLa and PC3 cells over 24 h to 48 h (Figure 3). Dual-color VLP633-GFP can be monitored for cellular internalization and distribution, particle integrity and cargo release. VLP633-GFP, with GFP confined in the VLP interior and DyLight 633 attached to the exterior, fluoresces at two different wavelengths (Figure 3). Yellow signal indicates intact VLP633-GFP particles, that emit both green (GFP) and red (DyLight 633). Green signal indicates GFP that has been released by the VLP, presumably upon VLP dissociation. Red signal indicates CP in isolation of GFP cargo.
Figure 3.
The internalization of dual-color VLP633-GFP (panels A–C, confocal microscopy) and VLP–GFP (panels D–F, fluorescence microscopy) by HeLa and PC3 cells. HeLa cells were incubated with VLP633-GFP (100 nM) for (A) 0 h, (B) 24 h and (C) 48 h. VLP633 are red. VLP–GFP are green. VLP633–GFP are yellow due to the co-localization of red and green. Green color at 24 h indicates that the VLPs have opened and released GFP. PC3 cells were incubated with VLP–GFP (360 nM) for 48 h show (D) nuclei, (E) GFP-containing VLPs and (F) merged fluorescent signals. Scale bar (A–C) = 20 μm or (D–F) = 300 μm. Nuclei are stained blue with DAPI.
The results show that VLP633-GFP particles are internalized by HeLa cells. The internalization of VLP633–GFP after 48 h of incubation is greater than after 24 h (Figure 3B and C). Yellow fluorescent particles within the cytoplasm partially resolve into points of distinct green and red emission, indicating that the GFP cargo is released. The particles appear to be excluded from the nucleus. Control experiments show that VLP internalization is not affected by attachment of fluorescent probes on the exterior (Figure 3B and C and Supplementary Figure S1) or by packaging of GFP into the interior of VLPs (Figure 3B and C, E and F). VLP-GFP are internalized by PC3 cells to roughly the same extent as VLP633–GFP in HeLa cells.
The RNAi scaffold is functional upon delivery by VLPs
VLP–RNAiGFP inhibits GFP expression in HeLa and U87 cells. A 22 base pair duplex containing a sequence from GFP mRNA was incorporated into the RNAi scaffold to form RNAiGFP (Figure 1A). RNAiGFP was co-expressed along with CP in E. coli to produce VLP–RNAiGFP. Purified VLP–RNAiGFP was incubated with U87 and HeLa cells expressing GFP.
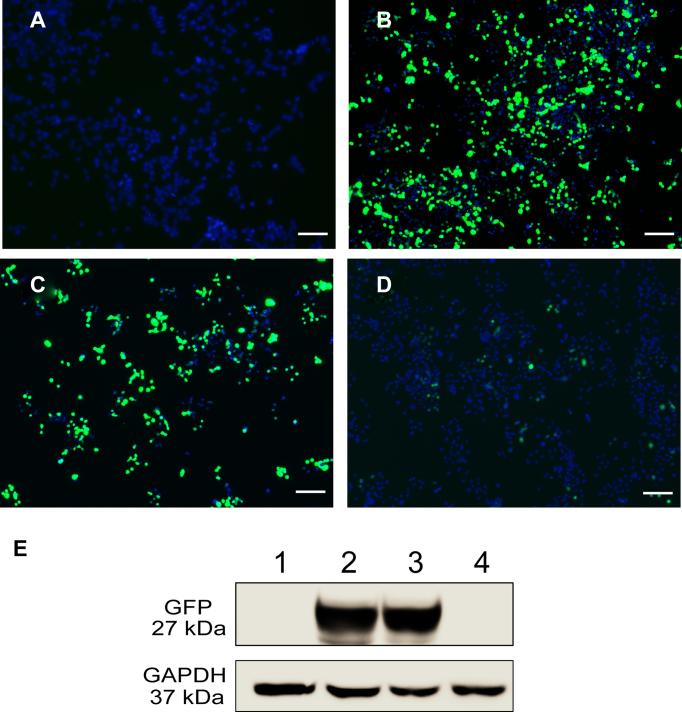
GFP expression is inhibited by VLP–RNAiGFP. Incubation of the transfected U87 cells with VLP–RNAiGFP decreases expression of GFP, as shown by fluorescence microscopy (Figure 4A–D) and Western blot analysis (Figure 4E). The results show incubation in 300 nM of VLP–RNAiGFP for 48 h efficiently inhibits expression of GFP (Figure 4D and E). High concentrations of VLPWT do not affect GFP expression. These experiments confirm that inhibition is caused by VLP–RNAiGFP, but not by control VLPs.
Figure 4.
Suppression of GFP expression by VLP–RNAiGFP in U87 cells. Cells were treated with (A) PBS, (B) a plasmid expressing GFP, (C) a plasmid expressing GFP followed, after 18 h, by treatment with 300 nM VLPWT, (D) a plasmid expressing GFP followed, after 18 h, by treatment with 300 nM of VLP–RNAiGFP. (E) Western blot analysis of GFP expression. Lane 1 shows the GFP expression of the cells corresponding to panel (A). Lane 2 shows expression of cells in panel (B). Lane 3 shows expression of cells in panel C. Lane 4 shows expression of cells in panel D. Green indicates GFP expression. Blue indicates nuclei. Images were obtained 48 h after transfection. Scale bar = 200 μm.
VLP–RNAiGFP shows similar inhibition of GFP in HeLa cells (Supplementary Figure S4). VLP–RNAiGFP induces dose-dependent and time-dependent inhibition of GFP expression in HeLa cells. Levels of inhibition of GFP expression in HeLa cells were analyzed quantitatively by flow cytometry (Supplementary Figure S5). For cells transfected with GFP-expressing plasmid in the absence of VLP–RNAiGFP, 23% of cells fluoresce after 24 h and 21% fluoresce after 48 h. In transfected cells treated with 45 nM of VLP–RNAiGFP, 15% fluoresce after 24 h and 10% fluoresce after 48 h. Fluorescence in 50% of the cells expressing GFP is inhibited by 360 nM of VLP–RNAiGFP after 48 h.
VLP–RNAiLet-7 targets the 3΄UTR of Ras mRNA in U87 cells
We constructed a second RNAi scaffold containing a sequence from Let-7 miRNA (Figure 1B). The RNAiLet-7 was transcribed and assembled in E. coli with CP to form VLP–RNAiLet-7.
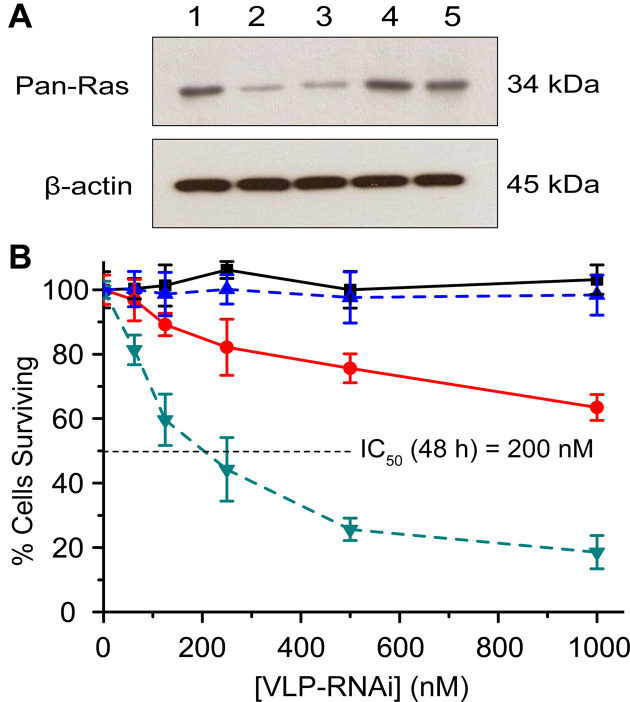
Pan-Ras expression is suppressed by VLP–RNAiLet-7 (Figure 5). U87 cells were treated with purified VLP–RNAiLet-7 and assayed for Pan-Ras expression. Expression levels of Pan-Ras were significantly reduced (>70%) by incubation in 750 nM of VLP–RNAiLet-7 for 48 h (Figure 5A, lane 1 and 3). The observed decrease in expression is similar to that observed after treatment with RNAiLet-7 and a transfection agent (Figure 5A, lane 2), as demonstrated previously (26). The results indicate that VLP-RNAiLet-7 is taken up by U87 cells and that functional RNAiLet-7 is released from the VLP to trigger RNAi. We have also performed a series of control experiments that minimize the chance that RNAi is caused by off-target effects. Pan-Ras expression levels from U87 cells are unaltered by VLPWT or by VLP–RNAiGFP (Figure 5A, lane 4 and 5). The results of these control experiments are consistent with a model in which activity of VLP–RNAiLet-7 arises from the expected mechanism.
Figure 5.
VLP–RNAiLet-7 inhibits Pan-Ras expression and demonstrates in vitro cytotoxicity in U87 cells. (A) Western blot analysis of Pan-Ras levels in U87 cells incubated for 48 h under various conditions. Lane 1: no treatment (negative control). Lane 2: Let-7 siRNA delivered by transfection with lipofectamine (positive control). Lane 3: VLP-RNAiLet-7. Lane 4: VLPWT (negative control). Lane 5: VLP-RNAiGFP (negative control). (B) Cell viability U87 cells treated with VLP–RNAiLet-7 and VLP–RNAiGFP. Cell viability was determined after treatment with various concentrations of VLP–RNAiLet-7 for 24 h ( ) and 48 h (
) and 48 h ( ) and VLP–RNAiGFP for 24 h (▪) and 48 h (
) and VLP–RNAiGFP for 24 h (▪) and 48 h ( ). The data are reported as percentage survival. Values are expressed as mean ± SD (n = 8).
). The data are reported as percentage survival. Values are expressed as mean ± SD (n = 8).
VLP–RNAiLet-7 attenuates U87 cell proliferation and promotes cell death
Cell viability decreases in a time and dose-dependent fashion consistent with delivery of RNAiLet-7 (Figure 5B). We confirm previous work (26) showing that chemically synthesized Let-7 siRNA alone was not functional in the absence of a transfection reagent.
We investigated the viability of U87 cells upon treatment with VLP–RNAiLet7 or VLP–RNAiGFP (control) for 24 h and 48 h. VLP–RNAiLet-7 promotes U87 cell death in a dose-dependent and time-dependent manner. The concentration required for 50% inhibition of growth (IC50) for VLP–RNAiLet-7 is around 1 μM after 24 h of incubation and is 200 nM when the incubation period is extended to 48 h. The time-dependence presumably arises in part from a lag in downstream effects of Ras expression. Control experiments confirm that cell death is caused by the VLP–RNAiLet-7. VLP–RNAiGFP does not promote mortality of U87 cells (Figure 5B).
DISCUSSION
We have designed a general utility RNAi scaffold that spontaneously folds and co-assembles with CP to form functional VLP–RNAi. The scaffold is a single RNA molecule containing the Qβ hairpin on the 5΄ end, a carrier strand, the miR-30 stem loop and a silencing strand. The RNAi scaffold is designed to promote unimolecular pairing of the carrier and silencing strands, and contains a 3΄ terminus appropriately placed to direct Dicer to release the single-stranded silencing strand. This RNAi scaffold could contain a broad variety of silencing sequences including miRNAs and small interfering RNAs. As a single RNA molecule, the scaffold folds efficiently and readily assembles with CP to form Qβ VLPs containing functional RNA. Advantages of the VLP–RNAi system we describe here include ease of vector construction, assembly and packaging, purification, targeting and delivery and in effectiveness of regulating gene expression.
Previous work identified a variant of human miR-30 that maximizes production of mature miRNA, and demonstrated function of RNAi in human cells by substituting the stems of miR-30 with artificial targeting sequences (13). Ramratnam incorporated an siRNA sequence targeting the mRNA of HIV-1 transactivator protein tat into the miR-30 stem (14). The resulting artificial miRNA is 80% more potent in reducing target p24 antigen production than conventional tat shRNA. The enhancement of gene silencing may be due in part to high stability of the miR-30 stem-loop (27). The miR-30 stem-loop provides a tool for intramolecular folding of shRNA, which is rapid and efficient, whereas formation of an intermolecular duplex by RNAs in unlinked strands is less efficient (28). We placed the Qβ hairpin on the 5΄ end of scaffold, following Stockley, who synthesized a bimolecular aptamer–siRNA (25). Our design of the RNAi scaffold presents a free 3΄ end for recognition by the PAZ domain of Dicer (19,29) and efficient transfer of the processed siRNA to Argonaute (29). To facilitate one-pot VLP–RNAi assembly in E. coli and to maximize activity, we co-express CP and cargo RNA, which contains the packaging aptamer (the Qβ hairpin) (25), and miR-30, which stabilizes the folded scaffold (13). The RNAi scaffold and Qβ CP were placed under control of T7 RNA polymerase promoters. The Qβ CP is transcribed and translated, and packages the transcribed RNAi scaffold, spontaneously forming functional VLPs in vivo. The assays and controls indicate that the RNAi scaffold is packaged within the VLPs.
We have incorporated several silencing sequences into the RNAi scaffold (Figure 1) and demonstrated function of these VLP–RNAi in a variety of human cell lines. Treatment of cells with VLP–RNAi particles leads to production of small RNAs. Expression of the Let-7 small RNA is observed by Northern blotting when Let-7-deficient U87 cells are treated with VLP–RNAiLet-7 particles (Figure 2). Let-7 small RNA is not observed in control treatments. Several independent experiments suggest that our RNAi scaffold is a substrate of Dicer, and that the RNA interference results from processed RNAi, not from full-length RNAi scaffold or off-target effects. The VLP–RNAi's inhibit expression of a reporter gene encoding GFP or expression of the Pan-Ras oncogene. GFP expression in HeLa and U87 cells is inhibited by VLP–RNAiGFP (Figure 4). The VLP-RNAiGFP induces dose-dependent and time-dependent inhibition of GFP expression. The effect is specific: high concentrations of VLPs containing other RNAs do not decrease the level of GFP expression (Figure 4D, G, Supplementary Figure S5).
Pan-Ras expression is suppressed by VLP–RNAiLet-7 (Figure 5). VLP–RNAiLet-7 attenuates U87 cell proliferation, promotes death in cell culture and down regulates Pan-Ras. Let-7 miRNAs function as tumor suppressors by decreasing expression of the Ras oncogene family at the level of translation (23,30), reducing cell proliferation and invasion. Down-regulation of Let-7 has been observed in lung cancer, colon cancer and melanoma and is correlated with poor patient survival (31–34). Restoration of Let-7 miRNA may be a useful cancer therapy (34). Ras proteins (K-Ras, N-Ras and Pan-Ras) are known to be up-regulated in these cancers (35). Human Ras expression is regulated by Let-7 miRNA by binding to the 3΄ UTR of Ras mRNA (23). In human brain tumor cells, overexpression of Let-7 miRNA leads to a decrease in Ras protein expression, inhibition of cell proliferation and reduced tumor growth, without harming normal astrocytes (36). Our control experiments confirm that Qβ VLPs containing non-specific RNAs are non-functional and benign. VLP–RNAi is active only when the RNAi scaffold contains functional RNAi sequences. In sum, the results show that the in vivo packaged VLP–RNAi containing the RNAi scaffold is efficient in inhibiting gene regulation and in promoting tumor cell death.
The combined results demonstrate that undecorated Qβ VLPs enter cell lines in the absence of transfection reagent and surface decoration. Our results, along with earlier studies, demonstrate that extended incubation times allow internalization of undecorated MS2 (25) and Qβ VLPs. These results extend prior work in which surface decoration facilitates uptake of MS2 VLPs and Qβ VLPs in mammalian cells (11,37–39). Previously, several groups showed that when surfaces of Qβ and the closely related MS2 VLPs are decorated with appropriate targeting ligands, the VLPs are quickly internalized by human cells via receptor-mediated endocytosis (11,24,37,38,40). Stockley showed that undecorated MS2 VLPs are also internalized, but more slowly (25). Once internalized, VLPs disassemble and release their cargo, which is functional in vivo (11,25,41).
Control and functional assays confirm that VLPs release their cargo intracellularly (Figures 4 and 5, Supplementary Figure S5). Previous work has reported similar cytoplasmic release from bacteriophage MS2 VLPs (11,25,37,39,42). The mechanism by which VLP-encapsidated RNA escapes from the endosome and enters cytoplasm remains unclear. Our results are consistent with previous work which show that single-stranded RNA animal viruses can pass their viral genomes to the cytoplasm from the endosomes (43).
The release mechanism of RNA from the VLP is most likely a combination of disulfide bond breakage and destabilization by low pH. The low pH in late endosomes and lysosomes may dissociate the coat proteins of the Qβ-VLPs. A combination of the above phenomena may result in the escape of encapsidated nucleic acid.
The efficiency of VLPs for delivering cargos to human cells depends on targeting, VLP uptake and endosome escape. Finn et al. and Stockley et al., both showed that covalent attachment of targeting peptides facilitates VLP uptake in human cell lines in short time periods at less than 50 nM (24,25,39). In addition, covalent attachment of a histidine-rich fusogenic peptide on the exterior surface VLPs can promote endosomal escape of internalized VLPs (11). This histidine-rich peptide acts by a ‘proton sponge’ effect. Histidine can sequester protons and buffer the acidification in the endosomes. The buffering induces osmotic swelling and bursting of endosomes (44,45). Impressively, the working concentration of siRNA-loaded VLPs with covalent attachment of both targeting and fusogenic peptides is less than 150 pM (11). Comparison of our work with the results obtained by several other groups who have decorated the MS2 or Qβ VLP external surface with different targeting ligands, suggests that undecorated VLP–RNAi requires higher concentrations and longer incubation times. Increasing the efficiency of VLP uptake and endosomal escape are future challenges for our one-step VLP–RNAi assembly method.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr M.G. Finn and Dr Mark Prausnitz for helpful discussions, and Justin Williams and Dr Brande Jones for technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Georgia Research Alliance [GRA.VL15.B11]; NASA [NNA09DA78A]; Ministry of Science and Technology of the Republic of China [MOST105-2314-B-110-001]; National Science Foundation Graduate Research Fellowship Program [DGE-1650044 to L.M.G.R. and S.H.]; Alfred P. Sloan & Georgia Tech UCEM Minority Doctoral Fellowship. Funding for open access charge: NASA [NNX16AJ29G].
Conflict of interest statement. None declared.
REFERENCES
- 1. Drude I., Dombos V., Vauleon S., Muller S.. Drugs made of RNA: development and application of engineered RNAs for gene therapy. Mini. Rev. Med. Chem. 2007; 7:912–931. [DOI] [PubMed] [Google Scholar]
- 2. Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C.. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis Elegans. Nature. 1998; 391:806–811. [DOI] [PubMed] [Google Scholar]
- 3. Meister G., Tuschl T.. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004; 431:343–349. [DOI] [PubMed] [Google Scholar]
- 4. Golmohammadi R., Fridborg K., Bundule M., Valegard K., Liljas L.. The crystal structure of Bacteriophage Q Beta at 3.5 angstrom resolution. Structure. 1996; 4:543–554. [DOI] [PubMed] [Google Scholar]
- 5. Machida K., Imataka H.. Production methods for viral particles. Biotechnol. Lett. 2015; 37:753–760. [DOI] [PubMed] [Google Scholar]
- 6. Bajaj S., Banerjee M.. Engineering virus capsids into biomedical delivery vehicles: structural engineering problems in nanoscale. J. Biomed. Nanotechnol. 2015; 11:53–69. [DOI] [PubMed] [Google Scholar]
- 7. Witherell G.W., Uhlenbeck O.C.. Specific RNA binding by Q beta coat protein. Biochemistry. 1989; 28:71–76. [DOI] [PubMed] [Google Scholar]
- 8. Ashcroft A.E., Lago H., Macedo J., Horn W.T., Stonehouse N.J., Stockley P.G.. Engineering thermal stability in RNA phage capsids via disulphide bonds. J. Nanosci. Nanotechnol. 2005; 5:2034–2041. [DOI] [PubMed] [Google Scholar]
- 9. Fiedler J.D., Brown S.D., Lau J.L., Finn M.G.. RNA-directed packaging of enzymes within virus-like particles. Angew. Chem. Int. Ed. 2010; 49:9648–9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lau J.L., Baksh M.M., Fiedler J.D., Brown S.D., Kussrow A., Bornhop D.J., Ordoukhanian P., Finn M.G.. Evolution and protein packaging of small-molecule RNA aptamers. ACS Nano. 2011; 5:7722–7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ashley C.E., Carnes E.C., Phillips G.K., Durfee P.N., Buley M.D., Lino C.A., Padilla D.P., Phillips B., Carter M.B., Willman C.L. et al. . Cell-specific delivery of diverse cargos by bacteriophage Ms2 virus-like particles. ACS Nano. 2011; 5:5729–5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Borodavka A., Tuma R., Stockley P.G.. Evidence that viral RNAs have evolved for efficient, two-stage packaging. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:15769–15774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zeng Y., Wagner E.J., Cullen B.R.. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell. 2002; 9:1327–1333. [DOI] [PubMed] [Google Scholar]
- 14. Boden D., Pusch O., Silbermann R., Lee F., Tucker L., Ramratnam B.. Enhanced gene silencing of Hiv-1 specific sirna using microrna designed hairpins. Nucleic Acids Res. 2004; 32:1154–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Silva J.M., Li M.Z., Chang K., Ge W., Golding M.C., Rickles R.J., Siolas D., Hu G., Paddison P.J., Schlabach M.R. et al. . Second-generation shrna libraries covering the mouse and human genomes. Nat. Genet. 2005; 37:1281–1288. [DOI] [PubMed] [Google Scholar]
- 16. Puglisi J.D., Tinoco I. Jr. Absorbance melting curves of RNA. Methods Enzymol. 1989; 180:304–325. [DOI] [PubMed] [Google Scholar]
- 17. Marky L.A., Breslauer K.J.. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987; 26:1601–1620. [DOI] [PubMed] [Google Scholar]
- 18. MacRae I.J., Zhou K., Li F., Repic A., Brooks A.N., Cande W.Z., Adams P.D., Doudna J.A.. Structural basis for double-stranded RNA processing by dicer. Science. 2006; 311:195–198. [DOI] [PubMed] [Google Scholar]
- 19. MacRae I.J., Zhou K., Doudna J.A.. Structural determinants of RNA recognition and cleavage by dicer. Nat. Struct. Mol. Biol. 2007; 14:934–940. [DOI] [PubMed] [Google Scholar]
- 20. Bowman J.C., Azizi B., Lenz T.K., Roy P., Williams L.D.. Conn GL. Recombinant and in Vitro RNA Synthesis: Methods and Protocols, Methods in Molecular Biology. 2012; 941, NY: Springer Science LLC; 19–41. [DOI] [PubMed] [Google Scholar]
- 21. Studier F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005; 41:207–234. [DOI] [PubMed] [Google Scholar]
- 22. Kozlovska T.M., Cielēns I., Dreilinņa D., Dišlers A., Baumanis V., Ose V., Pumpēns P.. Recombinant RNA phage Qβ capsid particles synthesized and self-assembled in Escherichia coli. Gene. 1993; 137:133–137. [DOI] [PubMed] [Google Scholar]
- 23. Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F.J.. Ras is regulated by the Let-7 microrna family. Cell. 2005; 120:635–647. [DOI] [PubMed] [Google Scholar]
- 24. Pokorski J.K., Hovlid M.L., Finn M.G.. Cell targeting with hybrid Q beta virus-like particles displaying epidermal growth factor. ChemBioChem. 2011; 12:2441–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Galaway F.A., Stockley P.G.. Ms2 viruslike particles: a robust, semisynthetic targeted drug delivery platform. Mol. Pharm. 2013; 10:59–68. [DOI] [PubMed] [Google Scholar]
- 26. Yang H.-W., Huang C.-Y., Lin C.-W., Liu H.-L., Huang C.-W., Liao S.-S., Chen P.-Y., Lu Y.-J., Wei K.-C., Ma C.-C.. Gadolinium-functionalized nanographene oxide for combined drug and microrna delivery and magnetic resonance imaging. Biomaterials. 2014; 35:6534–6542. [DOI] [PubMed] [Google Scholar]
- 27. Shan Z., Lin Q., Deng C., Li X., Huang W., Tan H., Fu Y., Yang M., Yu X.-Y.. An efficient method to enhance gene silencing by using precursor microrna designed small hairpin RNAs. Mol. Biol. Rep. 2009; 36:1483–1489. [DOI] [PubMed] [Google Scholar]
- 28. Yu J.-Y., DeRuiter S.L., Turner D.L.. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:6047–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Macrae I., Li F., Zhou K., Cande W., Doudna J.. Cold Spring Harbor Symp. Quant. Biol. 2006; 71, NY: Cold Spring Harbor Laboratory Press; 73–80. [DOI] [PubMed] [Google Scholar]
- 30. Roush S., Slack F.J.. The Let-7 family of micrornas. Trends Cell Biol. 2008; 18:505–516. [DOI] [PubMed] [Google Scholar]
- 31. Akao Y., Nakagawa Y., Naoe T.. Let-7 microrna functions as a potential growth suppressor in human colon cancer cells. Biol. Pharm. Bull. 2006; 29:903–906. [DOI] [PubMed] [Google Scholar]
- 32. Schultz J., Lorenz P., Gross G., Ibrahim S., Kunz M.. Microrna Let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with Anchorage-independent growth. Cell Res. 2008; 18:549–557. [DOI] [PubMed] [Google Scholar]
- 33. Trang P., Medina P.P., Wiggins J.F., Ruffino L., Kelnar K., Omotola M., Homer R., Brown D., Bader A.G., Weidhaas J.B. et al. . Regression of murine lung tumors by the Let-7 Microrna. Oncogene. 2010; 29:1580–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y. et al. . Reduced expression of the Let-7 Micrornas in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004; 64:3753–3756. [DOI] [PubMed] [Google Scholar]
- 35. Yu F., Yao H., Zhu P., Zhang X., Pan Q., Gong C., Huang Y., Hu X., Su F., Lieberman J. et al. . Let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007; 131:1109–1123. [DOI] [PubMed] [Google Scholar]
- 36. Lee S.-T., Chu K., Oh H.-J., Im W.-S., Lim J.-Y., Kim S.-K., Park C.-K., Jung K.-H., Lee S.K., Kim M. et al. . Let-7 microrna inhibits the proliferation of human glioblastoma cells. J. Neurooncol. 2011; 102:19–24. [DOI] [PubMed] [Google Scholar]
- 37. Pan Y., Zhang Y., Jia T., Zhang K., Li J., Wang L.. Development of a microrna delivery system based on bacteriophage Ms2 virus-like particles. FEBS J. 2012; 279:1198–1208. [DOI] [PubMed] [Google Scholar]
- 38. Wei B., Wei Y., Zhang K., Wang J., Xu R., Zhan S., Lin G., Wang W., Liu M., Wang L.. Development of an antisense RNA delivery system using conjugates of the Ms2 bacteriophage capsids and Hiv-1 tat cell penetrating peptide. Biomed. Pharmacother. 2009; 63:313–318. [DOI] [PubMed] [Google Scholar]
- 39. Wu M., Brown W.L., Stockley P.G.. Cell-specific delivery of bacteriophage-encapsidated ricin a chain. Bioconjug. Chem. 1995; 6:587–595. [DOI] [PubMed] [Google Scholar]
- 40. Wu M., Sherwin T., Brown W.L., Stockley P.G.. Delivery of antisense oligonucleotides to leukemia cells by RNA bacteriophage capsids. Nanomedicine. 2005; 1:67–76. [DOI] [PubMed] [Google Scholar]
- 41. Hovlid M.L., Lau J.L., Breitenkamp K., Higginson C.J., Laufer B., Manchester M., Finn M.G.. Encapsidated atom-transfer radical polymerization in Q beta virus-like nanoparticles. ACS Nano. 2014; 8:8003–8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ida R., Wu G.. Direct NMR detection of alkali metal ions bound to G-quadruplex DNA. J. Am. Chem. Soc. 2008; 130:3590–3602. [DOI] [PubMed] [Google Scholar]
- 43. Tuthill T.J., Bubeck D., Rowlands D.J., Hogle J.M.. Characterization of early steps in the poliovirus infection process: receptor-decorated liposomes induce conversion of the virus to membrane-anchored entry-intermediate particles. J. Virol. 2006; 80:172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nel A.E., Mädler L., Velegol D., Xia T., Hoek E.M., Somasundaran P., Klaessig F., Castranova V., Thompson M.. Understanding biophysicochemical interactions at the Nano–Bio interface. Nat. Mater. 2009; 8:543–557. [DOI] [PubMed] [Google Scholar]
- 45. Lächelt U., Kos P., Mickler F.M., Herrmann A., Salcher E.E., Rödl W., Badgujar N., Bräuchle C., Wagner E.. Fine-tuning of proton sponges by precise Diaminoethanes and histidines in pdna polyplexes. Nanomedicine. 2014; 10:35–44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.