Abstract
Online Mendelian Inheritance in Man (OMIM™) is a comprehensive, authoritative and timely knowledgebase of human genes and genetic disorders compiled to support human genetics research and education and the practice of clinical genetics. Started by Dr Victor A. McKusick as the definitive reference Mendelian Inheritance in Man, OMIM (http://www.ncbi.nlm.nih.gov/omim/) is now distributed electronically by the National Center for Biotechnology Information, where it is integrated with the Entrez suite of databases. Derived from the biomedical literature, OMIM is written and edited at Johns Hopkins University with input from scientists and physicians around the world. Each OMIM entry has a full-text summary of a genetically determined phenotype and/or gene and has numerous links to other genetic databases such as DNA and protein sequence, PubMed references, general and locus-specific mutation databases, HUGO nomenclature, MapViewer, GeneTests, patient support groups and many others. OMIM is an easy and straightforward portal to the burgeoning information in human genetics.
INTRODUCTION
Mendelian Inheritance in Man (MIM), which has been published in 12 print editions since 1966 (1), is a compendium of information on genetic disorders and genes. Its electronic counterpart, Online Mendelian Inheritance in Man (OMIMTM), is updated daily and is freely available on the World Wide Web (http://www.ncbi.nlm.nih.gov/omim/). OMIM is an authoritative full-text overview of genes and genetic phenotypes that can be used by students, researchers and clinicians. Curation of the database and editorial decisions take place at Johns Hopkins University School of Medicine. Authors are located at Johns Hopkins and around the world. Distribution of OMIM and software development are provided by the National Center for Biotechnology Information (NCBI) at the National Library of Medicine (NLM).
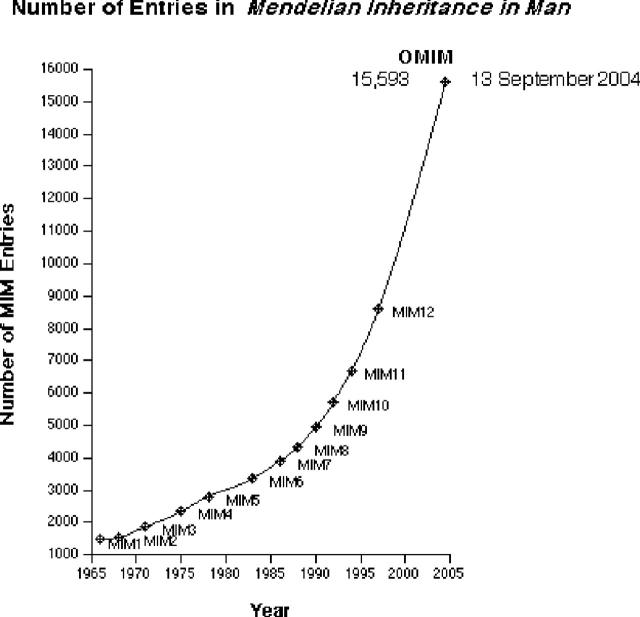
MIM has been computerized since 1964 and periodically published, first in 1966 and the 12th edition in 1998 (1). The print editions began as ‘Catalogs of Autosomal Dominant, Autosomal Recessive and X-linked Phenotypes’; since 1992 the subtitle of MIM has been ‘Catalogs of Human Genes and Genetic Disorders’. OMIM has been available online since 1987, first from Johns Hopkins University and since 1995 from the NCBI. Figure 1 illustrates the growth of the database in terms of numbers of entries in each edition of the book and in OMIM through September 13, 2004. OMIM averages at least 8500 unique users and 100 000 queries per day.
Figure 1.
Growth of the database in terms of number of entries in each edition of MIM and in OMIM as of September 13, 2004.
SEARCHING OMIM
OMIM can be searched from its homepage or from any page in the NCBI Entrez suite of databases. Information in OMIM can be retrieved by queries on MIM number, disorder, gene name and/or symbol, or plain English (e.g. ‘cryptorchidism webbed neck’). The limits function may be used to perform a restricted search of parts of a MIM entry (number, titles, references, etc.) and/or type of MIM entry (gene or phenotype). Regardless of the method used, the search engine ranks the entries that match the query so that the entries that are most relevant to the question are generally in the top 10 retrievals.
Each OMIM entry is assigned a unique six-digit number whose first digit indicates whether its inheritance is autosomal, X-linked, Y-linked or mitochondrial: 1, autosomal loci or phenotypes (entries created before May 15, 1994); 2, autosomal loci or phenotypes (entries created before May 15, 1994); 3, X-linked loci or phenotypes; 4, Y-linked loci or phenotypes; 5, mitochondrial loci or phenotypes; and 6, autosomal loci or phenotypes (created after May 15, 1994).
In addition, MIM entries are categorized by whether they contain information on genes, phenotypes or both. This is denoted by the symbol that precedes a MIM number. An asterisk (*) before an entry number indicates a gene of known sequence. A number symbol (#) before an entry number indicates that it is a descriptive entry, usually of a phenotype, and does not represent a unique locus. The reason for the use of the #-sign is given in the first paragraph of the entry. Discussion of any gene(s) related to the phenotype resides in appropriate entry(ies) as described in the first paragraph. A plus sign (+) before an entry number indicates that the entry contains the description of a gene of known sequence and a phenotype. A percentage sign (%) before an entry number indicates that the entry describes a confirmed Mendelian phenotype or phenotypic locus for which the underlying molecular basis is not known. No symbol before an entry number generally indicates a description of a phenotype for which the Mendelian basis, although suspected, has not been clearly established or that the distinctness of this phenotype from that in another entry is unclear. A caret symbol (^) before an entry number means the entry was removed from the database or moved to another entry as indicated. As of September 13, 2004, OMIM had 10 208 entries describing genes with known sequence and 5777 entries describing phenotypes (Table 1).
Table 1. Number of gene and phenotype entries in OMIM as on September 13, 2004.
| Autosomal | X-linked | Y-linked | Mitochondrial | Total | |
|---|---|---|---|---|---|
| * Gene with known sequence | 9318 | 414 | 47 | 37 | 9816 |
| + Gene with known sequence and phenotype | 356 | 36 | 0 | 0 | 392 |
| # Phenotype description, molecular basis known | 1462 | 134 | 1 | 25 | 1622 |
| % Mendelian phenotype or locus, molecular basis unknown | 1288 | 130 | 4 | 0 | 1422 |
| Other, mainly phenotypes with suspected mendelian basis | 2185 | 154 | 2 | 0 | 2341 |
| Total | 14 609 | 868 | 54 | 62 | 15 593 |
HOME PAGE
From the OMIM home page, one can review OMIM Statistics including the current count of the number of entries in OMIM (15 593 on September 13, 2004) organized by catalog (autosomal, X-linked, Y-linked and mitochondrial) as well as a count by chromosome of OMIM's Synopsis of the Human Gene Map (9098 loci on September 13, 2004). The Update Log allows a quick check of the latest additions and changes to OMIM. About 70 entries are created and 600 updated each month. Restrictions on Use informs the user that OMIM is copyrighted by the Johns Hopkins University and is made available to the general public subject to certain restrictions. Finally, the OMIM home page maintains links to many useful genetic resources including the Genetic Alliance, an umbrella organization of over 600 genetic support groups.
WITHIN AN ENTRY
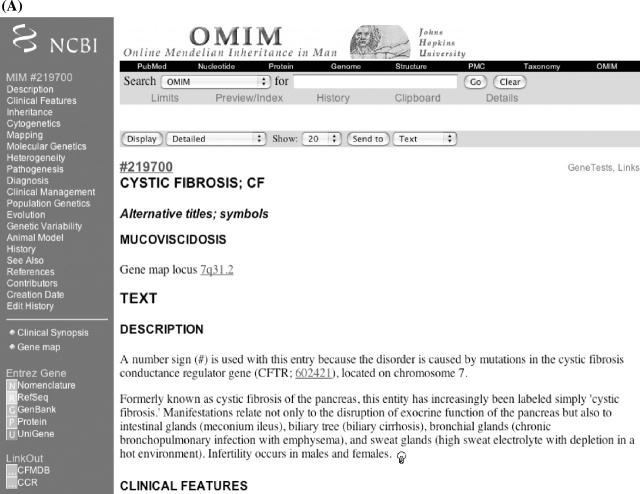
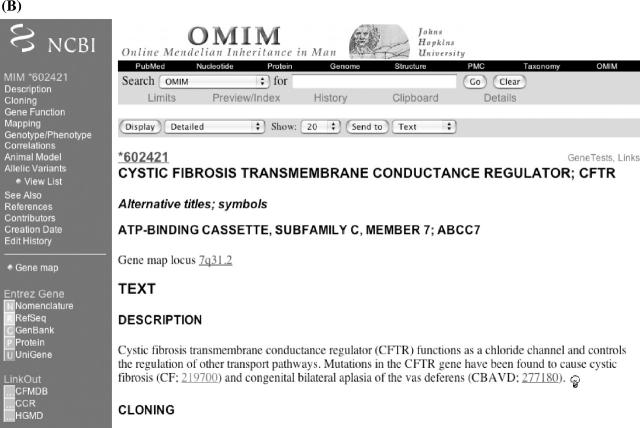
Within an OMIM entry, the blue bar along the left-hand side provides a menu for direct access to subheadings in the entry (Figure 2A and B). In addition, there are links to resources both within and outside OMIM. Clinical Synopsis takes one to a listing of the clinical features of the disorder. There are over 4500 clinical synopses in OMIM. Genemap takes the user to OMIM's Synopsis of the Human Gene Map. The links within a phenotype or gene entry are not necessarily the same. In the case of the cystic fibrosis (OMIM 219700) and cystic fibrosis transmembrane conductance regulator (OMIM 602421) entries, the links may include Entrez Gene (NCBI LocusLink database), Nomenclature (HUGO Nomenclature Committee), RefSeq (NCBI reference sequences), GenBank (NCBI nucleotide sequence database), Protein (NCBI protein sequence database) and UniGene (NCBI UniGene project). Additional external links include the cystic fibrosis locus-specific mutation database (CFMDB), the Coriell Cell Repository (CCR) and the Human Gene Mutation Database at Cardiff (HGMD). The CCR lists CF cell lines available for research. In all cases, the links take one directly to the relevant entry within the other database. When available, gene and phenotype entries contain links to GeneTests to the right-hand side of the entry title. The GeneTests link takes the user directly to the relevant gene entry in GeneTests, a database of information on genetic testing and its use in diagnosis, management and genetic counseling.
Figure 2.
(A) Entry for cystic fibrosis and (B) CFTR showing links to Entrez Gene, Nomenclature, RefSeq, GenBank, Protein and UniGene databases, as well as links to the CFTR mutation database (CFMDB), the Coriell Cell Repository (CCR) and the Human Gene Mutation Database (HGMD).
An OMIM entry includes the primary title and symbol, alternative titles and symbols, and ‘included’ titles (i.e. related but not synonymous information that is not addressed in another entry). The gene map locus displays the cytogenetic location of the gene or disorder. This information is derived from the OMIM gene map. Multiple map locations may be given if a disease is known to be genetically heterogeneous. The light bulb icons located at the end of every paragraph link to the NCBI's ‘neighboring’ feature. This feature takes keywords from the paragraph preceding the light bulb and searches PubMed to create a list of related articles. References within an OMIM entry are linked to the complete citation at the end of the entry. The PubMed ID at the end of the OMIM reference is linked to the PubMed abstract and in some instances to the full text of the article if the journal is online. Following the references is a list of credits and edit history: the Creation Date lists the date when the entry was created and the name of the author; authors who subsequently contribute additions or changes to the entry are given credit in the Contributors field; changes made by the editorial staff are documented in the Edit History field.
Allelic Variants with functional significance are maintained within the relevant gene entry. Allelic variants are given a 10-digit number: the six-digit number of the parent entry followed by a decimal point and a unique four-digit variant number. For most genes, only selected mutations are included as specific subentries. Criteria for inclusion are: first several mutations to be discovered, high population frequency, distinctive phenotype, historic significance, unusual mechanism of mutation, unusual pathogenetic mechanism and distinctive inheritance [e.g. dominant with some mutations, recessive with other mutations in the same gene (see OMIM entry 601079)]. Most of the allelic variants represent disease-producing mutations. A few polymorphisms are included, mainly those that show statistical association with particular common disorders. As of September 13, 2004, OMIM listed 12 715 allelic variants in 1651 entries.
THE OMIM GENE MAP
OMIM's Synopsis of the Human Gene Map is maintained as a convenience to users and focuses on the ‘Morbid Map’, i.e. the mapping of disorders. In chromosome-by-chromosome tabular form, the OMIM Synopsis of the Human Gene Map lists for each gene, the cytogenetic location, gene symbols, method(s) of mapping and disorder(s) related to the specific gene. Links to the sequence-based MapViewer are available from the cytogenetic location in the Gene Map. The Morbid Map is an alphabetical tabular listing of all mapped disorders. As of September 13, 2004, there were at least 3659 disorders spread across 2558 loci. In 2563 of these disorders, the molecular basis has been identified at the DNA level. These 2563 disorders of known molecular basis are distributed over the 1651 loci with at least one allelic variant; many genes are the site of more than one mutation causing phenotypically distinct disorders (e.g. cystic fibrosis and congenital bilateral absence of the vas deferens caused by different mutations in CFTR). The Morbid Map may be displayed from within NCBI's MapViewer program to allow integration with finer physical and sequence-based maps of the human genome.
As a continuously updated repository of intersecting data on genes and disorders, OMIM offers help to clinicians, students and researchers in unraveling the complex relationship between genes and disease. OMIM's priorities continue to be to provide timely information about single gene disorders and select complex diseases, along with the molecular bases underlying them, and to provide descriptions of genes with known function.
Acknowledgments
ACKNOWLEDGEMENTS
Funding is provided by the National Human Genome Research Institute (NHGRI) through a contract administered by the NLM (N01-LM-43504). Maintenance of OMIM is also supported by licensing fees, and the terms of these arrangements are being managed by the Johns Hopkins University in accordance with its conflict of interest policies.
REFERENCES
- 1.McKusick V.A. (1998) Mendelian Inheritance in Man. A Catalog of Human Genes and Genetic Disorders, 12th edn. Johns Hopkins University Press, Baltimore, MD. [Google Scholar]