Abstract
Entrez Gene (www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene) is NCBI's database for gene-specific information. It does not include all known or predicted genes; instead Entrez Gene focuses on the genomes that have been completely sequenced, that have an active research community to contribute gene-specific information, or that are scheduled for intense sequence analysis. The content of Entrez Gene represents the result of curation and automated integration of data from NCBI's Reference Sequence project (RefSeq), from collaborating model organism databases, and from many other databases available from NCBI. Records are assigned unique, stable and tracked integers as identifiers. The content (nomenclature, map location, gene products and their attributes, markers, phenotypes, and links to citations, sequences, variation details, maps, expression, homologs, protein domains and external databases) is updated as new information becomes available. Entrez Gene is a step forward from NCBI's LocusLink, with both a major increase in taxonomic scope and improved access through the many tools associated with NCBI Entrez.
INTRODUCTION
Entrez Gene is the gene-specific database at the National Center for Biotechnology Information (NCBI), a division of the National Library of Medicine (NLM), located on the campus of the US National Institutes of Health (NIH) in Bethesda, MD, USA. Entrez Gene provides unique integer identifiers for genes and other loci for a subset of model organisms. It tracks those identifiers, and is integrated with the Entrez system for interactive query, LinkOuts, and access by E-utilities (1). The information that is maintained includes nomenclature, chromosomal localization, gene products and their attributes (e.g. protein interactions), associated markers, phenotypes, interactions, and a wealth of links to citations, sequences, variation details, maps, expression reports, homologs, protein domain content and external databases.
Data in Entrez Gene result from a mixture of curation and automated analyses. Annotation in sequences from NCBI's Reference sequence project (2) or the International Nucleotide Sequence Database Collaboration (DDBJ, EMBL, GenBank) (3) is integrated with information from collaborating model organism databases, literature review (especially the Gene References into Function or GeneRIFs) (1), and public users, with curation by RefSeq staff as required.
Entrez Gene is an integral part of representation of gene-specific information at NCBI. The information conveyed by establishing a ‘gene-to-sequence’ relationship is used by other NCBI resources (1) such as BLAST, Geo, HomoloGene, Map Viewer, UniGene, UniSTS and NCBI's genome annotation pipeline. For example, the names associated with GeneIDs are used in HomoloGene, Map Viewer, UniGene and the Mammalian Gene Collection (4). Inconsistencies in representation of genes and their sequences are investigated, and resolved by NCBI staff in collaboration with external nomenclature authorities.
The content, display and bulk reporting from Entrez Gene continue to be developed. Users may be interested in subscribing to gene-announce@ncbi.nlm.nih.gov to receive information about modifications.
FUNCTION OF THE DATABASE
The primary goals of Entrez Gene are to provide tracked, unique identifiers for genes and to report information associated with those identifiers for unrestricted public use. The identifier that is assigned (GeneID) is an integer, and is species specific. In other words, the integer assigned to dystrophin in human is different from that in any other species. For genomes that had been represented in LocusLink, the GeneID is the same as the LocusID. The GeneID is reported in RefSeq records as a ‘db_xref’ (e.g. /db_xref=“GeneID:856646”, in GenBank format).
Entrez Gene provides multiple reports. For the interactive user, the defaults are the HTML summary display resulting from an Entrez query (Figure 1) or a gene-specific report achieved by clicking on the symbol in the summary page (Figure 2). The Gene Table display option is useful to obtain a report of the intron/exon organization of the gene as annotated on a RefSeq genomic sequence, and to navigate quickly to the sequence of any of those gene features. In addition to the multiple views from Entrez, Gene provides a complete database extraction in ASN.1 format as well as several tab-delimited reports for ftp transfer (ftp://ftp.ncbi.nlm.nih.gov/gene/). The data are also available from the programmatic interface to Entrez, namely e-utilities (1).
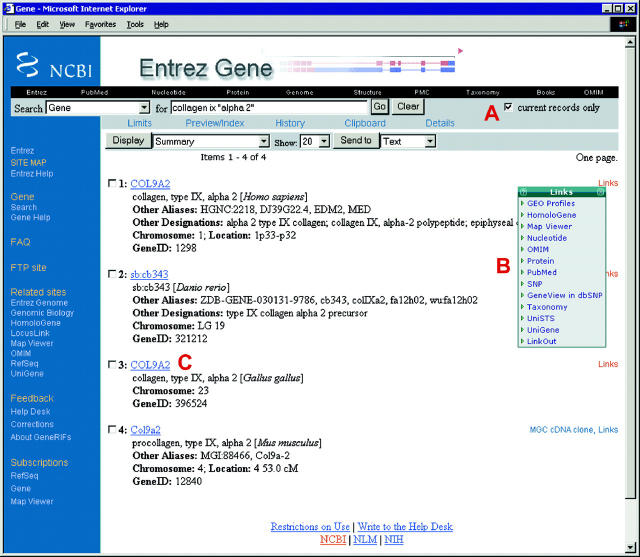
Figure 1.
Summary view of query results. The query is collagen ix “alpha 2”. The phrase alpha 2 is submitted in double quotes to restrict the query for alpha followed by 2. Note that the box ‘current records only’ (A) is checked automatically; to retrieve outdated records, this box must be unchecked. The default Summary display option allows gene-specific links to many other resources both within NCBI's Entrez system and not (LinkOut, MGC cDNA clone) via the Links menu (shown expanded, B). The summary includes the species of origin, preferred and alternate (Other aliases) symbols, preferred and other (Other designations) descriptive names, chromosome localization, the GeneID. Click on any symbol to the left of the check box (C) to link to the full report (Figure 2). The top black navigation bar and the blue sidebar at the left provide general links to other sites, including Genome-specific resource guides (Genomic Biology), the FTP site, forms to submit feedback (Feedback), and forms to subscribe to mail lists to be informed of changes (Subscriptions).
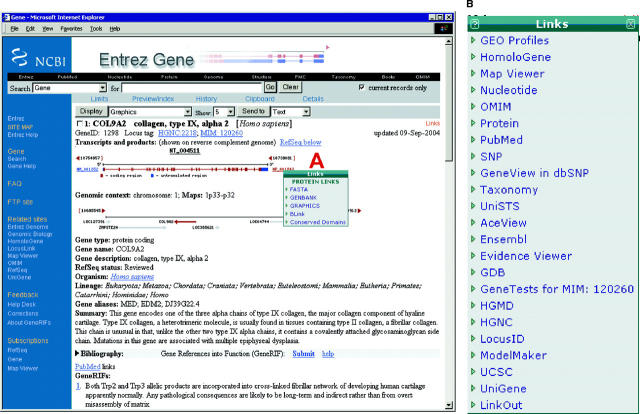
Figure 2.
Partial report page. The standard gene-specific report page starts with the preferred symbol (name) and description of the gene, the species and the ID. As appropriate, it provides links to key external resource, using the label ‘Locus tag’ consistent with feature annotation of a GenBank record. If the gene has been annotated on a RefSeq genomic sequence (NT_004511), a graphic is provided showing the placement of the gene and those of its neighbors. The accessions assigned to the mRNA and proteins are shown at the left and right respectively (NM_001852, NP_001843 in this case). Clicking on any of these accessions opens a menu of options to obtain the sequence. The menu attached to the protein accession (A) shows that you can link to the sequence in either FASTA, GenBank or graphical format, or use BLink (1) to display information about related proteins, or conserved domains. The Links menu at the upper right corner of the complete record (expanded in B) indicates what resources have information related specifically to this gene. Some of these links are the same as those seen on the summary display (Figure 1), but there are often additional Links to information not accessible directly from Entrez.
SCOPE OF THE DATABASE
When are GeneIDs assigned?
Identifiers are always assigned to what is annotated as a Gene on a RefSeq record. Records may also be created when an authoritative source for a genome assigns an identifier to a gene, mapped locus or trait, even though that entity is not yet defined by explicit sequence. Although this means that Entrez Gene is not restricted to what might be considered a gene biologically, the expectation is that some of these records will become more ‘gene-like’ as the molecular basis of traits or other loci is defined. Each Gene record is assigned a type from an enumerated list in the ASN.1 specification for Entrez Gene. (See the Gene Help document for more information.) This type, which is indexed in Entrez as a named property (e.g. genetype protein coding), can be changed without changing the GeneID.
Some current statistics
As of September 2004, there were more than 2400 taxa represented in Entrez Gene, with a total of approximately 958 000 current records. Not all the taxa are completely represented in Entrez Gene; most of the eukaryotes (∼600 total), for example, have Gene records only for their mitochondrial genomes. More than half of the taxa represented are viruses (∼1350). Next of those having genomes with comprehensive gene annotation are eubacteria and Archaea (∼200 and 20, respectively). About 95 per cent of all records are for protein-coding genes.
Record content
Table 1 summarizes the gene-specific information that can be retrieved through Entrez Gene, how the data are shown, and some aspects of how those data are processed. For example, GeneRIFs, contributed primarily by the public and the Index Section of the National Library of Medicine, provide an annotated bibliography of the function, discovery and mapping of genes from the current literature and are seen in the default report. Information about Clusters of Orthologous Groups (COGs) (5) is available via Links menus. This combination of text and connections is designed to provide sufficient descriptions, keywords and links to make Entrez Gene an effective starting place to retrieve information of interest.
Table 1. Categories of information in Entrez Gene.
| Subcategory | Showna | Comments |
|---|---|---|
| Nomenclature | ||
| Gene symbols and full descriptions | Report, Table | Sources: External authorities, GenBank, Publications. ‘LOC’+GeneID designation assigned if none of the above officially accepted nomenclature has precedence |
| Protein names | Report, Table | Often same as the gene name, but may be edited to make orthologs' names uniform |
| Gene structure and sequence | ||
| Gene structure | Report, Table | Based on annotation of the Reference sequence |
| Reference sequences | Report, Table, Links | The accessions are shown in the report page; the sequences are retrieved from Nucleotide or Protein |
| Related sequences | Report, Links | Based on cDNA or protein comparison, best genomic placement and curation. Accessions are shown in the report page; sequences are retrieved from Nucleotide or Protein |
| Genomic position | ||
| By sequence | Report, Links | Genomic annotation |
| By independent maps | Links | Shared markers or reports of cytogenetic position |
| Citations | ||
| Not annotated | Links | Sources: external authorities, RefSeq curation |
| Annotated | Report, Links | Sources: external databases, GeneRIFs |
| Functional annotation | ||
| Domain content | Report, Links | Conserved Domain Database (CDD) |
| GO terms | Report | GO Consortium |
| Pathways and interactions | Report, Links | KEGG, HIV Interactions Database |
| Disease and other phenotypes | Report, Links | External authorities such as OMIM, RefSeq curation |
| Homology | ||
| By Gene | Links | HomoloGene |
| By Protein | Links | COG |
| Conserved segments | Links | Map Viewer |
| Expression | ||
| ESTs | Links | UniGene |
| External resources | Links | External resource is named and used to anchor a Link. The expression data are available at that source |
| Arrays | Links | GEO |
| Related information | ||
| Integrated by Gene staff | Links | May be displayed on report as well as from Links menu |
| External sources | Links | LinkOut choice in Links menu |
aWhere information is shown: Report, Graphic Display; Table, Gene Table Display; Links, Links menu.
ACCESS TO ENTREZ GENE
The information in Entrez Gene can be accessed in multiple ways at NCBI (Table 2). The most direct is to submit a query to Entrez from the NCBI home page and display the results in Gene, or enter a query in any Entrez query bar and restrict the database search to Gene. Another way is to take advantage of the Links computed by the Entrez system. For example, you might find a PubMed record of interest, and from PubMed's Links menu discover that there is a record in Gene connected to the publication.
Table 2. Accessing Entrez Gene.
| Direct query—detailed instructions in the Query tips section of the online help documentation: | |
| http://www.ncbi.nlm.nih.gov/entrez/query/static/help/genehelp.html#query | |
| www.ncbi.nlm.nih.gov/Entrez/ or www.ncbi.nlm.nih.gov | Enter search term(s) and select results shown in the Gene section |
| www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene or select Gene as the search option from any Entrez query bar | Enter search term(s) |
| Record-specific connections in other NCBI databases | |
| Gene option in the Links menu at the upper right of a display in a non-Gene record | Click on Gene to find Gene records related to the record being displayed |
| Links called Gene or G | Map Viewer's annotation of Genes; BLAST retrieval of accessions connected to Gene records |
Many databases within NCBI take advantage of the GeneID<->sequence relationship maintained by Gene to make connections from a sequence of interest to the Entrez Gene record. For example, BLAST queries matching protein or mRNA accessions associated with an Entrez Gene record are identified by the blue G icon. Map Viewer provides links from annotated genes to Entrez Gene. And RefSeq records include the GeneID as a db_xref in the gene feature. Thus you can obtain gene-specific information not only by text queries but also by genomic position (Map Viewer), RefSeq annotation and related sequences (BLAST, Entrez Nucleotide, Entrez Protein).
LINKS TO EXTERNAL DATABASES FROM ENTREZ GENE
Entrez Gene can serve as a directory to gene-specific information for databases outside of NCBI. External databases can register with the LinkOut service (1) and submit information about how their database should be connected to any Gene record. Any user of Entrez Gene retrieving a record with LinkOuts will then be able to connect to the registered database according to the specification of the data provider.
FEEDBACK
We welcome your feedback with respect to the Entrez Gene interface, or any data contained therein. You may use any of the Feedback options on a Gene page (Figure 1).
REFERENCES
- 1.Wheeler D.L., Benson,D.A., Bryant,S., Canese,K., Church,D.M., Edgar,R., Federhen,S., Helmberg,W., Kenton,D., Khovayko,O. et al. (2005) Database resources of the National Center for Biotechnology Information: Update. Nucleic Acid Res, 33, D39–D45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pruitt K.D., Tatusova,T. and Maglott,D. (2005) NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts, and proteins. Nucleic Acids Res, 33, D501–D504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J., Wheeler,D.L. (2005) GenBank. Nucleic Acids Res, 33, D34–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strausberg R.L., Feingold,E.A., Grouse,L.H., Derge,J.G., Klausner,R.D., Collins,F.S., Wagner,L., Shenmen,C.M., Schuler,G.D., et al. and Mammalian Gene Collection Program Team. (2002) Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl Acad. Sci. USA, 99, 16899–16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tatusov R.L., Fedorova,N.D., Jackson,J.D., Jacobs,A.R., Kiryutin,B., Koonin,E.V., Krylov,D.M., Mazumder,R., Mekhedov,S.L., Nikolskaya,A.N. et al. (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics, 4, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]