Abstract
The SYSTERS project aims to provide a meaningful partitioning of the whole protein sequence space by a fully automatic procedure. A refined two-step algorithm assigns each protein to a family and a superfamily. The sequence data underlying SYSTERS release 4 now comprise several protein sequence databases derived from completely sequenced genomes (ENSEMBL, TAIR, SGD and GeneDB), in addition to the comprehensive Swiss-Prot/TrEMBL databases. The SYSTERS web server (http://systers.molgen.mpg.de) provides access to 158 153 SYSTERS protein families. To augment the automatically derived results, information from external databases like Pfam and Gene Ontology are added to the web server. Furthermore, users can retrieve pre-processed analyses of families like multiple alignments and phylogenetic trees. New query options comprise a batch retrieval tool for functional inference about families based on automatic keyword extraction from sequence annotations. A new access point, PhyloMatrix, allows the retrieval of phylogenetic profiles of SYSTERS families across organisms with completely sequenced genomes.
INTRODUCTION
The principal goal of the SYSTERS project is to automatically partition all the available protein space. Because the fully automated classification scheme does not rely on interventions and updates by experts, the SYSTERS approach is complementary to expert-curated protein domain or protein family classification schemes like Pfam (1), SMART (2) or PROSITE (3). The SYSTERS database is derived from rigorous all-against-all Smith–Waterman searches (4). The resulting pairwise sequence similarities are used in a refined two-step clustering approach that assigns each protein to a family and a superfamily (A. Krause, J. Stoye and M. Vingron, submitted for publication).
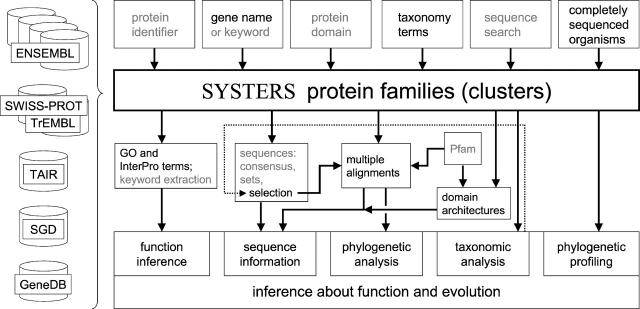
The SYSTERS web resource comprises a multitude of query access points, data retrieval options, pre-processed sequence analyses of individual families and comprehensive views on multiple families (Figure 1). The automatically derived protein families are augmented with expert-curated biological information from various resources. For the functional characterization of each cluster, keywords are extracted from annotations of source sequence databases and are assigned to each family. In SYSTERS release 4, Pfam domain assignments to sequences of Swiss-Prot/TrEMBL (5) help to visualize the domain architecture of a protein and to identify differences in domain composition within a protein family. A special focus of SYSTERS is to support phylogenetic studies of protein families. Sequences of SYSTERS families can be selected and downloaded in multiple ways. The users are offered pre-calculated multiple alignments and phylogenetic trees that can serve as a starting point for their own focused analyses.
Figure 1.
Information flow in SYSTERS. Left-hand side: publicly accessible protein sequence resources: input to SYSTERS. Four information levels in rows: top row, possible queries to the web server; second row, the SYSTERS database; third row, output features; and bottom row: analysis options. In black: new in SYSTERS release 4.
In this paper, we will describe the differences of SYSTERS release 4 compared to previous releases and highlight the recent developments of tools to access and view information on SYSTERS protein families and superfamilies.
INPUT DATA AND CLUSTERING RESULTS OF SYSTERS RELEASE 4
The underlying protein data for SYSTERS release 4 comprise more than 1.1 million sequences. The Swiss-Prot/TrEMBL database content was extended by several protein data sources with information from completely sequenced genomes (Figure 1): Saccharomyces cerevisiae (6), Schizosaccharomyces pombe (7), Arabidopsis thaliana (8), Drosophila melanogaster, Anopheles gambiae, Caenorhabditis elegans, Caenorhabditis briggsae, Takifugu rubripes, Mus musculus and Homo sapiens (9). After removal of redundant sequences, the results of more than 1011 pairwise Smith–Waterman comparisons were fed into the clustering procedure (Table 1).
Table 1. Characteristic data of SYSTERS all-against-all search and clustering procedure for the number of sequences obtained from source databases and used in the two pre-processing steps are given.
| Input | |
|---|---|
| Numbers of sequences | Sequence quality; usage in SYSTERS procedure |
| 1 168 498 | Redundant sequences from all source databases (for details see text) |
| −139 843 | Duplicated sequences: 100% identity, full length of both sequences |
| −59 076 | Included sequences: 100% identity, full length of shorter sequence |
| 969 579 | Non-redundant sequence set, used in Smith–Waterman all-against-all searches |
| −423 041 | Fragmental sequences: ≥80% identity and ≥80% of length of shorter sequence |
| 546 538 | Non-redundant sequence set, used in clustering procedure |
The non-redundant sequence set results from the subtraction of identical and fragmental sequences.
The resulting numbers of SYSTERS superfamilies and protein families are presented in Table 2. Only 11.8% of sequences remained as singletons. The majority (74%) of multi-sequence families are ‘perfect’, meaning that all sequences in a family match with each other. Only 6.5% of the families are classified as ‘overlapping’: these families might harbour protein pairs that do not share homologous regions, but are linked indirectly via an intermediate protein that has distinct homologous regions in common with both. The protein family size is power-law-like distributed (10). There are few families with many sequences and many families with only a few sequences. This result complements earlier findings (11,12) on the mode of protein evolution.
Table 2. Characteristic data of SYSTERS all-against-all search and clustering procedure for the two clustering steps result in SYSTERS superfamiliesand families.
| Output superfamilies | |||||||
|---|---|---|---|---|---|---|---|
| Number of superfamilies | Superfamily type | Number of sequences | Number of output families to protein families | Cluster graph type | Number of sequences | ||
| Non-redundant | Redundant | Non-redundant | Redundant | ||||
| 37 488 | Multi sequence | 436 230 | 1 030 969 | 35 345 | Perfect | 134 191 | 238 717 |
| 110 308 | Single sequence | 110 308 | 137 529 | 9355 | Nested | 127 036 | 265 357 |
| 147 796 | Superfamilies | 546 538 | 1 168 498 | 3131 | Overlapping | 174 989 | 526 877 |
| 110 322 | Single | 110 322 | 137 547 | ||||
| 158 153 | Protein families | 546 538 | 1 168 498 | ||||
Protein families are categorized according to the intra-family relationships between proteins: in perfect clusters all sequences match each other, in nested clusters at least one sequence matches all others, in overlapping clusters there is no sequence matching with all others.
NEW FEATURES AND SERVICES
Information characterizing a SYSTERS family
For each protein family, SYSTERS provides a comprehensive overview of its member proteins and their annotations. On the entry page, users have access to more detailed information on protein annotations, sequences, multiple alignments, phylogenetic analyses, protein domains, taxonomic distribution and gene structure-related data (Figure 1).
In addition to pre-calculated multiple alignments by MView (13), the SYSTERS web server now offers multiple alignments and UPGMA trees generated using DIALIGN (14). The DIALIGN alignment incorporates all sequences in full length, colour-coded information on alignment quality and Pfam domain positions. From MView alignments we derived consensus sequences for each family. The database of consensus sequences can be queried by the user via BLAST (15) interface. SYSTERS provides a new wizard-like tool that allows a flexible selection of user-defined sequences. In this way, users can compile sequences of different SYSTERS families or user-supplied sequences. Subsequently, multiple alignment and UPGMA trees can be constructed using DIALIGN and viewed online.
We extracted frequently occurring keywords from all original protein annotations of a SYSTERS family. The keyword list represents a succinct functional description of a family, thus helping to infer functions of hypothetical proteins. We integrated further Swiss-Prot/TrEMBL annotations such as Gene Ontology (16) terms, InterPro (17) terms and Enzyme Commission (EC) numbers (18) that support function inference. A new batch-retrieval tool allows fast annotation of large protein sets. Users can supply a list of sequence database identifiers, e.g. from SWISS-PROT, and are offered to download a list of associated SYSTERS protein family IDs, extracted keywords and GO terms.
Protein domain positions of all Swiss-Prot/TrEMBL proteins as annotated in the Pfam database are now integrated into SYSTERS. Domain architectures of all proteins in a SYSTERS family are visualized and can easily be compared. This allows to pinpoint differences in domain architectures within the family that might indicate lineage-specific domain acquisitions or losses.
Taxonomy and phylogenetic profiling
We have integrated the taxonomic system as maintained by the NCBI (19) into SYSTERS and offer to visualize the distribution of protein family members over the taxonomic tree. This now allows users to select sequences of a subfamily specified by internal nodes of the taxonomic tree for further analysis. Additionally, it is possible to select all SYSTERS protein families that have (at least one/exclusively) member protein(s) within a user-defined taxonomic range.
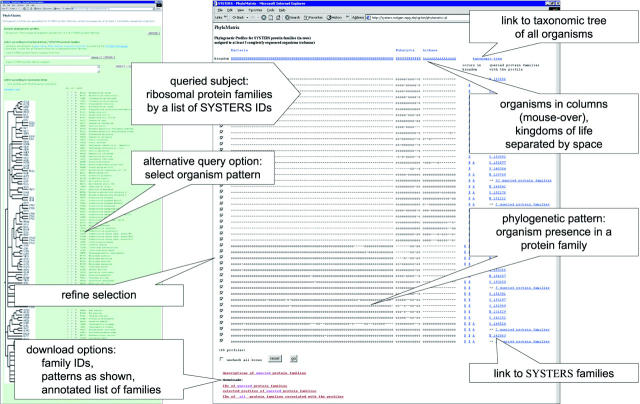
A special taxonomic view of a protein family focuses on the presence/absence patterns of member proteins across organisms, also known as phylogenetic profiles (20). Similar profiles often point to similar cellular function or a physical interaction. We set up PhyloMatrix, an extension of SYSTERS to phylogenetic profiling. PhyloMatrix profiles are based on the representation of 106 completely sequenced organisms in SYSTERS protein families, 78 bacteria, 12 eukaryota and 16 archaea. We found 7563 different profiles for 19 374 protein families under the constraint that at least three organisms be present in a family. Users can define a list of protein family IDs to retrieve a set of profiles. Alternatively, PhyloMatrix can be queried with a specific organism pattern to display profiles of matching families. PhyloMatrix is a helpful tool for the exploration of evolutionary events. For example, Figure 2 shows profiles of ribosomal protein families. These are complementary for mitochondrial and cytosolic forms reflecting the endosymbiotic origin of mitochondria.
Figure 2.
PhyloMatrix: phylogenetic profiling based on SYSTERS protein families. (i) On the left: PhyloMatrix entry page with several options to access phylogenetic profiles: (a) by browsing, (b) by family selection or (c) by specification of an organism pattern. (ii) On the right: 45 phylogenetic patterns describe 99 protein families comprising ribosomal proteins. For the given example, protein families were pre-selected via the second option (b). The order of organisms in each pattern follows the same order in taxonomic tree of the query page. Selected profiles are sorted according to a hierarchical clustering of all profiles.
Cross-references to external databases
The SYSTERS web server augments information on sequences and protein families by links to a multitude of data resources. We reference all protein source databases (Figure 1). In addition, SYSTERS can be queried with gene names, with accessions from the EMBL nucleotide database (21) or with identifiers of the specialized structure databases, such as PDB (22), MSD (23) and IMB (24). SYSTERS is embedded in the network of genomic database resources in the Computational Molecular Biology Department of the Max Planck Institute for Molecular Genetics, Berlin, including GeneNest, SpliceNest (25) and CORG (26).
Acknowledgments
ACKNOWLEDGEMENTS
We acknowledge funding from Bundesministerium für Bildung und Forschung (BMBF) through the Helmholtz Network for Bioinformatics (HNB).
REFERENCES
- 1.Bateman A., Coin,L., Durbin,R., Finn,R.D., Hollich,V., Griffiths-Jones,S., Khanna,A., Marshall,M., Moxon,S., Sonnhammer,E.L. et al. (2004) The Pfam protein families database. Nucleic Acids Res., 32, D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Letunic I., Copley,R.R., Schmidt,S., Ciccarelli,F.D., Doerks,T., Schultz,J., Ponting,C.P. and Bork,P. (2004) SMART 4.0: towards genomic data integration. Nucleic Acids Res., 32, D142–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hulo N., Sigrist,C.J., Saux,V.L., Langendijk-Genevaux,P.S., Bordoli,L., Gattiker,A., Castro,E.D., Bucher,P. and Bairoch,A. (2004) Recent improvements to the PROSITE database. Nucleic Acids Res., 32, D134–D137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith T.F. and Waterman,M.S. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
- 5.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M.C., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O'Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christie K.R., Weng,S., Balakrishnan,R., Costanzo,M.C., Dolinski,K., Dwight,S.S., Engel,S.R., Feierbach,B., Fisk,D.G., Hirschman,J.E. et al. (2004) Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res., 32, D311–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hertz-Fowler C., Peacock,C.S., Wood,V., Aslett,M., Kerhornou,A., Mooney,P., Tivey,A., Berriman,M., Hall,N., Rutherford,K. et al. (2004) GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res., 32, D339–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhee S.Y., Beavis,W., Berardini,T.Z., Chen,G., Dixon,D., Doyle,A., Garcia-Hernandez,M., Huala,E., Lander,G., Montoya,M. et al. (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res., 31, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birney E., Andrews,D., Bevan,P., Caccamo,M., Cameron,G., Chen,Y., Clarke,L., Coates,G., Cox,T., Cuff,J. et al. (2004) Ensembl 2004. Nucleic Acids Res., 32, D468–D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meinel T., Vingron,M. and Krause,A. (2003) The SYSTERS Protein Family Database: taxon-related protein family size distributions and singleton frequencies. In Proceedings of the German Conference on Bioinformatics. Belleville, Munich, Germany, pp. 103–108. [Google Scholar]
- 11.Koonin E.V., Wolf,Y.I. and Karev,G.P. (2002) The structure of the protein universe and genome evolution. Nature, 420, 218–223. [DOI] [PubMed] [Google Scholar]
- 12.Huynen M.A. and van Nimwegen,E. (1998) The frequency distribution of gene family sizes in complete genomes. Mol. Biol. Evol., 15, 583–589. [DOI] [PubMed] [Google Scholar]
- 13.Brown N.P., Leroy,C. and Sander,C. (1998) MView: a web-compatible database search or multiple alignment viewer. Bioinformatics, 14, 380–381. [DOI] [PubMed] [Google Scholar]
- 14.Morgenstern B. (1999) DIALIGN2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics, 15, 211–218. [DOI] [PubMed] [Google Scholar]
- 15.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris M.A., Clark,J., Ireland,A., Lomax,J., Ashburner,M., Foulger,R., Eilbeck,K., Lewis,S., Marshall,B., Mungall,C. et al. (2004) The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res., 32, D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mulder N.J., Apweiler,R., Attwood,T.K., Bairoch,A., Barrell,D., Bateman,A., Binns,D., Biswas,M., Bradley,P., Bork,P. et al. (2003) The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res., 31, 315–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bairoch A. (2000) The ENZYME database in 2000. Nucleic Acids Res., 28, 304–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wheeler D.L., Church,D.M., Edgar,R., Federhen,S., Helmberg,W., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Sequeira,E. et al. (2004) Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res., 32, D35–D40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pellegrini M., Marcotte,E.M., Thompson,M.J., Eisenberg,D. and Yeates,T.O. (1999) Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA, 96, 4285–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulikova T., Aldebert,P., Althorpe,N., Baker,W., Bates,K., Browne,P., van den Broek,A., Cochrane,G., Duggan,K., Eberhardt,R. et al. (2004) The EMBL nucleotide sequence database. Nucleic Acids Res., 32, D27–D30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golovin A., Oldfield,T.J., Tate,J.G., Velankar,S., Barton,G.J., Boutselakis,H., Dimitropoulos,D., Fillon,J., Hussain,A., Ionides,J.M. et al. (2004) E-MSD: an integrated data resource for bioinformatics. Nucleic Acids Res., 32, D211–D216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reichert J. and Sühnel,J. (2002) The IMB Jena Image Library of biological macromolecules: 2002 update. Nucleic Acids Res., 30, 253–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krause A., Haas,S.A., Coward,E. and Vingron,M. (2002) SYSTERS, GeneNest, SpliceNest: exploring sequence space from genome to protein. Nucleic Acids Res., 30, 299–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dieterich C., Wang,H., Rateitschak,K., Luz,H. and Vingron,M. (2003) CORG: a database for COmparative Regulatory Genomics. Nucleic Acids Res., 31, 55–57. [DOI] [PMC free article] [PubMed] [Google Scholar]