ABSTRACT
Sumoylation is an essential post-translational modification in Arabidopsis thaliana, which entails the conjugation of the SUMO protein onto lysine residues in target proteins. In Arabidopsis, 2 closely related genes, SUMO1 and SUMO2, act redundantly and are in combination essential for plant development, i.e. the combined loss of SUMO1 and SUMO2 results in embryo-lethality. To circumvent this lethality, SUMO2 was previously knocked down in a sumo1 knockout background by expressing an artificial microRNA that targets SUMO2 (amiR-SUMO2). This sumo1/2KD line with low SUMO2 levels represents a valuable genetics tool to investigate SUMO function in planta. Here, we re-sequenced the whole-genome of this sumo1/2KD line and identified 2 amiR-SUMO2 insertions in this line, which were confirmed by PCR-genotyping. Identification of these 2 insertions enables genetics with this tool.
KEYWORDS: Arabidopsis, SUMO, T-DNA insertions, whole genome resequencing
Introduction
Sumoylation is a post-translational modification resulting in conjugation of SUMO (Small Ubiquitin-like Modifier) proteins onto targets through the side chain of lysine residues. SUMO is encoded by a single copy gene in many eukaryotes like budding yeast (Saccharomyces cerevisiae), Caenorhabditis elegans and fruit fly (Drosophila melanogaster).1 In contrast to these species, the genome of Arabidopsis (Arabidopsis thaliana) contains 8 SUMO genes. SUMO1 and SUMO2 are the main isoforms used for sumoylation.2-5 They act redundantly and are essential in Arabidopsis, as both the sumo1–1 and sumo2–1 single null mutants do not display any aberrant development phenotype, while the corresponding double mutant is embryo-lethal.4 To understand the function of sumoylation in planta, we created a transgenic line where SUMO1 is knocked out (KO) and SUMO2 knocked down (KD).6 These lines were obtained by crossing the sumo1–1 null mutant with the SUMO2KD line B, a line silenced for SUMO2 using an artificial microRNA (amiR) targeting SUMO2 transcripts (amiR-SUMO2); this amiR-SUMO2 was engineered according to the instructions of WMD MicroRNA Designer: http://wmd3.weigelworld.org/cgi-bin/webapp.cgi.7 The sumo1–1 SUMO2KD mutant (hereafter called sumo1/2KD) displays a strong phenotype characterized by enhanced accumulation of salicylic acid (SA), accumulation of the Pathogenesis-Related proteins 1 and 2 (PR1/2), spontaneous cell death in leaves, early flowering, partial sterility and a dwarf stature.6 Although SUMO2 conjugation levels are strongly suppressed in this line, the low levels of SUMO2 protein are apparently sufficient to maintain plant viability.
As the insertion site of the amiR-SUMO2 construct is unknown, genotyping of ‘SUMO2KD allele’ was till now based on the assessment of the presence of the amiR-SUMO2 construct using PCR and on segregation for kanamycin-resistance in seedlings (the plant selection marker that was co-integrated with the amiR-SUMO2 construct). Genetics with sumo1/2KD is, therefore, tedious: homozygous lines can only be found by examining the segregation for kanamycin resistance in the next generation.
While out-crossing the sumo1/2KD line to different mutant backgrounds, we noted that the dwarf phenotype segregated in the resulting F2 generation, albeit the F2 plants were genotyped as homozygous for the sumo1–1 and amiR-SUMO2 alleles. As stable transformation of Arabidopsis can result in multiple T-DNA insertions,8 we reasoned that the original sumo1/2KD line might contain multiple amiR-SUMO2 integration sites. Variation in the number of insertions potentially leads to different SUMO2 silencing levels and could explain the heterogeneous phenotype of the F2 progenies. Identification of the insertion sites is, therefore, needed for reverse genetics with the original sumo1/2KD line. Here we report on the amiR-SUMO2 insertion locations in the genome of the sumo1/2KD line B21, which facilitates classical genetics with this line. We have identified both the number of insertions and their localization based on whole genome re-sequencing of the sumo1/2KD line using next-generation sequencing. By mapping the generated sequencing reads, we found 2 genomic insertions. Using PCR-based genotyping, we could confirm the location of both insertions in the sumo1/2KD line. Using these PCR primers, the presence of both amiR-SUMO2 insertions can now be quickly assessed in the offspring of out-crosses with this sumo1/2KD line.
Materials and methods
Genomic DNA extraction, re-sequencing and short read mapping
We isolated gDNA from pools of seedlings of sumo1/2KD from van den Burg et al.6 using the Nucleospin II plant kit (Macherey-Nagel). The gDNA isolation yielded 38.8 ng/uL, with A260/280 ratios of 1.87 and A260/230 ratios of 2.47. The gDNA was sequenced according to the manufacturer instructions on the Ion Torrent platform (ThermoFischer). The obtained short sequencing reads (average length 150 bp) were then mapped onto both Arabidopsis genomic sequence (TAIR10) and the amiR-SUMO2 plasmid using the CLC workbench v6.5 software by applying the strategy outlined in Fig. 1A. The parameters used for mapping were: miss match cost = 2; insertion cost = 3; deletion cost = 3.
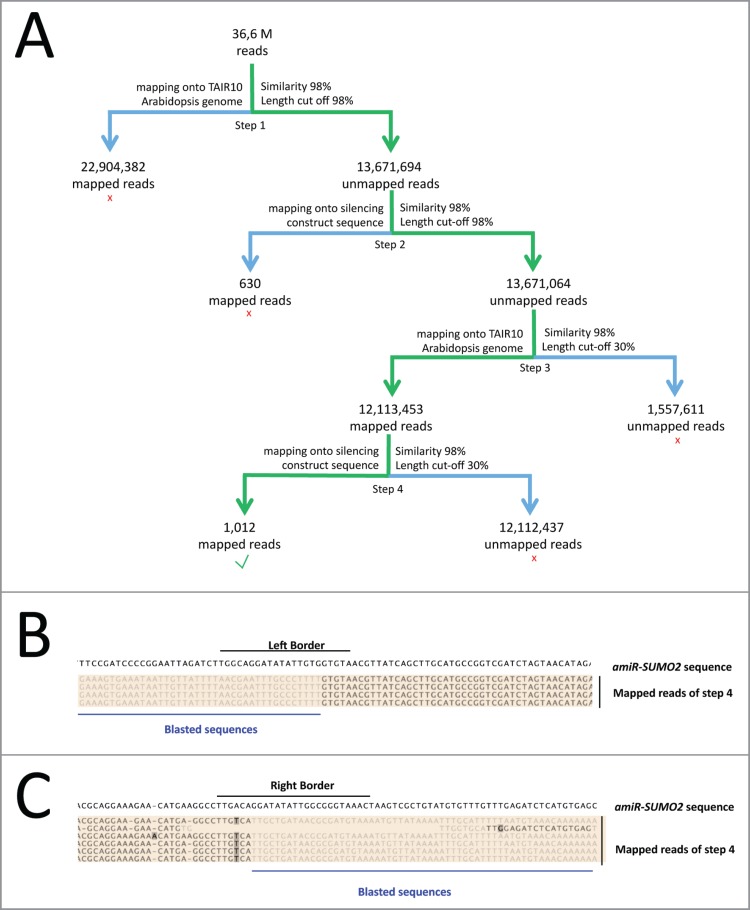
Figure 1.
Identification of the 2 amiR-SUMO2 insertion sites using NGS sequencing. (A) Pipeline used to identify the T-DNA insertion sites. (Step 1) Reads were mapped onto the Arabidopsis genome assembly (TAIR10), using a sequence similarity cut-off of > 98% and read length cut-off of > 98% sequence overlap. (Step 2) To remove the reads that fully matched to the amiR-SUMO2 construct, the unmapped reads were then mapped to the amiR-SUMO2 construct using similar parameters. (Step 3) We then selected in the remaining set of unmapped reads, the reads that partially mapped to the Arabidopsis genome, using >98% similarity and a length cut-off of > 30%. (Step 4) The retained reads (from Step 3) were then mapped to the amiR-SUMO2 construct, with >98% similarity and a length cut-off of > 30% to obtain the reads that map across single integration site boundaries with at least 30 bp. We found 1,012 reads, which mapped to 2 different genomic sites. The insertions were identified by blast searches with these latter reads using the part of the reads that did not map onto the amiR-SUMO2 construct. (B) and (C) Visualization of the mapped reads of Step 4 from panel A (shown on color background) on the SUMO2 silencing construct sequence at the Left Border (B) and Right Border (C). Within the mapped reads of step 4, black sequences indicates the regions of the reads that map onto the amiR-SUMO2 construct, while gray sequences indicate the regions of the reads from Step 4 that do not map on the amiR-SUMO2 construct.
Primer design and PCR genotyping
PCR genotyping was performed on sumo1–1, SUMO2KD line B (i.e., parental lines), and 2 sumo1/2KD lines: sumo1/2KD line B21 and sumo1/2KD line B22#1. Both are lines obtained from the same cross between sumo1–1 and SUMO2KD. Primer sequences and primer combinations used for genotyping are summarized in Table 1. SUMO1 genotyping was done with the primers 3039 and 6541 for the wild type SUMO1 (SUMO1WT) allele, and primers 6541 and 3249 for sumo1–1. PFK7 genotyping was done with primers 4904 and 4980 for PFK7 wild type (PFK7WT), 4904 and 4719 for amiR-SUMO2 in PFK7 (PFK7amiR-SUMO2). proCYP98A3 genotyping was done with the primers 5733 and 5578 for proCYP98A3 wild type (proCYP98A3WT), 5733 and 4714 for amiR-SUMO2 in proCYP98A3 (proCYP98A3amiR-SUMO2). The fragments were amplified using a touch-down PCR (35 cycles): (i) a melting temperature of 95°C for 30s, (ii) an annealing temp of 60°C with -1°C each cycle for 10 cycles and then 25 additional cycles at 50°C, (iii) an elongation time of 1m 15s at 72°C, and back to (i).
Table 1.
(A) Primer combinations used for genotyping of the different alleles and (B) sequences of the primers used for PCR genotyping.
| A | ||
|---|---|---|
| Locus | Allele | primer name |
| SUMO1 | SUMO1WT | 3039 |
| 6541 | ||
| sumo1–1 | 6541 | |
| 6249 | ||
| PFK7 | PFK7WT | 4904 |
| 4980 | ||
| PFK7amiR-SUMO2 | 4904 | |
| 4719 | ||
| proCYP98A3 | proCYP98A3WT | 5733 |
| 5578 | ||
| proCYP98A3amiR-SUMO2 | 5733 | |
| 4714 | ||
| B | ||
| primer name |
primer sequence (5′ to 3′) |
|
| 3039 | TCTGCAAACCAGGAGGAAG | |
| 4714 | CATTAATGAATCGGCCAACGCGCG | |
| 4719 | TCGCCTTCTTGACGAGTTCTTCTGA | |
| 4904 | AGTTTCTTGGGGCCTAAGGATACA | |
| 4980 | AGTGTGAAAAAACATATACAAGAAC | |
| 5578 | CACCGCTATTAGAAACCACGAC | |
| 5733 | CAGCAGACGAAACCAACAACACT | |
| 6249 | TGGTTCACGTAGTGGGCCATCG | |
| 6541 | TAGGATCCGATACCAAACGAACAA | |
Results
After extraction of the gDNA from the sumo1/2KD line B21 used in van den Burg et al.6 samples were sequenced using next-generation sequencing. We obtained 36.6 M reads with a median length of 177 bp. To localize the genome insertion sites of the amiR-SUMO2 T-DNA, we identified the reads that partially (i.e., >30%) mapped to both the Arabidopsis genome and the amiR-SUMO2 construct (Fig. 1). Briefly, we first removed the reads that align for >98% with either the Arabidopsis genome or with the amiR-SUMO2 construct. From the remaining reads, we then selected the reads that partially mapped onto the amiR-SUMO2 construct (minimum overlap of 30%). These reads were then mapped onto the Arabidopsis genome allowing again a min. match of 30%. With this method 1,012 reads remained (Fig. 1A). Some of these reads partially mapped onto the amiR-SUMO2 construct at the Left (LB) or Right Border (RB) sequence. The sequence fragment of these latter reads, which could not be mapped to the plasmid, was then blasted to NCBI to identify their location in the Arabidopsis genome. Considering the reads that mapped onto the LB (Fig. 1B) or RB (Fig. 1C) of the amiR-SUMO2 construct, we identified 2 locations: (i) the 3’UTR of PFK7 (AT5G56630), a gene coding for PHOSPHOFRUCTOKINASE 7 located on chromosome 5, and (ii) the promoter region of CYP98A3 (proCYP98A3; AT2G40890) located on chromosome 2. Our sumo1/2KD line contains, therefore, 2 different insertions located on 2 different chromosomes. Apparently, both T-DNA integration events were retained after crossing with sumo1–1. The first site is 587 bp upstream of the start codon (-578) of the gene CYP98A3, while the second site is 2,531 bp downstream of the start codon (+2,531) of the gene PFK7.
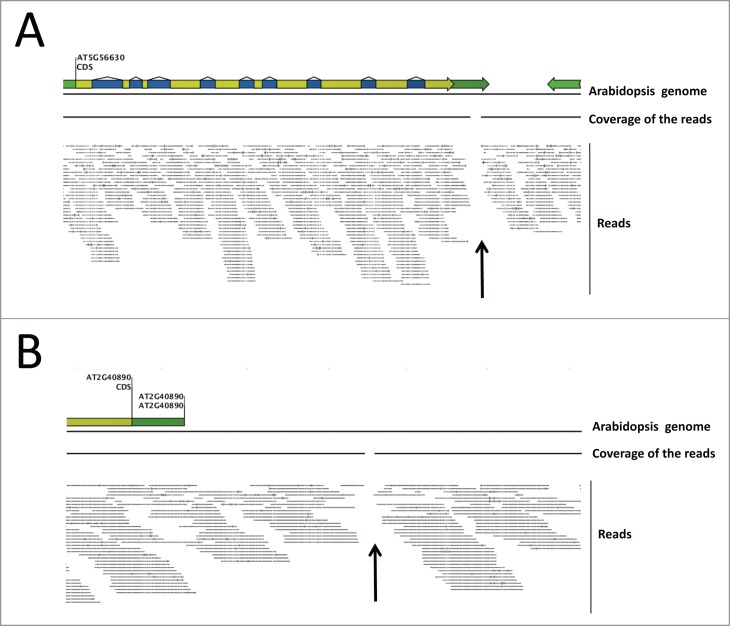
The 36,6 M reads were then mapped onto the TAIR10 Arabidopsis genome assembly with a similarity of 98% and a length cut-off of 98%. When we visualized the reads onto the Arabidopsis genome using CLC workbench (Figs. 2A and B), we observed a gap in the read coverage (black arrows) exactly at the expected PFK7 and proCYP98A3 insertion locations, while the surrounding gDNA is nicely covered by reads. Both insertions are, therefore, present in a homozygous state in the previously reported sumo1/2KD line.6
Figure 2.
The SUMO2 silencing construct is homozygous at both the PFK7 (AT5G56630) and proCYP98A3 (AT2G40890) integration site in the sumo1/2KD line (B21). The 36.6 M short sequencing reads were mapped onto the Arabidopsis genome assembly (TAIR10), with a similarity match of > 98% and length cut-off of > 98%. Visualization of the reads on (A) PFK7 genomic and (B) CYP98A3 promoter (proCYP98A3) sequences shows the gap in read coverage in the 2 identified insertion sites (black arrows).
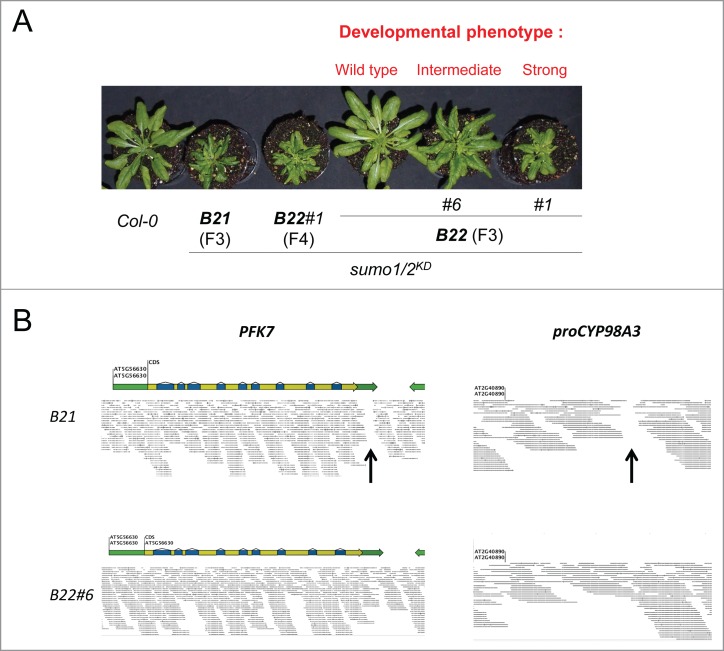
The original cross between sumo1–1 and SUMO2KD yielded a F2 population, which included 2 F2 sister plants: B21 (the original line detailed in van den Burg et al.6) and B22. Whereas selfings of B21 show no segregation for the reported strong ‘sumo1/2KD developmental’ phenotype, selfings of the B22 F2 plant (i.e., F3 generation) displayed 3 different phenotypes: normal plants, plants with an intermediate rosette size (e.g., B22#6) and ‘B21-like’ plants (e.g., B22#1) (Fig. 3A). The developmental phenotype of B22#1 proofed to be stable in the next generation. However, the phenotype of B22#6 continued segregating in normal, intermediate, and ‘B21-like’ plants. Using next-generation sequencing, we then also sequenced a pool of the B26#6 progeny that all had the intermediate phenotype. Using our bioinformatics pipeline, we could confirm the presence of the silencing construct at both integration sites (PFK7 and proCYP98A3), meaning that both insertions were still present in the parental plant B22#6. The obtained sequencing reads were then mapped onto the TAIR10 Arabidopsis genome assembly with a similarity of 98% and a length cut-off of 98%. Upon visualization of the reads onto the Arabidopsis genome, we found individual reads that span across either of the 2 insertion sites, meaning that both insertions were still heterozyogous in B22#6 (Fig. 3B). Combined with the result of outcrossing the B21 line, we conclude that both amiR-SUMO2 integration events need to be present in a homozygous state to obtain a strong developmental phenotype as seen with the SUMO1/2KD B21 line.
Figure 3.
Both the amiR-SUMO2 integration events (i.e., at PFK7 and proCYP98A3) need to be present in a homozygous state to obtain a strong sumo1/2KD phenotype. (A) Seedlings from the self-pollination offspring of sumo1/2KD lines B21 (F3 generation); B22 (F3 generation); and B22#1 (F4 generations) were grown for 4 weeks in short day conditions (11 hours in day light / 13 hours in dark). The lines B21 and B22 represent 2 sister plants obtained from the same cross between sumo1–1 and SUMO2KD line B; B22#1 and B22#6 exemplify selfings from the plant B22. Whereas the strong developmental phenotype did not segregate for the progeny of B21, the progeny of B22 displayed 3 different phenotypes: normal, intermediate, and strong (i.e., ‘B21-like’). A set of B22#6 intermediate plants was pooled and their combined gDNA was sequenced using next-generation sequencing. (B) Visualization of the reads obtained from re-sequencing of the pool of the B22#6 at the PFK7 and proCYP98A3 promoter (proCYP98A3) genomic regions reveals no gap in the read coverage for both insertion sites (bottom row), while a gap in the read coverage is visible for both genomic regions in the case of the B21 plant (top row, black arrows).
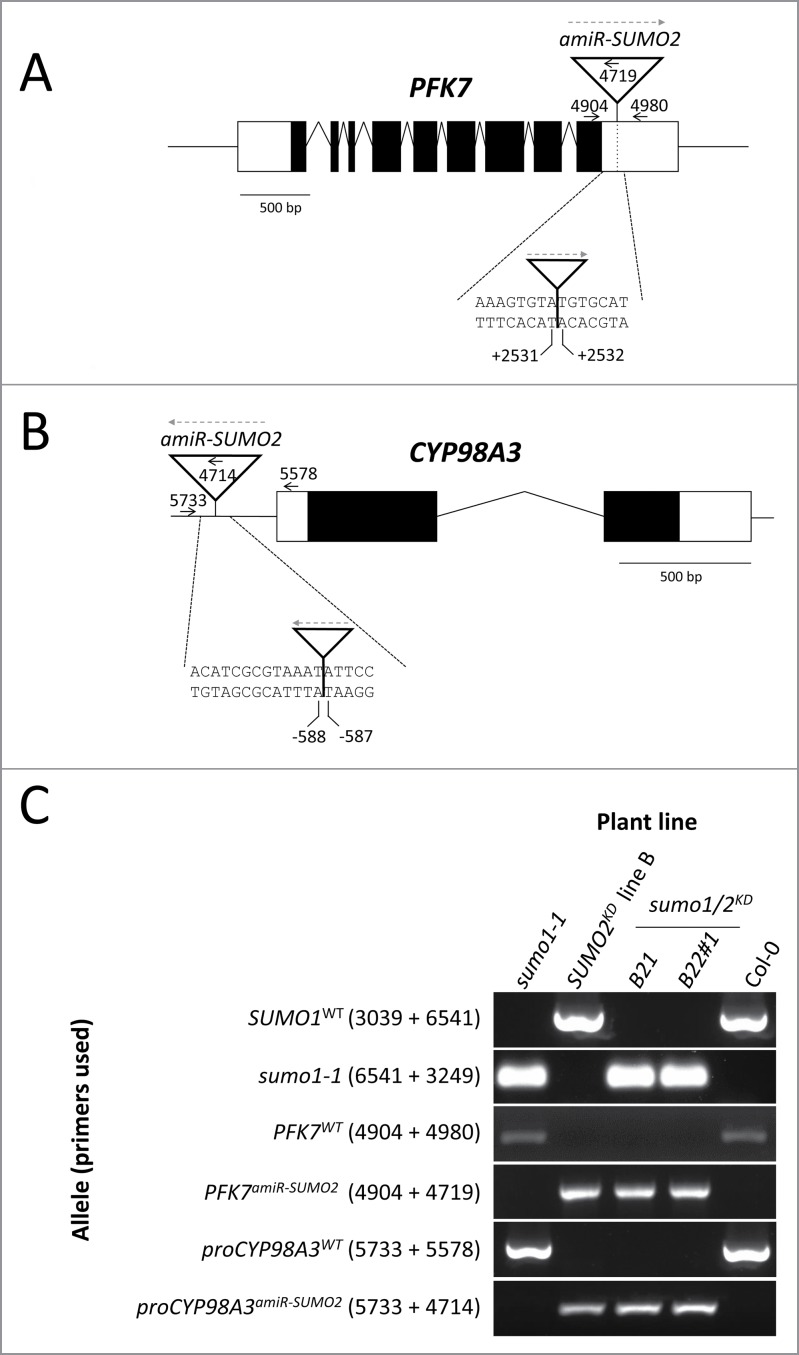
Subsequently, we developed primers to genotype for both insertion sites. These primer pairs either (i) amplify the genomic region surrounding the T-DNA integration site or (ii) amplify a fragment that encompasses both the amiR-SUMO2 T-DNA and the flanking genomic region (Fig. 4A; B and Table 1). Using the primer pairs 4904+4980, 4904+4719, 5733+5578 and 5733+4714, we could genotype for the PFK7WT, PFK7amiR-SUMO2, proCYP98A3WT and proCYP98A3amiR-SUMO2 alleles. Using these primers, we then confirmed our next generation sequencing result that both insertion sites are homozygous in the 2 sumo1/2KD lines: sumo1/2KD B21 and sumo1/2KD B22#1 (Fig. 4C).
Figure 4.
PCR-based genotyping of the PFK7 and proCYP98A3 alleles in the sumo1/2KD line. A and B. True to scale diagrams of the PFK7 and CYP98A3 genes. The amiR-SUMO2 integration sites (arrowheads) are located in (A) PFK7 at +2,531 bp and (B) in CYP98A3 –587 bp, calculated from the start codon (+1). The exons and introns are presented by boxes and broken lines, respectively. The white boxes reflect the 5’- and 3’-untranslated regions, while the black boxes refer to the coding regions. The primers used for genotyping are indicated by small black arrows with their ID numbers given (not to scale; see also Table 1). The orientation of the amiR-SUMO2 constructs is indicated using gray-dash arrows (from the Left Border to the Right Border). (C) PCR-based genotyping using the primers represented in (A) and (B) of the SUMO2KD line B and sumo1/2KD lines B21 and B22#1, 2 lines obtain from the same cross between sumo1–1 and SUMO2KD line B. See also Table 1 for primer sequences.
Discussion
Traditionally, Southern-blotting is used to reveal the number of T-DNA insertions, while TAIL-PCR (Thermal asymmetric interlaced-PCR) is used to identify the integration sites.9 However, TAIL-PCR does not guarantee identification of all integration sites. Here, we identified by whole genome re-sequencing followed by the mapping of sequencing reads using bioinformatics that the sumo1/2KD line contains 2 amiR-SUMO2 constructs, and we identified their exact genomic locations. Finally, we established a PCR-based genotyping approach for both amiR-SUMO2 integration sites. Knowing that the cost of whole genome (re-)sequencing has dramatically decreased over the last years, this constitutes a powerful and rapid method to localize and genotype T-DNA insertions in transgenic lines of e.g., Arabidopsis.
Prior this study, the genotyping of the sumo1/2KD line relied on the assessment of the presence of the amiR-SUMO2 construct by PCR, and on the segregation for the kanamycin-resistance, which was co-integrated with the amiR-SUMO2 construct. Repeatedly, we observed intermediate phenotypes (similar to the intermediate phenotype observed in the line B22#6) for the sumo1/2KD transgenic plants (curled leaves, reduced rosette size) when we out-crossed this mutant to other genetic backgrounds,6 despite the fact that the F2 plants were found to be homozygous for both sumo1–1 and kanamycin-resistance and that they contained the amiR-SUMO2 construct. Hence, the silencing of SUMO2 does not only show semi-dominance, but we also noted that lines homozygous for kanamycin-resistance segregated for the morphological phenotype. The here presented data indicates that both amiR-SUMO2 insertions contribute to the original sumo1/2KD phenotype. Genetics with the sumo1/2KD line must consequently take into consideration that both amiR-SUMO2 integrations (at the PFK7 and proCYP98A3 loci) are needed for phenotypic comparisons when using this line.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Jeffrey Ham for his help with genotyping. We are also thankful to Peter van Dam for the valuable inputs in the bioinformatics works.
References
- 1.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem 2013; 82:357-385; PMID:23746258; http://dx.doi.org/ 10.1146/annurev-biochem-061909-093311 [DOI] [PubMed] [Google Scholar]
- 2.Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung DY, Vierstra RD. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem 2003; 278:6862-6872; PMID:12482876; http://dx.doi.org/ 10.1074/jbc.M209694200 [DOI] [PubMed] [Google Scholar]
- 3.Lois LM, Lima CD, Chua NH. Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. Plant cell 2003; 15:1347-1359; PMID:12782728; http://dx.doi.org/ 10.1105/tpc.009902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saracco SA, Miller MJ, Kurepa J, Vierstra RD. Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol 2007; 145:119-134; PMID:17644626; http://dx.doi.org/ 10.1104/pp.107.102285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammoudi V, Vlachakis G, Schranz ME, van den Burg HA. Whole-genome duplications followed by tandem duplications drive diversification of the protein modifier SUMO in Angiosperms. New Phytologist 2016; 211:172-85; PMID:26934536; http://dx.doi.org/ 10.1111/nph.13911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van den Burg HA, Kini RK, Schuurink RC, Takken FLW. Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell 2010; 22:1998-2016; PMID:20525853; http://dx.doi.org/ 10.1105/tpc.109.070961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant cell 2006; 18:1121-1133; PMID:16531494; http://dx.doi.org/ 10.1105/tpc.105.039834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castle LA, Errampalli D, Atherton TL, Franzmann LH, Yoon ES, Meinke DW. Genetic and molecular characterization of embryonic mutants identified following seed transformation in Arabidopsis. Mol Gen Genet 1993; 241:504-514; PMID:8264525; http://dx.doi.org/7550382 10.1007/BF00279892 [DOI] [PubMed] [Google Scholar]
- 9.Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis-thaliana T-DNA insert junctions by thermal asymmetric interlaced PRC. Plant J 1995; 8:457-463; PMID:7550382; http://dx.doi.org/ 10.1046/j.1365-313X.1995.08030457.x [DOI] [PubMed] [Google Scholar]