Abstract
We recently discovered that the Warburg effect, defined by the dramatically enhanced metabolism of glucose to pyruvate, even in well-oxygenated cancer cells, can occur as a consequence of mutations that enhance lipid biosynthesis at the expense of respiratory capacity. Specifically, mutations in the E1 subunit of either of two respiratory enzymes, pyruvate dehydrogenase (PDC) or α-ketoglutarate dehydrogenase (KGDC), change substrate specificity from the 3-carbon α-ketoacid pyruvate, or the 5-carbon α-ketoacid α-ketoglutarate, to a 4-carbon α-ketoacid oxaloacetate (OADC). These mutations result in OADC-catalyzed synthesis of malonyl-CoA (MaCoA), the essential precursor of all fatty acids. These mutants arose as spontaneous suppressors of a yeast acc1cs cold-sensitive mutation encoding an altered form of AcCoA carboxylase (Acc1) that fails to produce MaCoA at the restrictive temperature (16°C). Notably, these suppressors are respiratory defective as a result of the same nuclear mutations that suppress acc1cs. These mutants also suppress sensitivity to Soraphen A, a potent inhibitor of Acc1 activity, at normal temperature (30°C). To our knowledge, OADC activity has never been identified in eukaryotic cells. Our results offer a novel perspective on the Warburg effect: the reprogramming of energy metabolism in cancer cells as a consequence of mutational impairment of respiration to meet the fatty acid requirements of rapidly proliferating cells. We suggest OADC activity is a common feature of cancer cells and represents a novel target for the development of chemotherapeutics.
Keywords: AcCoA carboxylase, Cancer cell metabolism, α-Ketoacid dehydrogenase, Warburg effect
Introduction
Enormous strides have been made over the past several decades in our understanding of cancer biology. Hanahan and Weinberg published a comprehensive review of the “hallmarks” of cancer in 2000 (Hanahan and Weinberg, 2000). These hallmarks, however, addressed exclusively regulatory and signaling processes. The Warburg effect was not mentioned in this review. Only in the past decade or so has there been a resurgence of interest in the Warburg effect and its role in cancer. In their updated review, Hanahan and Weinberg included two additional hallmarks of cancer: (i) defense against immune destruction and (ii) modifications in energy metabolism (Hanahan and Weinberg, 2011). The latter – aberrant energy metabolism – is a trait common to all cancers that was initially described in the 1920s (reviewed in (Warburg, 1956b)). Understanding the Warburg effect, specifically how and why cancer cells metabolize glucose by “aerobic glycolysis,” rather than by respiration, is critically important to reach a fundamental understanding of cancer. Efforts directed toward this goal are imperative, as cancer remains a major health problem worldwide. Indeed, cancer recently surpassed heart disease as the leading cause of death in the United States (Twombly, 2005).
Warburg recognized that aerobic glycolysis is not a consequence of oxygen deprivation, but occurs even in well-oxygenated tumor cells (Warburg, 1956a, b). Indeed, the Warburg effect has been exploited clinically since 1976 as the basis of the PET (positron emission tomography) scan that detects uptake of the radiolabelled, glucose analog 2-18F-2-deoxyglucose (FDG) by cancer cells (Fowler and Ido, 2002). The Warburg effect seems to be a common metabolic thread that runs through all or nearly all cancers. Nonetheless, despite having been identified more than 90 years ago, the biochemical basis of the Warburg effect remains poorly understood and largely unexplained.
The Warburg effect established that the limiting factor in cancer cell growth is not energy production in the form of ATP, but metabolic intermediates derived from glucose that serve as building blocks for cell proliferation (Jiang and Deberardinis, 2012). During the past decade, attempts have been made to rationalize the Warburg effect based on diversion of carbon metabolism away from energy production and toward the synthesis of metabolic intermediates. As a notable example, Cantley and colleagues identified a splice site isoform of pyruvate kinase (PK M2) that is important for tumor metabolism and proliferation (Christofk et al., 2008). They proposed that PK M2 restricts the terminal reaction in glycolysis, thereby facilitating exit of upstream glycolytic intermediates into other pathways essential for cell growth (Vander Heiden et al., 2009). However, the pathways and metabolites critical for rapid cell growth and division remain undefined.
α-Ketoacid dehydrogenase complexes
All cells contain α-ketoacid dehydrogenase complexes that catalyze the irreversible oxidative decarboxylation of α-ketoacids yielding CoA derivatives and reduced NADH. The most well-characterized of these enzymes is the pyruvate dehydrogenase complex (PDC), an extraordinary complex composed of three enzymatic activities (E1, decarboxylase; E2, transacetylase; and E3 dehydrogenase) and five cofactors (thiamine, CoA, NAD+, FAD and lipoid acid) that link glycolysis with the citric acid cycle by catalyzing oxidation of the 3-carbon α-ketoacid, pyruvate, to produce AcCoA (Patel et al., 2014):
| [1] |
The citric acid cycle includes an analogous reaction catalyzed by the α-ketoglutarate dehydrogenase complex (KGDC). KGDC catalyzes oxidation of the 5-carbon α-ketoacid, α-ketoglutarate, to succinyl-CoA:
| [2] |
An analogous α-ketoacid dehydrogenase complex recognizes 5-carbon and 6-carbon α-ketoacids involved in branched chain amino acid metabolism (Kumaran et al., 2013). Remarkably, no analogous 4-carbon α-ketoacid dehydrogenase has ever been described in eukaryotic cells; indeed, there is only a single, unsubstantiated report of a 4-carbon α-ketoacid dehydrogenase complex in bacteria (Laakel et al., 1994). Why have cells evolved dehydrogenase complexes that metabolize 3-, 5- and 6-carbon α-ketoacids, while essentially no organisms have acquired a 4-carbon α-ketoacid dehydrogenase complex? This question underlies our hypothesis regarding the Warburg effect (below).
Fatty acid biosynthesis
The first step in fatty acid biosynthesis is the carboxylation of AcCoA to yield MaCoA, catalyzed by AcCoA carboxylase (Acc1) in a reaction that requires the energy of ATP hydrolysis:
| [3] |
This reaction is irreversible and the first committed step in fatty acid biosynthesis. This is the only known reaction that catalyzes the synthesis of MaCoA. As such, it is the principal site of fatty acid regulation and is subject to extraordinary regulatory parameters, including hormonal response to insulin/glucagon, Acc1 phosphorylation status, ATP energy charge, CoA:AcCoA ratio, citrate, and feedback inhibition by the pathway end product, palmitate (Natter and Kohlwein, 2013). Activation of Acc1 even involves polymerization of Acc1 into filaments, an effect that might underlie the cold-sensitivity of the acc1cs mutation that is central to this study (Guthrie et al., 1969). Synthesis of MaCoA by OADC, however, would circumvent Acc1 regulation, yielding MaCoA in a manner unregulated by normal parameters.
Synthesis of MaCoA is followed by the elongation phase of fatty acid synthesis, catalyzed by the multienzyme fatty acid synthase complex (FAS) (Wu et al., 2014). The first step is transfer of acetyl- and malonyl-groups from AcCoA and MaCoA to ACP, forming AcACP and MaACP. The FAS condensing enzyme then generates acetoacetyl-ACP:
| [4] |
AcACP is the two-carbon backbone on which fatty acid synthesis occurs with AcACP, a three-carbon compound serving as the two-carbon donor in a cyclic series of reactions (reaction 4). This cycle continues until the 16-carbon compound, palmitoyl-ACP, is formed. This compound is then a substrate for the thioesterase that hydrolyzes palmitoyl-ACP to yield the fatty acid palmitate. In effect, the thioesterase acts as a ruler to determine fatty acid chain length. All other fatty acids are derived from palmitate.
Hypothesis
We propose that OADC arises as a new enzyme by mutation(s) that alters the substrate specificity of the E1 subunit of either PDC (pyruvate) or KGDC (α-ketoglutarate), such that oxaloacetate is now recognized as substrate and MaCoA is generated as product:
| [5] |
The apparent absence of OADC in normal cells is especially intriguing as oxaloacetate is not only plentiful, but is a “crossroad” metabolite produced and consumed in several metabolic pathways including gluconeogenesis and the citric acid cycle. However, OADC would yield MaCoA, potentially bypassing the strict regulation imposed on fatty acid biosynthesis by circumventing the rate-limiting Acc1-catalyzed reaction.
An appealing aspect of this hypothesis is that it accounts for two key aspects of the Warburg effect: (i) enhanced uptake of glucose by cancer cells; and (ii) impaired respiratory capacity. Although many of the glucose metabolites would exit the glycolytic pathway to be used as substrates to produce carbon building blocks essential for cell division (Jiang and Deberardinis, 2012; Vander Heiden et al., 2009), much of the glucose would be metabolized via glycolysis to pyruvate and subsequently be converted to oxaloacetate by pyruvate carboxylase, the first enzyme in the pathway to gluconeogenesis:
| [6] |
Mutations in the genes encoding the E1 subunit of either pyruvate dehydrogenase or α-ketoglutarate dehydrogenase would account for both the impaired respiratory capacity of cancer cells, and the ability to produce the massive amount of fatty acids, estimated to be as much as 40% of the biomass of replicating cells, as a consequence of OADC bypassing the normal regulation of MaCoA synthesis (Wu et al., 2014).
Experimental Approach
Isolation and genetic analysis of acc1cs suppressors
We are taking an unbiased genetic approach to address our hypothesis, similar to the strategy that led to our identification of gene loops (Hampsey et al., 2011). Specifically, we selected for suppressors of the cold-sensitive growth phenotype of an acc1cs mutant (H-470) that fails to grow on glucose medium at 16°C (Csm− phenotype) as the result of a missense mutation encoding a G1783A amino acid substitution in Acc1 (Schneiter et al., 2000). This mutation depletes MaCoA due to impaired Acc1 activity at the restrictive temperature. To date, we have isolated 53 independent, spontaneous (no mutagen) suppressors of the Csm− phenotype. Remarkably, 51 of the 53 strains spontaneously fail to respire, defined by the inability to grow on medium containing 3% glycerol as the sole carbon source (Gly− phenotype).
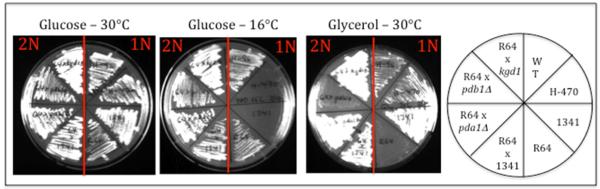
When backcrossed to an acc1cs mutant of opposite mating type (strain YMH1341), the resulting diploid strains grow at 16°C, indicating that the acc1cs suppressors are dominant gain-of-function mutation(s) with respect to MaCoA synthesis, yet these strains recover the ability to respire, indicating that the pleiotropic Gly− phenotype is the result of recessive loss-of-function mutation(s). This is precisely what our hypothesis predicts: bypass of the acc1cs defect by a novel pathway to MaCoA synthesis (dominant, retained in the presence of wild type allele of the suppressor gene) comes at the expense of respiratory capacity (recessive, restored in the presence of the wild type allele of the suppressor gene). These phenotypes are depicted for one of our suppressor (R64) suppressor in Figure 1.
Fig. 1.
Growth phenotypes for the R64 suppressor of acc1cs. The right panel depicts growth of strains on 2% glucose medium at 30°C (2 days); the middle panel depicts growth on 2% glucose at 16°C (10 days); the right panel depicts growth on 3% glycerol medium (3 days). The red lines divide diploid (2N) and haploid (1N) strains. Key: WT (ACC1+); H-470 (MATα acc1cs); 1341 (MATa acc1cs); R64 (acc1cs pdb1sup). Haploid strains show the tight Csm− phenotype of acc1cs, and the effective suppression of Csm− by the R64 suppressor (middle panel). The R64 × 1341 backcross shows that the R64 suppressor is recessive with respect to Gly−, but dominant with respect to Csm+. The other three diploid strains show that the R64 Gly− phenotype is not complemented when crossed with a pdb1Δ deletion strain, but is complemented with crossed with pda1Δ and kgd1Δ strains. This is compelling genetic evidence that the R64 suppressor is a pdb1 allele.
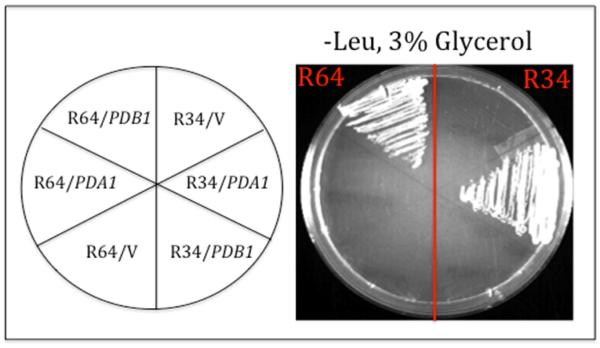
Similar complementation results demonstrate that the R34 suppressor is an allele of PDA1. Complementation data using the pda1Δ and pdb1Δ deletions mutants were confirmed by a reciprocal genetic experiment. The Gly-phenotypes of the haploid repressor strains were rescued when plasmid-borne wild-type copies of PDA1 and PDB1 were introduced into the suppressor strains: PDA1 rescued the Gly-phenotype of R34, but not R64, whereas PDB1 rescued R64, but not R34 (Fig. 2). Taken together, these results demonstrate that acc1cs is bypassed in R34 by a pda1 suppressor allele, whereas acc1cs is bypassed in R64 by a pdb1 allele.
Fig. 2.
Complementation of acc1cs suppressor strains R34 and R64. Growth phenotypes of acc1cs suppressor strains R34 and R64 into which a LEU2 vector alone (V) or the same vector carry the wild type PDA1 or PDB1 gene was introduced. Transformants were selected on –Leu, 2% glucose medium, then scored for respiratory competence on –Leu, 3% glycerol medium. Growth was photographed after two days incubation at 30°C.
We have currently identified 14 of the 51 acc1cs suppressors as alleles of either PDA1 or PDB1, encoding altered forms of the E1 subunit of either PDC or KGD. We have begun to sequence the suppressor genes from these strains. We predict that the pda1 and pdb1 suppressor mutations encode amino acid replacements at the interface of the α (pda1) and β (pdb1) subunits of the E1 decarboxylase component of PDC, and that this interface comprises the catalytic domain. Similarly, the kgd1 alleles would encode amino acid replacement(s) within the catalytic center of the E1 decarboxylase component of KDC.
Suppressors of the acc1cs relieve Soraphen toxicity
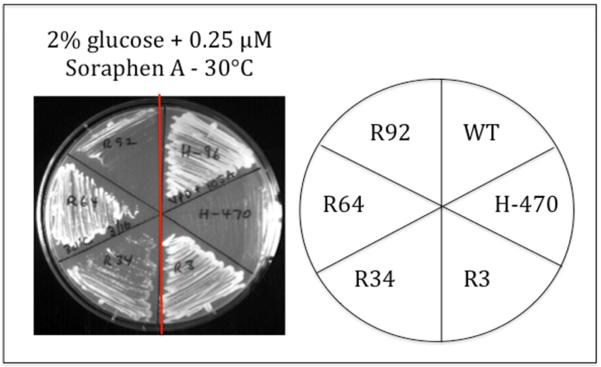
Wild type ACC1 strains are extremely sensitive to the antifungal polyketide Soraphen A, defined by failure to grow in the presence of as little as 1 μM Soraphen A. Soraphen A is an Acc1 inhibitor and effectively depletes cells of MaCoA (Schneiter et al., 2000). This phenotype is especially dramatic for the acc1cs mutant (H-470), which is normally permissive for growth at 30°C (Fig. 3). However, in the presence of as little as 250 nM Soraphen A, the acc1cs mutant fails to grow even at permissive temperature.
Fig. 3.
Suppression of Soraphen A toxicity. The acc1cs mutant (H-470) is extremely sensitive to Soraphen A, a potent inhibitor of AcCoA carboxylase, even at the permissive temperature of 30°C. This growth defect is effectively suppressed by the acc1cs suppressor strains R3, R34, R64 and, to a lesser extent, R92. The plate was photographed after 2 days incubation at 30°C. Note: The H-470 (acc1cs) sensitivity to Soraphen A is at 30°C, identical to the Csm− phenotype of H-470 in the absence of Soraphen A at 16°C (Fig. 1).
If our acc1cs suppressors are bypassing Acc1 to synthesize MaCoA by the novel OADC activity, then acc1cs suppressors should exhibit resistance to Soraphen A. Remarkably, all 14 of our defined acc1cs suppressors are resistant to Soraphen A, even at 4-fold the concentration required for inhibition of growth of the acc1cs mutant. These phenotypes are depicted for suppressor strains R3, R34, R64 and R92 (Fig. 3). We conclude that diminished cellular levels of MaCoA, due either to genetic (acc1cs) or to phenotypic (Soraphen A) inhibition of Acc1, are enhanced by OADC-catalyzed decarboxylation of oxaloacetate, rather than by Acc1-catalyzed carboxylation of AcCoA. These conclusions are consistent with a novel explanation for the Warburg effect: cancer cells enhance fatty acid biosynthesis by sacrificing respiratory capacity, and this physiological change is permanent, the result of mutations, rather than a potentially reversible regulatory response to physiological signals.
The Cancer Genome Atlas
If the Warburg effect is the result of mutations that encode amino acid replacements in respiratory enzymes, homologous to those that we have identified in yeast, then comparable mutations should be prevalent in tumor cells. We are screening The Cancer Genome Atlas (TCGA; NIH) (http://cancergenome.nih.gov), as well as the Catalog of Somatic Mutations in Cancer (COSMIC) (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/), two continually updated repositories of mutations in cancer cells, for mutations in genes encoding E1 subunit of PDC or KGD. Our initial scan of these two resources reveals that mutations do indeed occur at loci encoding α-ketoacid dehydrogenases in the genome of cancer cell. Moreover, these mutations are not randomly distributed, as might be expected for mutations that simply diminish respiratory capacity, but cluster in limited exonic regions, consistent with the possibility that these mutations not only diminish respiratory capacity, but also alter substrate specificity. Moreover, alignment of the encoded amino acid replacements in the E1 subunit of human KGD with the crystal structure of bacterial KGD, is consistent with these mutations lying in the catalytic center. Interestingly, the x-ray structure of bacterial E1 KGD includes a molecule of oxaloacetate within the active center of the enzyme (Frank et al., 1995). Although oxaloacetate is not a substrate of the normal E1 enzyme, this structure establishes that oxaloacetate can be accommodated within the E1 catalytic center. We are especially interested to know if the human E1 amino acid replacement(s), modeled onto the bacterial enzyme, would physically contact oxaloacetate in a manner that would suggest oxaloacetate decarboxylase activity.
Much more extensive sequence analysis of cancer cells is available from targeted sequence analysis of ~330 established oncogenes or tumor suppressor genes. These genes, however, do not include genes encoding subunits of PDA, PDB or KGD as they have not (yet) been identified as target cancer genes. Using our yeast sequence data and sequence analysis from TCGA and COSMIC we plan to sequence the specific exonic regions of these genes that are predicted to comprise their catalytic centers. We are especially interested to know if these mutations are likely, based on 3D structural alignments, to alter the catalytic activity such that oxaloacetate might be the substrate and MaCoA the product.
Biochemical evidence for OADC activity
All genetic data will be confirmed by biochemical data. We are using established assays for α-ketoacid dehydrogenase complexes from crude yeast extracts (Repetto and Tzagoloff, 1989, 1990). If our hypothesis is correct, we will detect PDC and KGD activity using pyruvate and α-ketoglutarate as substrates, respectively, from wild type and acc1cs cell extracts (reaction [1] and [2] above) by assaying α-ketoacid- and CoA-dependent reduction of NAD+ to NADH. If, however, acc1cs suppressor mutations altered substrate specificity, we anticipate diminished activity in the presence of the normal substrates, and enhanced activity in the presence of oxaloacetate.
Precedent for novel enzymatic activity in cancer cells
There is precedent for a cancer-associated mutation changing the catalytic specificity of a respiratory enzyme. Cantley and colleagues discovered mutations in the human gene encoding isocitrate dehydrogenase (IDH), resulting in loss of the enzymatic activity that converts isocitrate to α-ketoglutarate, and acquisition of a new enzymatic activity that catalyzes NADPH-dependent generation of 2-hydroxyglutarate (2HG) from α-ketoglutarate (Dang et al., 2009). These mutations encode a single amino acid replacement (R132H) within the catalytic center of isocitrate dehydrogenase. 2HG is associated with formation and progression of malignant gliomas, although the biochemical basis of this effect remains to be established. This is in contrast to our studies, where we propose a change in the catalytic specificity of a respiratory enzyme that in altered form synthesizes the rate-limiting metabolite (MaCoA) in fatty acid biosynthesis. We propose that bypassing the highly regulated Acc1 activity by an alternative pathway for MaCoA synthesis is important, or even essential, for the formation and/or progression of some (perhaps many) cancers, especially those undergoing rapid proliferation, or in clinical terms, exhibiting a high mitotic index.
Summary
The yeast Saccharomyces cerevisiae is a model organism often touted as offering the “awesome power of yeast genetics” not only to elucidate fundamental processes in molecular biology, but to provide mechanistic insight into human diseases (e.g., (Folkmann et al., 2014)). The physiology of a yeast cell and a proliferating cancer cell are remarkably similar, with comparable metabolic fluxes in these two cell systems (Natter and Kohlwein, 2013). The Crabtree effect, which describes the preferential metabolism of glucose by fermentation rather than respiration, is comparable to the Warburg effect, except that the Crabtree effect is reversible in yeast, whereas the Warburg effect is irreversible, the consequence of genetic mutations (Vander Heiden et al., 2009). It is also noteworthy that the incidence of cancer and mortality rates correlate in many cases with blood glucose levels (Jee et al., 2005; Kellenberger et al., 2010; Krone and Ely, 2005; Liu et al., 2011; Sieri et al., 2012). Hence, increased glucose levels appear to be associated with the switch of neoplastic cells from respiration to aerobic glycolysis, reciprocal to the effect that occurs when yeast reach the diauxic shift and switch from fermentation of glucose to metabolism of ethanol by respiration.
Although mammalian cells are able to take up all major biomass constituents – including glucose, amino acids, fatty acids and cholesterol from the bloodstream – proliferation of cancer cells seems to rely primarily on the endogenous synthesis of these components, as indicated by the irreversible genetic switch to fermentation (Warburg, 1956b). Furthermore, several steps in lipid synthesis have been recognized as being crucial for rapidly growing cancer as well as for yeast cells, emphasizing the metabolic similarities between both types of cells (Natter and Kohlwein, 2013).
The recent resurgence of interest in the Warburg effect and cancer cell metabolism in general is remarkable in that the effect was first discovered in 1923. Warburg knew that this effect is not due simply to oxygen deprivation in poorly vascularized tumors, but also occurs even in well-oxygenated tumor cells, including hematologic malignancies. The biochemical basis of the Warburg effect could not have been understood in 1923. But in 1956, subsequent to the elucidation of the glycolytic pathway and citric acid cycle, the genetic and biochemical basis of the Warburg effect remained unresolved (Warburg, 1956a). This irony is further underscored by the clinical importance of the Warburg effect in detection of tumors by PET scans, a clinical technique based on the massive uptake of glucose by cancer cells, detected as 18F-2-deoxyglucose (FDG) in whole body scans (Vander Heiden et al., 2009).
Several innovative concepts underlie our hypothesis: (i) the realization that no 4-carbon α-ketoacid dehydrogenase complex has been reported in eukaryotic cells; (ii) that such an activity would generate MaCoA, the rate-limiting substrate in fatty acid biosynthesis; (iii) that cells rely on up-regulation of fatty acid synthesis for membrane genesis in rapidly proliferating cells; (iv) that the need for MaCoA could be met by a novel reaction, mechanistically different from the reaction catalyzed by Acc1; and (v) that the substrate for this reaction, oxaloacetate, can readily be produced in a single step from pyruvate (catalyzed by pyruvate carboxylase), independent of respiration. Furthermore, the NADPH essential for fatty acid synthesis can be generated by oxidation of glucose in the pentose phosphate pathway (Patra and Hay, 2014). Integration of these concepts led to the principal hypothesis in this proposal: cancer cells acquire mutations in the E1 subunits of α-ketoacid dehydrogenase complexes, resulting in recognition of oxaloacetate as substrate and synthesis of MaCoA as product.
Acknowledgments
We are indebted to Steve Brill (Rutgers University), Yuh-Hwa Wang (University of Virginia Medical School) and Liang Tong (Columbia University) for valuable discussion during the course of this work and for strains and reagents. This work was supported by NIH grant R01 GM39484 and by NIH IMSD Award – Rutgers University Pipeline Program (R25 GM55145).
Abbreviations
- Acc1
AcCoA carboxylase
- FAS
fatty acid synthase complex
- KGD
pyruvate dehydrogenase complex
- PDC
pyruvate dehydrogenase complex
- OADC
oxaloacetate dehydrogenase complex
- MaCoA
malonyl-CoA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors declare no competing conflicts of interest.
References
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkmann AW, Dawson TR, Wente SR. Insights into mRNA export-linked molecular mechanisms of human disease through a Gle1 structure-function analysis. Advances in biological regulation. 2014;54:74–91. doi: 10.1016/j.jbior.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler JS, Ido T. Initial and subsequent approach for the synthesis of 18FDG. Seminars in nuclear medicine. 2002;32:6–12. doi: 10.1053/snuc.2002.29270. [DOI] [PubMed] [Google Scholar]
- Frank DJ, Tyree CM, George CP, Kadonaga JT. Structure and function of the small subunit of TFIIF (RAP30) from Drosophila melanogaster. Journal of Biological Chemistry. 1995;270:6292–6297. doi: 10.1074/jbc.270.11.6292. [DOI] [PubMed] [Google Scholar]
- Hampsey M, Singh BN, Ansari A, Laine JP, Krishnamurthy S. Control of eukaryotic gene expression: gene loops and transcriptional memory. Advances in enzyme regulation. 2011;51:118–125. doi: 10.1016/j.advenzreg.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Jee SH, Ohrr H, Sull JW, Yun JE, Ji M, Samet JM. Fasting serum glucose level and cancer risk in Korean men and women. JAMA : the journal of the American Medical Association. 2005;293:194–202. doi: 10.1001/jama.293.2.194. [DOI] [PubMed] [Google Scholar]
- Jiang L, Deberardinis RJ. Cancer metabolism: When more is less. Nature. 2012;489:511–512. doi: 10.1038/489511a. [DOI] [PubMed] [Google Scholar]
- Kellenberger LD, Bruin JE, Greenaway J, Campbell NE, Moorehead RA, Holloway AC, Petrik J. The role of dysregulated glucose metabolism in epithelial ovarian cancer. Journal of oncology. 20102010:514310. doi: 10.1155/2010/514310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krone CA, Ely JT. Controlling hyperglycemia as an adjunct to cancer therapy. Integrative cancer therapies. 2005;4:25–31. doi: 10.1177/1534735404274167. [DOI] [PubMed] [Google Scholar]
- Kumaran S, Patel MS, Jordan F. Nuclear magnetic resonance approaches in the study of 2-oxo acid dehydrogenase multienzyme complexes--a literature review. Molecules (Basel, Switzerland) 2013;18:11873–11903. doi: 10.3390/molecules181011873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laakel M, Lebrihi A, Khaoua S, Schneider F, Lefebvre G, Germain P. Relationship between valine, fatty acids, and spiramycin biosynthesis in Streptomyces ambofaciens. Can J Microbiol. 1994;40:672–676. doi: 10.1139/m94-106. [DOI] [PubMed] [Google Scholar]
- Liu H, Ma Q, Li J. High glucose promotes cell proliferation and enhances GDNF and RET expression in pancreatic cancer cells. Molecular and cellular biochemistry. 2011;347:95–101. doi: 10.1007/s11010-010-0617-0. [DOI] [PubMed] [Google Scholar]
- Natter K, Kohlwein SD. Yeast and cancer cells - common principles in lipid metabolism. Biochimica et biophysica acta. 2013;1831:314–326. doi: 10.1016/j.bbalip.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MS, Nemeria NS, Furey W, Jordan F. The Pyruvate Dehydrogenase Complexes: Structure-based Function and Regulation. The Journal of biological chemistry. 2014;289:16615–16623. doi: 10.1074/jbc.R114.563148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra KC, Hay N. The pentose phosphate pathway and cancer. Trends in biochemical sciences. 2014;39:347–354. doi: 10.1016/j.tibs.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repetto B, Tzagoloff A. Structure and regulation of KGD1, the structural gene for yeast alpha-ketoglutarate dehydrogenase. Molecular and cellular biology. 1989;9:2695–2705. doi: 10.1128/mcb.9.6.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repetto B, Tzagoloff A. Structure and regulation of KGD2, the structural gene for yeast dihydrolipoyl transsuccinylase. Molecular and cellular biology. 1990;10:4221–4232. doi: 10.1128/mcb.10.8.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R, Guerra CE, Lampl M, Tatzer V, Zellnig G, Klein HL, Kohlwein SD. A novel cold-sensitive allele of the rate-limiting enzyme of fatty acid synthesis, acetyl coenzyme A carboxylase, affects the morphology of the yeast vacuole through acylation of Vac8p. Molecular and cellular biology. 2000;20:2984–2995. doi: 10.1128/mcb.20.9.2984-2995.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieri S, Muti P, Claudia A, Berrino F, Pala V, Grioni S, Abagnato CA, Blandino G, Contiero P, Schunemann HJ, et al. Prospective study on the role of glucose metabolism in breast cancer occurrence. International journal of cancer Journal international du cancer. 2012;130:921–929. doi: 10.1002/ijc.26071. [DOI] [PubMed] [Google Scholar]
- Twombly R. Cancer surpasses heart disease as leading cause of death for all but the very elderly. Journal of the National Cancer Institute. 2005;97:330–331. doi: 10.1093/jnci/97.5.330. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On respiratory impairment in cancer cells. Science. 1956a;124:269–270. [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956b;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Wu X, Qin L, Fako V, Zhang JT. Molecular mechanisms of fatty acid synthase (FASN)-mediated resistance to anti-cancer treatments. Advances in biological regulation. 2014;54:214–221. doi: 10.1016/j.jbior.2013.09.004. [DOI] [PubMed] [Google Scholar]