Abstract
The Immuno Polymorphism Database (IPD) (http://www.ebi.ac.uk/ipd/) is a set of specialist databases related to the study of polymorphic genes in the immune system. IPD currently consists of four databases: IPD-KIR, contains the allelic sequences of Killer-cell Immunoglobulin-like Receptors; IPD-MHC, a database of sequences of the Major Histocompatibility Complex of different species; IPD-HPA, alloantigens expressed only on platelets; and IPD-ESTAB, which provides access to the European Searchable Tumour Cell-Line Database, a cell bank of immunologically characterized melanoma cell lines. The IPD project works with specialist groups or nomenclature committees who provide and curate individual sections before they are submitted to IPD for online publication. The IPD project stores all the data in a set of related databases. Those sections with similar data, such as IPD-KIR and IPD-MHC share the same database structure. The sharing of a common database structure makes it easier to implement common tools for data submission and retrieval. The data are currently available online from the website and ftp directory; files will also be made available in different formats to download from the website and ftp server. The data will also be included in SRS, BLAST and FASTA search engines at the European Bioinformatics Institute.
INTRODUCTION
The Immuno Polymorphism Database (IPD) is a set of specialist databases related to the study of polymorphic genes in the immune system. The IPD project works with specialist groups or nomenclature committees, which each curate a different section of the project. IPD currently consists of four databases: IPD-KIR, contains the allelic sequences of Killer-cell Immunoglobulin-like Receptors (KIRs); IPD-MHC, a database of sequences of the Major Histocompatibility Complex (MHC) of different species; IPD-HPA, alloantigens expressed only on platelets; and IPD-ESTAB, which provides access to the European Searchable Tumour Cell-Line Database (ESTDAB), a cell bank of immunologically characterized melanoma cell lines.
The study of the immune system constitutes many different complex areas of research. By providing a centralized resource for the work of different groups, it is hoped that we can bring together similar data to aid in the study and analysis of this area. This will be done by contacting various expert groups, such as the nomenclature committees for certain genes, and inviting them to contribute their data to a central resource. The individual nomenclature committees are all established within their own field, their continuing role being the identification and naming of new alleles based on the submission of new sequences to generalist databases like the European Molecular Biology Laboratory's nucleotide sequence database (EMBL), the National Center for Biotechnology Information's GenBank and the DNA DataBank of Japan (DDBJ). The main difference between the data held within a specialist system like IPD and the generalist databases is the further curation of the files by experts in the relevant field. This additional step allows improvements in data quality and the addition of more specialized information. This may result in there being differences in the entries kept in the different databases; in such a case the entry in IPD should be considered as the most accurate. The inclusion of some nomenclature committees has meant the online publication of sequence alignments for the first time. This is particularly important when it comes to the sequences of polymorphic genes. Also because the IPD project is able to provide the work of different committees in a common format, it is easier to compare the sequences of different species.
The IPD project stores all the data in a set of related databases. Those sections with similar data, IPD-KIR and IPD-MHC share the same database structure. The sharing of a common database structure makes it easier to implement common tools for data submission and retrieval. Other unrelated sections like IPD-ESTDAB currently have their own unique structure.
IPD-KIR
The KIRs are members of the immunoglobulin super family (IgSF) formerly called Killer-cell Inhibitory Receptors. KIRs have been shown to be highly polymorphic both at the allelic and haplotypic levels (1). They are composed of two or three Ig-domains, a transmembrane region and cytoplasmic tail, which can in turn be short (activatory) or long (inhibitory). The Leukocyte Receptor Complex (LRC), which encodes KIR genes, has been shown to be polymorphic, polygenic and complex in a manner similar to the MHC. This complexity in sequences has led to the formation of the KIR nomenclature committee. The nomenclature committee is responsible for the naming of new allele sequences, and produced its first report in 2002 (2); this was complemented by the inclusion of the KIR data into IPD. The IPD-KIR Sequence Database contains the most up-to-date nomenclature and sequence alignments. Also available is an online submission tool that allows the submission of new and confirmatory KIR sequences directly to the KIR nomenclature committee. Sequences submitted to IPD as part of the work of the individual nomenclature committees are based on sequences currently found in the EMBL Nucleotide Sequence Database (EMBL) (3), GenBank (4) and the DDBJ (5). Indeed a requirement of all submissions to IPD is that they have been submitted to these more general databases. Future developments of the IPD-KIR section will involve working with the KIR nomenclature committee to provide nomenclature and tools for study of the complex haplotypes and genotypes currently seen in KIR research.
IPD-MHC
The MHC sequences of many different species have been reported previously (6–9), along with different nomenclature systems used in the naming and identification of new genes and alleles in each species. The sequences of the MHC from a number of different species are highly conserved between species (10). By bringing the work of different nomenclature committees and the sequences of different species together it is hoped to provide a central resource that will facilitate further research on the MHC of each species and on their comparison.
The first release of the IPD-MHC database involved the work of groups specializing in non-human primates, canines (DLA) and felines (FLA) and incorporated all data previously available in the IMGT/MHC database (11). This release included data from 5 species of ape, 16 species of new world monkey, 17 species of old world monkey, as well as data on different canines and felines. Since the first release, we have been able to add sequences from cattle (BoLA), and are now working to include the MHC sequences from swine (SLA), chickens, horses (ELA) and rats (RT1).
For each species, there are some differences in the spectrum of data covered but all sections provide the core nomenclature pages and sequence alignments. The nomenclature and alignments follow a similar structure to that of the IPD-KIR section, and the same basic tools are used in both sections. Currently, the IPD-MHC sequence alignments are limited to species-specific alignments; however, we are working to allow cross-species alignments and the inclusion of human sequences from the IMGT/HLA database (12) for comparative purposes. The IPD-MHC Sequence Databases will also contain a submission tool for online submission of new and confirmatory sequences to the appropriate nomenclature committee.
COMMON TOOLS FOR IPD-KIR AND IPD-MHC
Some sections of the project are clearly more closely related, namely the IPD-KIR and IPD-MHC sections, both deal with sequences from polymorphic genes and as such they share a number of features. As each section is based on the work of a nomenclature committee, the website includes or links to a portable document format (PDF) file of recent nomenclature reports. The website also provides a recognized location for updates to the nomenclature between published reports. These pages contain the official allele name, any previous designations, the EMBL, GenBank or DDBJ accession number(s) and a reference linked wherever possible to the PubMed abstract. If possible additional details on the source of sequence are also provided. Some nomenclature committees may provide additional information but the core components of any nomenclature reported are the allele names, accession numbers and publications.
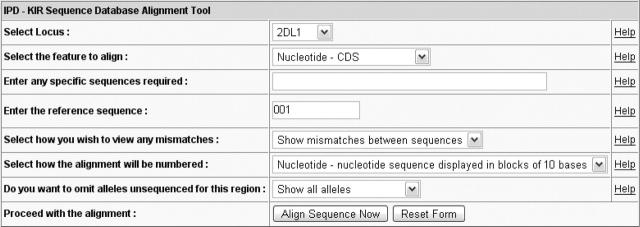
The second common feature is the sequence alignment tool, which is provided using a common interface for both sections. Figure 1 shows the alignment tool used for the IPD-KIR section. The main difference seen in the IPD-MHC interface is the locus listing, which is also preceded by a species listing. The selection of a locus, automatically updates the list of features, which can be aligned, as well as the default reference sequence used for the alignment. The remaining options cover display and formatting options. The alignment tool uses standard formatting conventions for the display of sequence alignments; these are based on those currently used by the IMGT/HLA Sequence Database (11–14) and published recommendations for Human Gene Mutations (15,16). The alignment tool options allow the user to display a subset of alleles of a particular locus, omit alleles unsequenced for a particular region and align against a particular reference or consensus sequence. In addition, the sequences can be displayed as complete nucleotide sequence, partial sequences of single exons or the amino acid sequence of the encoded protein (Figure 2). The alignment tool does not perform a sequence alignment each time it is used, but it extracts pre-aligned sequences, allowing for faster access. For some of the sections included, this is the first time users have been able to view sequence alignments online.
Figure 1.
The alignment interface provides a user-friendly method of viewing sequence alignments with output options easily selected.
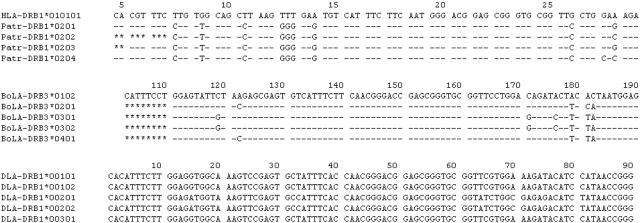
Figure 2.
Alignment formats available from IPD. The examples shown are all alignments of DRB alleles from different species. In these alignments a dash (-) indicates identity to the reference sequence and an asterisk (*) denotes an unsequenced base. The first alignment shows the nucleotide sequence of Chimpanzee (Pan troglodytes) DRB1 alleles with the human HLA-DRB1*010101 allele. The nucleotide sequence is also displayed in amino acid codons; the first base of the initial codon is encoded in exon 1 and so not displayed in this alignment. The second set of sequences shows DRB alleles from cattle. The final alignment uses canine DRB alleles, and demonstrates how the full sequence can be displayed.
IPD-HPA
Human platelet antigens (HPAs) are alloantigens expressed only on platelets, specifically on platelet membrane glycoproteins. These platelet-specific antigens are immunogenic and can result in pathological reactions to transfusion therapy. The HPA nomenclature system was adopted in 1990 (17,18) to overcome problems with the previous nomenclature. Since then more antigens have been described and the molecular basis of many has been resolved. As a result the nomenclature was revised in 2003 (19) and included in the IPD project. The IPD-HPA section contains nomenclature information and additional background material. The different genes in the HPA system have not been sequenced to the same level as some of the other projects and so currently only single nucleotide polymorphisms (SNPs) are used to determine alleles. This information is presented in a grid of SNP for each gene. The IPD and HPA nomenclature committee hope to expand this to provide full sequence alignments when possible.
IPD-ESTDAB
IPD-ESTDAB is a database of immunologically characterized melanoma cell lines. The database works in conjunction with the European Searchable Tumour Cell Line Database (ESTDAB) cell bank, which is housed in Tübingen, Germany and provides immunologically characterized tumour cells. The IPD-ESTDAB section of the website provides an online search facility for cells stored in this cell bank. This enables investigators to identify cells possessing specific parameters important for studies of immunity, immunogenetics, gene expression, metastasis, response to chemotherapy and other tumour biological experimentation. The search tool allows for searches based on a single parameter, or clusters of parameters on over 250 different markers for each cell. The detailed reports produced can then be used to identify cells of interest, which in turn can then be obtained from the cell bank.
DISCUSSION
The IPD project provides a new resource for those interested in the study of polymorphic sequences in the immune system. By accommodating related systems in a single database, data can be made available in common formats aiding use and interpretation. As the projects grow and more sections are added, the benefit of having expertly curated sequences from related areas stored in a single location will become more apparent. This is particularly true of the IPD-MHC project, where cross-species studies will be able to utilize the high-quality sequences provided by the different nomenclature committees in a common format, ready for use. The initial release of the IPD Database contained only four sections and a small number of tools; however, as the database grows and more sections and species are added, more tools will be added to the website. We plan to use the existing database structures to house data for new sections of the IPD project as they become available. The files will also be made available in different formats for downloading from the website, and ftp server, and the data will be included in SRS, BLAST and FASTA search engines at the European Bioinformatics Institute (20).
ACCESS AND CONTACT
IPD Homepage: http://www.ebi.ac.uk/ipd/
IPD-KIR Homepage: http://www.ebi.ac.uk/ipd/kir/
IPD-MHC Homepage: http://www.ebi.ac.uk/ipd/mhc/
IPD-HPA Homepage: http://www.ebi.ac.uk/ipd/hpa/
IPD-ESTDAB Homepage: http://www.ebi.ac.uk/ipd/estdab/
Contact: ipd@ebi.ac.uk
If you are interested in contributing to the project, there are specific guidelines for the inclusion of new sections, and interested parties should contact Dr S. G. E. Marsh, marsh@ebi.ac.uk for further information.
REFERENCES
- 1.Garcia C.A., Robinson,J., Guethlein,L.A., Parham,P., Madrigal,J.A. and Marsh,S.G.E. (2003) Human KIR sequences 2003. Immunogenetics, 55, 227–239. [DOI] [PubMed] [Google Scholar]
- 2.Marsh S.G.E., Parham,P., Dupont,B., Geraghty,D.E., Trowsdale,J., Middleton,D., Vilches,C., Carrington,M., Witt,C., Guethlein,L.A. et al. (2003) Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Immunogenetics, 55, 220–226. [DOI] [PubMed] [Google Scholar]
- 3.Kanz C., Aldebert,P., Althorpe,N., Baker,W., Baldwin,A., Bates,K., Browne,P., van den Broek,A., Castro,M., Cochrane,G. et al. (2005) The EMBL Nucleotide Sequence Database. Nucleic Acids Res., 33, D29–D33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J. and Wheeler,D.L. (2005) GenBank. Nucleic Acids Res., 33, D34–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyazaki S., Sugawara,H., Ikeo,K., Gojobori,T. and Tateno,Y. (2004) DDBJ in the stream of various biological data. Nucleic Acids Res., 32, D31–D34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kennedy L.J., Angles,J.M., Barnes,A., Carter,S.D., Francino,O., Gerlach,J.A., Happ,G.M., Ollier,W.E., Thomson,W. and Wagner,J.L. (2001) Nomenclature for factors of the dog major histocompatibility system (DLA), 2000: second report of the ISAG DLA Nomenclature Committee. Tissue Antigens, 58, 55–70. [DOI] [PubMed] [Google Scholar]
- 7.Kennedy L.J., Altet,L., Angles,J.M., Barnes,A., Carter,S.D., Francino,O., Gerlach,J.A., Happ,G.M., Ollier,W.E., Polvi,A. et al. (1999) Nomenclature for factors of the dog major histocompatibility system (DLA), 1998. First report of the ISAG DLA Nomenclature Committee. International Society for Animals Genetics. Tissue Antigens, 54, 312–321. [DOI] [PubMed] [Google Scholar]
- 8.Davies C.J., Joosten,I., Bernoco,D., Arriens,M.A., Bester,J., Ceriotti,G., Ellis,S., Hensen,E.J., Hines,H.C., Horin,P. et al. (1994) Polymorphism of bovine MHC class I genes. Joint report of the Fifth International Bovine Lymphocyte Antigen (BoLA) Workshop, Interlaken, Switzerland, 1 August 1992. Eur. J. Immunogenet., 21, 239–258. [DOI] [PubMed] [Google Scholar]
- 9.Klein J., Bontrop,R.E., Dawkins,R.L., Erlich,H.A., Gyllensten,U.B., Heise,E.R., Jones,P.P., Parham,P., Wakeland,E.K. and Watkins,D.I. (1990) Nomenclature for the major histocompatibility complexes of different species: a proposal. Immunogenetics, 31, 217–219. [DOI] [PubMed] [Google Scholar]
- 10.Parham P. (1999) Virtual reality in the MHC. Immunol. Rev., 167, 5–15. [DOI] [PubMed] [Google Scholar]
- 11.Robinson J., Waller,M.J., Parham,P., de Groot,N., Bontrop,R., Kennedy,L.J., Stoehr,P. and Marsh,S.G.E. (2003) IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res., 31, 311–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson J., Malik,A., Parham,P., Bodmer,J.G. and Marsh,S.G.E. (2000) IMGT/HLA database—a sequence database for the human major histocompatibility complex. Tissue Antigens, 55, 280–287. [DOI] [PubMed] [Google Scholar]
- 13.Robinson J., Waller,M.J., Parham,P., Bodmer,J.G. and Marsh,S.G.E. (2001) IMGT/HLA Database—a sequence database for the human major histocompatibility complex. Nucleic Acids Res., 29, 210–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson J. and Marsh,S.G.E. (2000) The IMGT/HLA sequence database. Rev. Immunogenet., 2, 518–531. [PubMed] [Google Scholar]
- 15.den Dunnen J.T. and Antonarakis,S.E. (2001) Nomenclature for the description of human sequence variations. Hum. Genet., 109, 121–124. [DOI] [PubMed] [Google Scholar]
- 16.Antonarakis S.E. (1998) Recommendations for a nomenclature system for human gene mutations. Nomenclature Working Group. Hum. Mutat., 11, 1–3. [DOI] [PubMed] [Google Scholar]
- 17.von dem Borne A.E. and Decary,F. (1990) ICSH/ISBT Working Party on platelet serology. Nomenclature of platelet-specific antigens. Vox. Sang., 58, 176. [DOI] [PubMed] [Google Scholar]
- 18.von dem Borne A.E. and Decary,F. (1990) Nomenclature of platelet-specific antigens. Hum. Immunol., 29, 1–2. [DOI] [PubMed] [Google Scholar]
- 19.Metcalfe P., Watkins,N.A., Ouwehand,W.H., Kaplan,C., Newman,P., Kekomaki,R., De Haas,M., Aster,R., Shibata,Y., Smith,J. et al. (2003) Nomenclature of human platelet antigens. Vox. Sang., 85, 240–245. [DOI] [PubMed] [Google Scholar]
- 20.Harte N., Silventoinen,V., Quevillon,E., Robinson,S., Kallio,K., Fustero,X., Patel,P., Jokinen,P. and Lopez,R. (2004) Public web-based services from the European Bioinformatics Institute. Nucleic Acids Res., 32, W3–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]