Abstract
ProDom is a comprehensive database of protein domain families generated from the global comparison of all available protein sequences. Recent improvements include the use of three-dimensional (3D) information from the SCOP database; a completely redesigned web interface (http://www.toulouse.inra.fr/prodom.html); visualization of ProDom domains on 3D structures; coupling of ProDom analysis with the Geno3D homology modelling server; Bayesian inference of evolutionary scenarios for ProDom families. In addition, we have developed ProDom-SG, a ProDom-based server dedicated to the selection of candidate proteins for structural genomics.
INTRODUCTION
The ProDom protein domain family database originates from the early recognition that automated methods are needed to reach comprehensiveness of protein domain analysis (1–3). This comprehensiveness makes ProDom a unique resource usefully complementing expert derived databases, such as PROSITE (4), PFAM (5) or SMART (6), thereby helping to sustain the rapid growth of InterPro (7,8). In recent years, we have developed relationships between ProDom and three-dimensional (3D) structural information, both for ProDom construction and in the ProDom user interface.
METHODS USED TO CONSTRUCT ProDom
ProDom2004.1 was built anew from the SWISS-PROT41.23 and TrEMBL24.11 databases, essentially as described previously (2,7). In the first stage, well-characterized domain families were used to recruit homologous domains using PSI-BLAST. Among these families, 21 families were derived from in-house expertise and 1352 structural domain families were selected from SCOP release 1.63 (9) using the ASTRAL compendium (10) on the basis of the following criteria: (i) length homogeneity (the shortest sequence should be at most 25% shorter than the longest); (ii) sequence homogeneity (family diameter below 450 PAM); (iii) the family should contain at least two different domains; (iv) domains should not contain internal repeats; and (v) they should not be longer than 500 amino acids. In the second stage, domain families were automatically clustered using the MKDOM2 program (2). The resulting protein domain families were aligned using an improved parallelized program called ProDomAlign, developed in C++ using OpenMP. ProDomAlign is based on MultAlin (11), a program well suited to align very large sequence families (thousands of sequences). Multiple alignments were assessed using the norMD objective function proposed by Thompson et al. (12). Other consistency indicators include family diameter and radius of gyration as proposed previously (3). Each family is identified by a unique accession number, which is followed across successive ProDom releases using the MatchDom program (7). Links among ProDom, InterPro (8) and Pfam-A (5) were calculated using MatchDom. Links with PROSITE were calculated using pftools (4). Matches with the Protein Data Bank (PDB) (13) were more difficult to maintain because of sequence numbering inconsistencies. We, therefore, searched the PDB for sequence identity with ProDom domains using the PDB ATOM record rather than SEQRES, in order to superimpose sequence and structural information. The current ProDom2004.1 release covers 726 272 protein sequences and contains 186 303 domain families with two or more domains.
THE GRAPHICAL USER INTERFACE ON THE ProDom WEBSITE
The ProDom website was completely redesigned in order to get a more ergonomic user interface. The main ProDom form consists of two parts. The first part (ProDom Browsing) allows querying of ProDom in a variety of ways: (i) by accession number (Display a ProDom entry); (ii) by the display of all proteins belonging to one or several ProDom families with logical AND/OR operators (All proteins in ProDom families); (iii) by related databases (InterPro, PROSITE, PFAM or PDB); (iv) by SWISS-PROT/TrEMBL identifier or accession number; and (v) by keyword search with AND/OR operators. The output is either information on a given domain family (Figure 1) or cartoons displaying the domain arrangements of all proteins matching the query (Figure 2). The number of different cartoons available for domain display was increased from 14 160 to 237 888 with the use of 64 colours, providing for more legible outputs while preserving consistency across different displays.
Figure 1.
Information, tools and links available for each ProDom domain family. (A) Tools and links, from left to right: domain arrangements for all proteins belonging to this family; corresponding family in ProDom-CG; family in the MSF or FASTA file formats; download PSI-BLAST generated profile; run Predict Protein on family (20); and family display enhanced with ESPript (21). (B) General information on domain family: most frequent protein names and descriptors, length of multiple sequence alignment, number of domains, consistency indicators (3) and norMD assessment of alignment quality (12). (C) Links to InterPro, PROSITE, PFAM-A and PDB. (D) Visualization of ProDom domains on 3D structure.
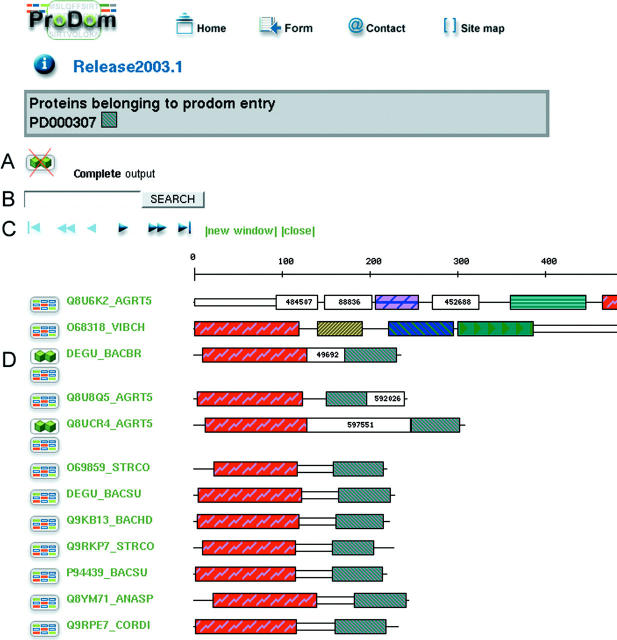
Figure 2.
Domain arrangements of proteins in a ProDom family. (A) Proteins sharing the same domain architecture are grouped in order to simplify the output. This feature can be disabled to display a complete output. (B) String search in SWISS-PROT identifiers or accession numbers allows to highlight the corresponding proteins in the display. (C) Navigation bar to browse the output, with at most 200 different domain arrangements displayed in a single page. (D) The left icon on each line is linked to a display of all proteins sharing at least one homologous domain with the current protein. When relevant, another icon points to the list of all proteins sharing the same domain architecture. Domain cartoons are hypertext-linked to the corresponding ProDom domain families (Figure 1) whose accession number is also available on mouse-over together with a short description.
The ProDom graphical interface also provides for the display of ProDom domains on 3D structures (Figure 1). It is possible to display one or all ProDom domains, either on one polypeptide chain or on all chains, with different colour codes for different domain families. These domain-enhanced structures can be displayed with Rasmol (14), MDL Chime or in VRML (Virtual Reality Modeling Language), provided the corresponding helper applications have been installed. Alternatively, they can be rendered as static images from three different angles generated with the help of DSSP (15) and MOLSCRIPT (16). Users may also choose to define particular viewing angles and opt for stereo display.
The second part of the main ProDom form allows for BLAST searches in ProDom (Compare your sequence with ProDom), suggesting a possible domain arrangement for any query protein. When 3D structures are available for target domains, the output is directly linked to both SWISS-MODEL (17) and Geno3D (18) servers for homology-based domain modelling.
ProDom-CG: RESTRICTION OF ProDom TO COMPLETED GENOMES
ProDom-CG is a subset of ProDom, restricted to sequences derived from completely sequenced genomes. Bacterial protein sets (19) were retrieved from the ExPASy server (ftp://www.expasy.org/databases/hamap/complete_proteomes), while eukaryotic protein sets were retrieved from the EBI server (http://www.ebi.ac.uk/integr8). All relevant multiple alignments and characteristics were recalculated on the resulting families. In order to provide insight into the evolution of domain families, we used a Bayesian network methodology to infer the most probable evolutionary scenario for each family. Such a scenario may be complex, including domain loss or horizontal transfer events. The taxonomy tree encompassing completely sequenced genomes was colour-coded so as to indicate ancestral nodes predicted to contain domains in a given ProDom-CG family. These colour-coded trees are available for each ProDom-CG entry on the ProDom website.
ProDom-SG FOR STRUCTURAL GENOMICS
In the framework of structural genomics projects, it is extremely useful to identify potential targets unlikely to share homology to already known structures. We, therefore, developed the ProDom-SG (Structural Genomics) server, designed to assist in the selection of protein domain families corresponding to potentially new folds on the basis of lack of detected homology. The server also allows for the identification of favourable protein candidates for crystallization studies. ProDom-SG was built in three steps. In the first step, only ProDom families with norMD values above 0.5 were considered. In the second step, potential homology relationships between ProDom families were identified using PSI-BLAST with family specific, position-specific scoring matrices. When applicable, the existence of such related families is indicated using a specific logo appearing at the top of the family information sheet (in field A, Figure 1). In the third step, both direct and indirect links to the PDB were recorded for each family. A direct link implies that an experimental structure is available for at least one domain in the ProDom family, whereas an indirect link reflects the existence of structural information for at least one domain in a related family. These relationships are stored in a PostgreSQL database that can be accessed on the ProDom-SG website and can be queried by keyword or species of interest. The user can retrieve ProDom families either linked or not linked to the PDB, directly or indirectly. Thus ProDom-SG provides for inspection of domain families on the basis of structure availability, which allows to characterize protein families containing a known fold. Conversely, ProDom-SG provides a quick handle on candidate domain families for which no structural information is available nor can be readily inferred. Such families are indicated by a ProDom-SG logo appearing at the top of the family information sheet (in field A, Figure 1). ProDom families retrieved can be further filtered on the basis of sequence homogeneity (family diameter) and the number of domains in the family. It is also possible to restrict the search to ProDom families containing at least one mono-domain protein, thus obviating the need for engineering individual domains separately before crystallization attempts.
AVAILABILITY AND LICENSING
The ProDom database is copyrighted by INRA and CNRS. ProDom is freely accessible at http://www.toulouse.inra.fr/prodom.html but commercial users need to sign a license agreement for download and local usage.
Acknowledgments
ACKNOWLEDGEMENTS
The ProDom project was supported by the ‘Programme de Bio-Informatique Inter-Organismes’, the ‘Réseau des Génopoles’ and the European Union.
REFERENCES
- 1.Sonnhammer E.L.L. and Kahn,D. (1994) Modular arrangement of proteins as inferred from analysis of homology. Protein Sci., 3, 482–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gouzy J., Corpet,F. and Kahn,D. (1999) Whole genome protein domain analysis using a new method for domain clustering. Comput. Chem., 23, 333–340. [DOI] [PubMed] [Google Scholar]
- 3.Corpet F., Servant,F., Gouzy,J. and Kahn,D. (2000) ProDom and ProDom-CG: tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res., 28, 267–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hulo N., Sigrist,C.J., Le Saux,V., Langendijk-Genevaux,P.S., Bordoli,L., Gattiker,A., De Castro,E., Bucher,P. and Bairoch,A. (2004) Recent improvements to the PROSITE database. Nucleic Acids Res., 32, D134–D137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bateman A., Coin,L., Durbin,R., Finn,R.D., Hollich,V., Griffiths-Jones,S., Khanna,A., Marshall,M., Moxon,S., Sonnhammer,E.L. et al. (2004) The Pfam protein families database. Nucleic Acids Res., 32, D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Letunic I., Copley,R.R., Schmidt,S., Ciccarelli,F.D., Doerks,T., Schultz,J., Ponting,C.P. and Bork,P. (2004) SMART 4.0: towards genomic data integration. Nucleic Acids Res., 32, D142–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Servant F., Bru,C., Carrere,S., Courcelle,E., Gouzy,J., Peyruc,D. and Kahn,D. (2002) ProDom: automated clustering of homologous domains. Brief Bioinformatics, 3, 246–251. [DOI] [PubMed] [Google Scholar]
- 8.Mulder N.J., Apweiler,R., Attwood,T.K., Bairoch,A., Barrell,D., Bateman,A., Binns,D., Biswas,M., Bradley,P., Bork,P. et al. (2003) The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res., 31, 315–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andreeva A., Howorth,D., Brenner,S.E., Hubbard,T.J., Chothia,C. and Murzin,A.G. (2004) SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res., 32, D226–D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandonia J.M., Hon,G., Walker,N.S., Lo Conte,L., Koehl,P., Levitt,M. and Brenner,S.E. (2004) The ASTRAL Compendium in 2004. Nucleic Acids Res., 32, D189–D192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corpet F. (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res., 16, 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson J.D., Plewniak,F., Ripp,R., Thierry,J.C. and Poch,O. (2001) Towards a reliable objective function for multiple sequence alignments. J. Mol. Biol., 314, 937–951. [DOI] [PubMed] [Google Scholar]
- 13.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sayle R.A. and Milner-White,E.J. (1995) RASMOL: biomolecular graphics for all. Trends Biochem. Sci., 20, 374. [DOI] [PubMed] [Google Scholar]
- 15.Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- 16.Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst., 24, 946–950. [Google Scholar]
- 17.Schwede T., Kopp,J., Guex,N. and Peitsch,M.C. (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res., 31, 3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Combet C., Jambon,M., Deleage,G. and Geourjon,C. (2002) Geno3D: automatic comparative molecular modelling of protein. Bioinformatics, 18, 213–214. [DOI] [PubMed] [Google Scholar]
- 19.Gattiker A., Michoud,K., Rivoire,C., Auchincloss,A.H., Coudert,E., Lima,T., Kersey,P., Pagni,M., Sigrist,C.J., Lachaize,C. et al. (2003) Automated annotation of microbial proteomes in SWISS-PROT. Comput. Biol. Chem., 27, 49–58. [DOI] [PubMed] [Google Scholar]
- 20.Rost B. and Liu,J. (2003) The PredictProtein server. Nucleic Acids Res., 31, 3300–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gouet P., Robert,X. and Courcelle,E. (2003) ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res., 31, 3320–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]