ABSTRACT
The cyanobacterium Fremyella diplosiphon possesses 3 genes encoding homologs of the tryptophan-rich sensory protein (TSPO). TSPO proteins are membrane proteins implicated in stress responses across a range of organisms from bacteria to humans. Diverse TSPO proteins appear to generally bind tetrapyrrole ligands. Previously, we reported that one of these homologs, FdTSPO1, is involved in salt-, osmotic- and oxidative stress responses in F. diplosiphon. Here, we show distinct regulation of cellular mRNA levels of all 3 FdTSPO homologs by different abiotic stresses. Given the prior finding that all 3 FdTSPO proteins are capable of binding tetrapyrroles of functional relevance in F. diplosiphon and the observation of a ligand-dependent functional role for FdTSPO1 in vivo, FdTSPO1, FdTSPO2, and FdTSPO3 appear to have distinct, yet overlapping, roles in vivo. We propose that these proteins regulate tetrapyrrole homeostasis and/or tetrapyrrole-modulated functions in F. diplosiphon in response to multiple environmental stresses.
KEYWORDS: Abiotic stress, cyanobacteria, diurnal, light, nitrogen availability, stress regulation, temperature, tryptophan-rich sensory protein (TSPO)
Introduction
Fremyella diplosiphon is a filamentous, freshwater cyanobacterium that serves as a model organism for complementary chromatic acclimation (CCA). CCA is a process by which the organism exhibits distinct physiologic and morphological changes primarily in response to green light (GL) and red light (RL), an acclimation that allows maximum light absorption and promotes productivity in different depths where either RL or GL is more abundant.1-3 F. diplosiphon carries 3 genes encoding proteins with homology to the nearly ubiquitous tryptophan-rich sensory protein/translocator protein 18kDa (TSPO).4 TSPO homologs have largely been studied in mammals and non-oxygenic photosynthetic bacteria, and the general role of TSPO proteins appears to be in organismal responses to stress where determined.5 We previously established a role for TSPO1 in F. diplosiphon (FdTSPO1) in stress- and light-regulated processes.6 We confirmed GL-dependent upregulation of FdTSPO1 and established physiologic roles for FdTSPO1 in stress-regulated processes including salt-, oxidative- and osmotic-dependent stresses6 and in ligand-dependent organismal responses.4 Based on quantitative real-time polymerase chain reaction (qRT-PCR) analyses, FdTSPO1 transcript was more abundant transiently after treatment with the salt sodium chloride, the osmoticum sorbitol, and the oxidative stress-inducing agent methyl viologen (MV).6 Recently, we reported the presence of 2 additional F. diplosiphon TSPO genes (FdTSPO2 and FdTSPO3) that are more similar in sequence to each other and smaller in size than the first identified TSPO in F. diplosiphon.4 Such a diverse complement of TSPO genes is relatively rare and among cyanobacteria appears to occur primarily in filamentous, nitrogen-fixing strains.4 All 3 encoded FdTSPO proteins bind functionally relevant tetrapyrroles, although with apparently distinct binding efficiencies.4 This observation, together with the observed disparate light-dependent regulation of the expression of FdTSPO genes, suggests potentially distinct, if partially overlapping, functions of these 3 homologs in vivo. Here, we describe novel stresses, including abiotic and nutrient stresses, that influence TSPO mRNA abundance in growing cells and describe distinctions in the impact of these factors on the regulation of the 3 distinct FdTSPO genes in F. diplosiphon. This survey of the environmental factors that impact FdTSPO expression is a first appraisal of the potentially distinct roles these genes may play in fine-tuning organismal responses to stress.
Various environmental cues have different impacts on distinct FdTSPO homologs
Light-dependent regulation of TSPOs
FdTSPO1 is upregulated under GL compared with RL conditions.4,6,7 A ∼6.5-fold induction under GL compared with RL was observed in microarray analyses,7 a ∼2.5-fold induction under GL was reported in RNA-sequencing based analysis6 and a ∼2-fold induction under GL observed by qRT-PCR.6 We recently determined that neither FdTSPO2 nor FdTSPO3 appear to be significantly regulated by these light qualities, as levels of mRNA accumulating in GL vs. RL was not significantly different for either gene.4 However, cellular levels of FdTSPO2 and FdTSPO3 mRNA were significantly higher than those observed for FdTSPO1,4 potentially indicating distinct regulation and functions for FdTSPO1 vs. FdTSPO2/FdTSPO3.
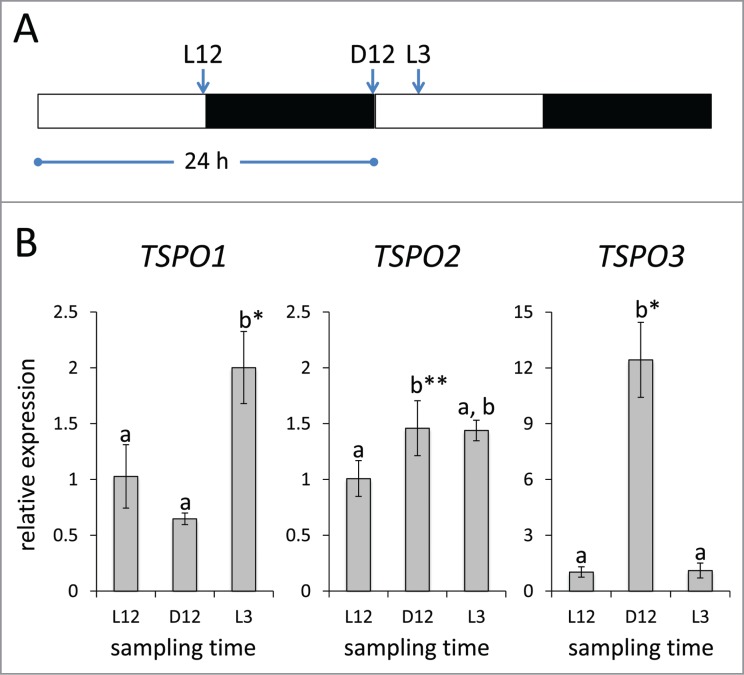
Recent research in Synechocystis sp. PCC 6803, a single-celled non-chromatically acclimating cyanobacterium, reported oscillation of the expression of the Synechocystis TSPO homolog with peak expression from very early (i.e., from 1 to 3 h) to near the middle (i.e., at 5 h) of the light phase under a 12 h diurnal cycle.8 We, thus, grew F. diplosiphon cells in diurnal cycles (12 h of white light [WL] at ∼10 µmol m−2 s−1 and 12 h of dark) and tested TSPO transcript accumulation. As an initial survey of the potentially distinct regulation of these genes under diurnal conditions, we determined TSPO mRNA levels 12 h into the light period (L12), 12 h into the dark period (D12) and 3 h into the light period (L3) (for schematic see Fig. 1A). FdTSPO1 mRNA levels increased (∼2-fold) at L3 compared with L12 or D12. FdTSPO2 was slightly, yet significantly, increased (∼1.5-fold) at D12 compared with L12, but was not significantly different from either L12 or D12 at L3 (Fig. 1B). FdTSPO3 exhibited the strongest response of all 3 homologs. Whereas there was no significant difference between mRNA levels for L3 vs. L12, FdTSPO3 transcript abundance was over 12-fold increased at the end of the dark period (D12) compared with light conditions (Fig 1). Although a more extensive analysis is necessary to confirm diurnal regulation of TSPO, together these results show that TSPO mRNA levels peak early during the light period (FdTSPO1) or directly before the onset of light (FdTSPO2/ FdTSPO3), suggesting a role for these proteins during dark/light transitions. A role for TSPO in the switch from oxygenic to anaerobic photosynthetic growth in Rhodobacter sphaeroides has been reported previously.9 Together, these data support roles for TSPO proteins in temporal or transitory responses to changing environmental conditions.
Figure 1.
Light-dependent regulation of FdTSPO transcript levels under diurnal cycles. Samples for quantitative real-time reverse transcription PCR (qRT-PCR) were taken from white light (WL; ∼10 µ mol−2 s−1) grown wild-type Fremyella diplosiphon cells acclimated to a 12 h diurnal cycle at 12 h into the light (L12) or dark period (D12) and 3 h into the light period (L3) [see schematic in (A)]. (B) qRT-PCR analysis for FdTSPO genes, including primers used, was as described in Busch et al, 20164. The relative expression level compared with ORF10B gene is shown as average values ( ± SD, n = 3). Expression of each FdTSPO is shown relative to expression levels at L12 sampling time. Identical letters over bars represent homogenous mean groups (p > 0.05), whereas asterisks indicate a significant difference (*p < 0.01, **p < 0.05) between expression level at 0 h relative to each time point, as determined by ANOVA and Tukey test.
Additional abiotic stresses alter TSPO transcript abundance
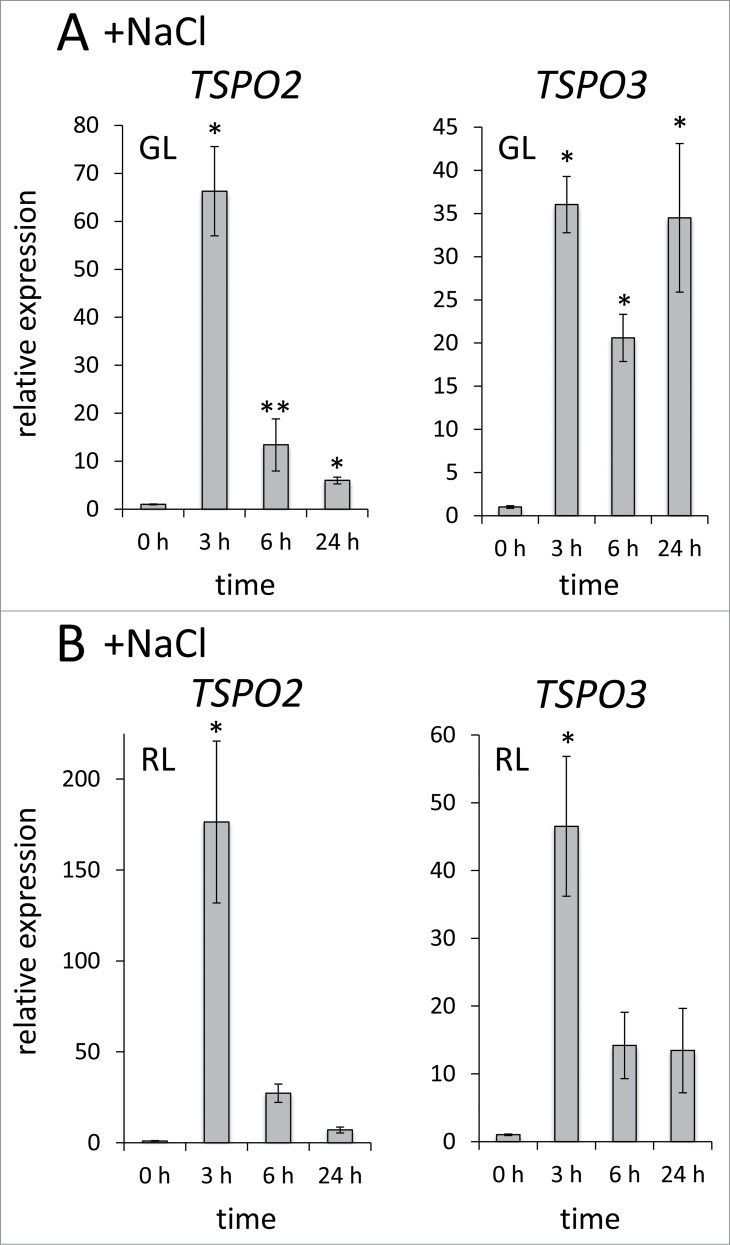
In addition to a role for light in regulation of FdTSPO expression, we previously assessed effects of several abiotic stresses on FdTSPO1 levels and found that transcript levels are transiently upregulated under multiple stress conditions, including salt stress, osmotic stress and oxidative stress.6 We therefore chose to further analyze the impact of select abiotic stresses on the family of FdTSPO genes. We first assessed salt stress-induced expression of FdTSPO2 and FdTSPO3 (Fig. 2). The mRNA levels of both FdTSPO2 and FdTSPO3 were upregulated in the presence of salt, with the highest average transcript levels observed 3 h after adding 200 mM salt (fold change: ∼66 in GL, ∼176 in RL for FdTSPO2 and ∼36 in GL and ∼47 in RL for FdTSPO3). As observed for FdTSPO1, this upregulation was transient in all cases except for FdTSPO3 where levels at 24 h in GL are similar to those at 3 h (Fig. 2A). The overall increase of transcript levels upon salt stress for FdTSPO2 and FdTSPO3 were substantially higher than those observed for FdTSPO1 (compare Fig. 2 with results for TSPO1 in ref. 6 in which a 2-fold induction of FdTSPO is reported), implying potentially distinct roles for these 2 groups of TSPOs in response to a common stressor, i.e., salt.
Figure 2.
Salt-dependent regulation of FdTSPO2 and FdTSPO3 transcript levels. Samples for quantitative real-time reverse transcription PCR (qRT-PCR) were taken before (0 h) and after (3 h, 6 h, 24 h) addition of 200 mM NaCl (w/v) in either (A) green light (GL) or (B) red light (RL) from wild-type F. diplosiphon cells adapted to the respective light color to assess FdTSPO2 and FdTSPO3 transcript accumulation in response to salt stress. Relative expression level compared with ORF10B gene is shown as average values ( ± SD, n = 3). Expression of FdTSPO2 or FdTSPO3 is shown relative to expression levels in WT cells at time 0 h in the respective light condition. Asterisk indicates a significant difference (*p < 0.01, **p < 0.05) between expression level at 0 h relative to each time point, as determined by ANOVA and Tukey test.
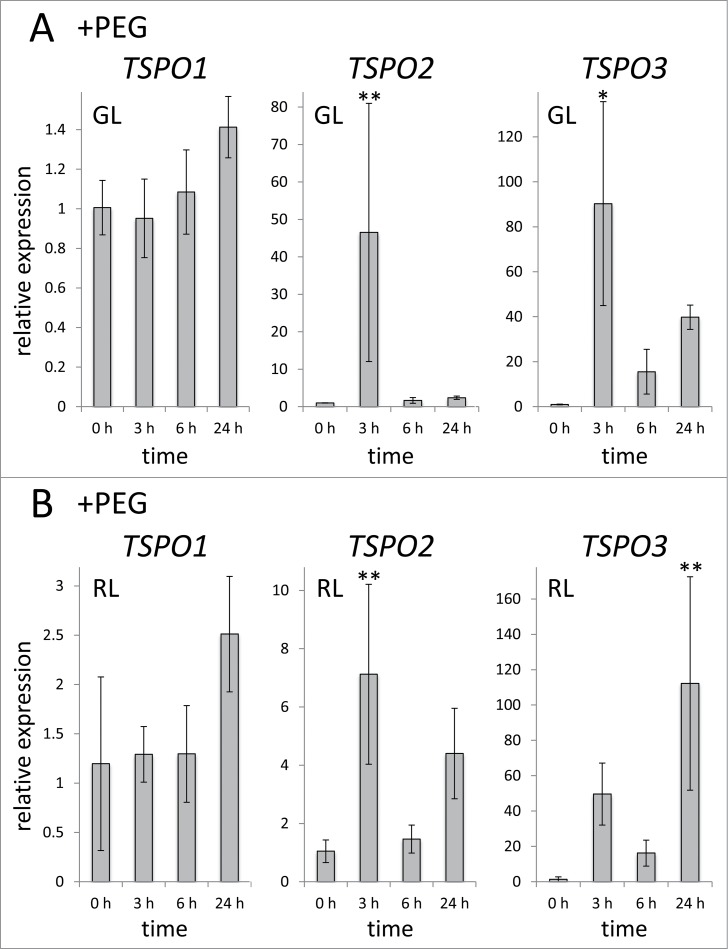
As salt stress involves both ionic stress and osmotic stress10 and TSPO has a role in salt stress in plants and cyanobacteria and has been also been implicated in drought stress in plants,6,11,12 we investigated the role of drought stress on FdTSPO mRNA accumulation. To mimick drought stress, we treated F. diplosiphon cells with the osmoticum PEG6000 (20% w/v) (Fig 3). A similar level of PEG (i.e., 20%) has been previously used to induce osmotic or drought stress in filamentous cyanobacteria.13,14 We observed distinct impacts of induced drought on the 3 FdTSPO homologs. FdTSPO1 mRNA levels exhibited a limited response to induced drought in GL and a ∼2.5-fold induction at 24 h in RL (Fig. 3). By contrast, both FdTSPO2 and FdTSPO3 were transiently upregulated at 3 h in GL (Fig. 3A), whereas the 2 genes showed distinct time points of significantly higher mRNA accumulation, i.e., early (3 h) for FdTSPO2 and late (24 h) for FdTSPO3, compared with 0 h in RL (Fig. 3B). These results distinctly implicate FdTSPO2 and FdTSPO3 in temporal cellular responses to induced drought stress in F. diplosiphon.
Figure 3.
Effect of PEG-induced drought on FdTSPO transcript abundance. Samples for quantitative real-time reverse transcription PCR (qRT-PCR) were taken before (0 h) and after (3 h, 6 h, 24 h) addition of 20% PEG6000 (w/v) in (A) green light (GL) and (B) red light (RL) to assess FdTSPO transcript accumulation in response to induced stress. Relative expression level compared with ORF10B gene is shown as average values ( ± SD, n = 3). Expression of each FdTSPO is shown relative to expression levels in wild-type F. diplosiphon cells at time 0 h in the respective light condition. Asterisks indicate a significant difference (*p < 0.01, **p < 0.05) between expression level at 0 h relative to each time point after treatment with PEG, as determined by ANOVA and Tukey test.
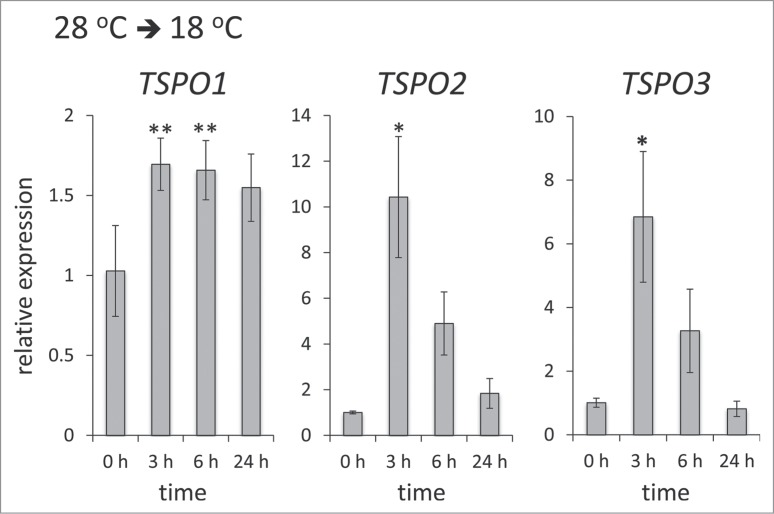
Given that changes in temperature impacts organismal fitness and are associated with diurnal environmental conditions, we investigated temperature effects on the regulation of FdTSPO homologs. To test the effect of a temperature drop, F. diplosiphon cells were initially acclimated under white light (WL) in standard temperature conditions of 28°C. Acclimated cells were then switched to a lower temperature of 18°C (Fig. 4). FdTSPO1 was ∼1.7-fold upregulated after 3 h of the temperature shift. Of the measured time points, TSPO mRNA levels were highest after 3 h for all 3 TSPO homologs. Notably, the more similar TSPOs, i.e., FdTSPO2 and FdTSPO3, showed a stronger increase than FdTSPO1 at ∼10.4-fold and ∼6.8-fold increases in mRNA levels at 3 h for FdTSPO2 and FdTSPO3, respectively. The increase in transcript abundance was transient, especially for FdTSPO2 and FdTSPO3 (Fig. 4).
Figure 4.
Temperature-dependent regulation of FdTSPO transcript levels. Samples for quantitative real-time reverse transcription PCR (qRT-PCR) were taken before (0 h) and after (3 h, 6 h, or 24 h) transferring cells acclimated to 28°C in white light (WL) to 18°C to assess transcript accumulation in response to cold stress for FdTSPO1, FdTSPO2 and FdTSPO3. The relative expression level compared with ORF10B gene is shown as average values ( ± SD, n = 3). Expression of each FdTSPO is shown relative to expression levels in wild-type F. diplosiphon cells at time 0 h before the temperature shift. Asterisks indicate a significant difference (*p < 0.01, **p < 0.05) between expression level at 0 h relative to each time point after the shift from 28°C to 18°C, as determined by ANOVA and Tukey test.
Nutrient stress affects TSPO transcript abundance
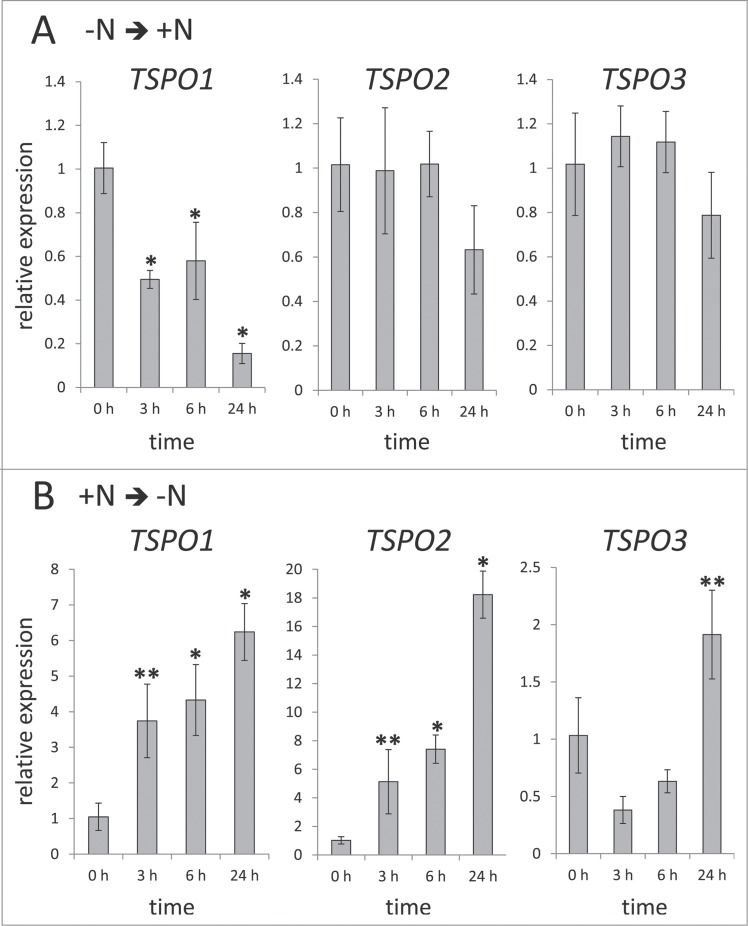
Changes in nitrogen availability previously have been shown to affect the abundance of a putative F. diplosiphon (aka Tolypothrix sp PCC 760115) TSPO-containing cotranscript.16 Therefore, the effect of changes in nitrogen availability on TSPO mRNA levels was assessed before and after exposing F. diplosiphon cells to changes in nitrogen availability (Fig. 5). Nitrogen-limited cells switched to nitrogen-replete medium exhibited a decrease in TSPO transcript abundance over the course of a day, albeit results were only significant for FdTSPO1 among the 3 homologs (Fig. 5A). When cells grown in nitrogen-replete medium were moved into nitrogen-deficient medium TSPO abundance was highest after 24 h for all 3 homologs (Fig. 5B). The highest fold change was observed for FdTSPO2 (∼18-fold), followed by FdTSPO1 (∼6-fold) and then FdTSPO3 (∼2-fold). Notably, FdTSPO1 and FdTSPO2 exhibited significant upregulation early (i.e., at 3 h) compared with FdTSPO3, which was only significantly higher at 24 h.
Figure 5.
Nitrogen-dependent regulation of FdTSPO transcript levels. Samples for quantitative real-time reverse transcription PCR (qRT-PCR) were taken before (0 h) and after (3 h, 6 h, or 24 h) exposing white light (WL) grown cells acclimated to either (A) nitrogen-limited (-N) conditions to nitrogen-replete (+N) medium [-N ➔ +N] or (B) grown under nitrogen-replete conditions and transferred to medium lacking added nitrogen [+N ➔ -N]. The relative expression level compared with ORF10B gene is shown as average values ( ± SD, n = 3). Expression of each FdTSPO is shown relative to expression levels in wild-type F. diplosiphon cells at time 0 h before changing nitrogen availability. Asterisks indicate a significant difference (*p < 0.01, **p < 0.05) between expression level at 0 h relative to each time point before the growth medium exchange, as determined by ANOVA and Tukey test.
Conclusion
Multiple abiotic stresses impact mRNA levels for all 3 TSPO genes found in F. diplosiphon. In most cases, FdTSPO1, FdTSPO2, and FdTSPO3 respond to similar stresses, but with distinct patterns and/or magnitudes. Frequently, FdTSPO2 and FdTSPO3 show the more similar stress-induced responses (e.g., induced drought, temperature shift, nitrogen stress), which is not surprising given their higher sequence similarity.4 Distinct responses for each is observed in some cases, including growth in diurnal light cycles and light-dependent responses to salt. Together with apparently distinct tetrapyrrole-binding profiles, these results indicate potentially distinct regulation and functions for these 3 FdTSPO homologs. Furthermore, distinct functions may be supported by the distinct location of these genes in 3 distinct phylogenetic clades.4 Definitive confirmation of the distinct roles of these homologs will require additional genetic experiments.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We also thank Dr. Ferguson-Miller and her laboratory members (Michigan State University) for helpful discussions about TSPO.
Funding
This work was supported by the National Science Foundation under Grant MCB–1243983 to B.L.M.
References
- 1.Bennett A, Bogorad L. Complementary chromatic adaptation in a filamentous blue-green alga. J Cell Biol 1973; 58:419-35; PMID:4199659; http://dx.doi.org/ 10.1083/jcb.58.2.419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gutu A, Kehoe DM. Emerging perspectives on the mechanisms, regulation, and distribution of light color acclimation in cyanobacteria. Mol Plant 2012; 5:1-13; PMID:21772031; http://dx.doi.org/ 10.1093/mp/ssr054 [DOI] [PubMed] [Google Scholar]
- 3.Montgomery BL. Mechanisms and fitness implications of photomorphogenesis during chromatic acclimation in cyanobacteria. J Exp Bot 2016; 67(14):4079-90; PMID:27217547; http://dx.doi.org/ 10.1093/jxb/erw206 [DOI] [PubMed] [Google Scholar]
- 4.Busch AWU, WareJoncas Z, Montgomery BL. Tryptophan-rich sensory protein/translocator protein (TSPO) from cyanobacterium Fremyella diplosiphon binds a broad range of functionally relevant tetrapyrroles. Biochemistry 2016; 56(1):73-84; PMID:27990801; http://dx.doi.org/ 10.1021/acs.biochem.6b01019 [DOI] [PubMed] [Google Scholar]
- 5.Li F, Liu J, Liu N, Kuhn LA, Garavito RM, Ferguson-Miller S. Translocator Protein 18 kDa (TSPO): An old protein with new functions? Biochemistry 2016; 55:2821-31; PMID:27074410; http://dx.doi.org/ 10.1021/acs.biochem.6b00142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busch AWU, Montgomery BL. The tryptophan-rich sensory protein (TSPO) is involved in stress-related and light-dependent processes in the cyanobacterium Fremyella diplosiphon. Front Microbiol 2015; 6:1393; PMID:26696996; http://dx.doi.org/ 10.3389/fmicb.2015.01393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stowe-Evans EL, Ford J, Kehoe DM. Genomic DNA microarray analysis: identification of new genes regulated by light color in the cyanobacterium Fremyella diplosiphon. J Bacteriol 2004; 186:4338-49; PMID:15205436; http://dx.doi.org/ 10.1128/JB.186.13.4338-4349.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saha R, Liu D, Hoynes-O'Connor A, Liberton M, Yu J, Bhattacharyya-Pakrasi M, Balassy A, Zhang F, Moon TS, Maranas CD, et al.. Diurnal regulation of cellular processes in the cyanobacterium Synechocystis sp. Strain PCC 6803: Insights from transcriptomic, fluxomic, and physiological analyses. mBio 2016; 7:e00464-16; PMID:27143387; http://dx.doi.org/ 10.1128/mBio.00464-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeliseev AA, Kaplan S. A sensory transducer homologous to the mammalian peripheral-type benzodiazepine receptor regulates photosynthetic membrane complex formation in Rhodobacter sphaeroides 2.4.1. J Biol Chem 1995; 270:21167-75; PMID:7673149; http://dx.doi.org/20618868 10.1074/jbc.270.36.21167 [DOI] [PubMed] [Google Scholar]
- 10.Hagemann M. Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol Rev 2011; 35:87-123; PMID:20618868; http://dx.doi.org/ 10.1111/j.1574-6976.2010.00234.x [DOI] [PubMed] [Google Scholar]
- 11.Hachez C, Veljanovski V, Reinhardt H, Guillaumot D, Vanhee C, Chaumont F, Batoko H. The Arabidopsis abiotic stress-induced TSPO-related protein reduces cell-surface expression of the Aquaporin PIP2;7 through protein-protein interactions and autophagic degradation. Plant Cell 2014; 26:4974-90; PMID:25538184; http://dx.doi.org/ 10.1105/tpc.114.134080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guillaumot D, Guillon S, Deplanque T, Vanhee C, Gumy C, Masquelier D, Morsomme P, Batoko H. The Arabidopsis TSPO-related protein is a stress and abscisic acid-regulated, endoplasmic reticulum-Golgi-localized membrane protein. Plant J 2009; 60:242-56; PMID:19548979; http://dx.doi.org/ 10.1111/j.1365-313X.2009.03950.x [DOI] [PubMed] [Google Scholar]
- 13.Close TJ, Lammers PJ. An osmotic stress protein of cyanobacteria is immunologically related to plant dehydrins. Plant Physiol 1993; 101:773-9; PMID:8310057; http://dx.doi.org/ 10.1104/pp.101.3.773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandes TA, Apte SK. Differential regulation of nitrogenase activity by ionic and osmotic stresses and permeable sugars in the cyanobacterium Anabaena sp. strain L-31. Plant Sci 2000; 150:181-9; http://dx.doi.org/ 10.1016/S0168-9452(99)00183-1 [DOI] [Google Scholar]
- 15.Yerrapragada S, Shukla A, Hallsworth-Pepin K, Choi K, Wollam A, Clifton S, Qin X, Muzny D, Raghuraman S, Ashki H, et al.. Extreme sensory complexity encoded in the 10-Megabase draft genome sequence of the chromatically acclimating cyanobacterium Tolypothrix sp. PCC 7601. Genome Announc 2015; 3:e00355-15; PMID:25953173; http://dx.doi.org/ 10.1128/genomeA.00355-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luque I, Ochoa De Alda JA, Richaud C, Zabulon G, Thomas JC, Houmard J. The NblAI protein from the filamentous cyanobacterium Tolypothrix PCC 7601: Regulation of its expression and interactions with phycobilisome components. Mol Microbiol 2003; 50:1043-54; PMID:14617160; http://dx.doi.org/ 10.1046/j.1365-2958.2003.03768.x [DOI] [PubMed] [Google Scholar]