ABSTRACT
The root hair development of vascular plants can be divided into 2 major processes, fate determination and hair morphogenesis, and the latter should be governed by the former so as to express the morphogenetic toolkits in a root hair-specific manner. Vascular plants, depending on taxa, show different fate-determining mechanisms for hair cell/non-hair cell fates, which leads to a question whether the downstream mophogenetic regulatory module is diverged accordingly to the upstream fate determiners or not. Our study demonstrates that the module of a transcription factor and a root hair-specific cis-element (RHE) for root hair-specific expression of morphogenetic toolkit genes is conserved in spite of different fate-determing mechanisms.
KEYWORDS: Evolution of root hair, RHD6-like 4 (RSL4), root hair, root hair-specific cis-element (RHE), root hair-specific gene expression, tip growth
Abbreviations
- bHLH
basic helix-loop-helix
- LP
left part
- RHE
Root Hair-specific cis-Element
- RHS
ROOT HAIR SPECIFIC
- RP
ight part
- RSL
ROOT HAIR DEFECTIVE SIX-LIKE
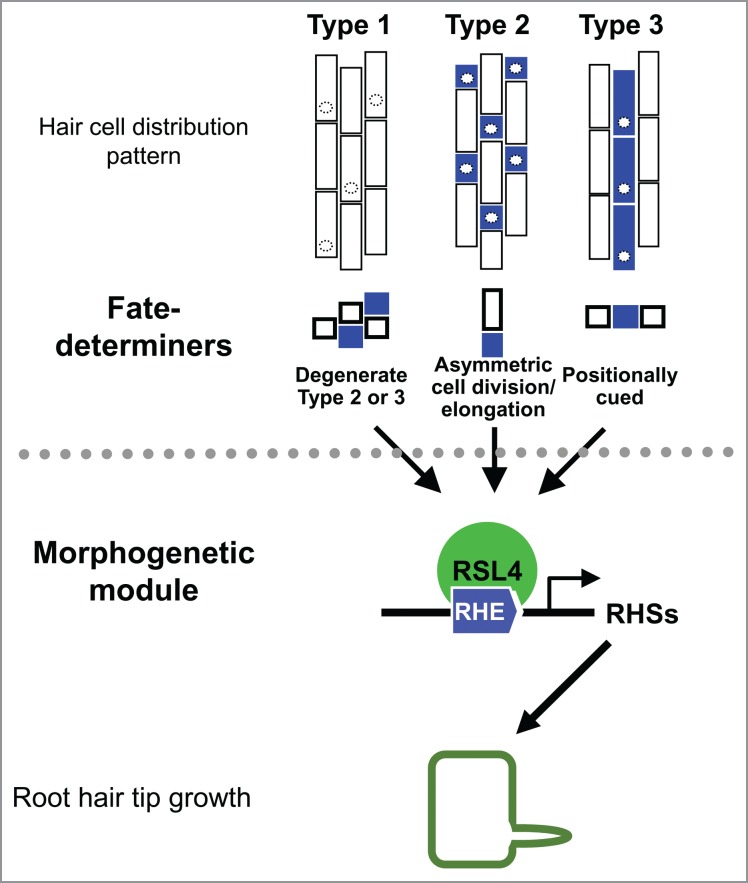
The root hair has been a fascinating model to study cell fate determination and cellular tip growth. Epidermal cells in vascular plants show 3 distinctive distribution patterns of hair cell/non-hair cell which are thought to be determined by different cell fate-determining mechanisms; random (type 1), asymmetric cell division/elongation (type 2), and position dependent (type 3) (Fig. 1).1,2,3,4 The position-dependent type 3 mechanism has been well characterized in Arabidopsis,5 but the mechanisms for other types have yet to be understood.
Figure 1.
The conserved RSL4-RHE regulatory module for root hair tip growth. The root hair-morphogenetic module for root hair-specific gene expression is under control of the upstream fate-determining machineries that varies depending on the evolutionary course of vascular plants. In spite of the different upstream fate determiners, the morphogenetic module (RSL4-RHE) has been conserved throughout the evolution of vascular plants.
A phylogenetic analysis based on the survey2 of root hair cell distribution pattern in diverse vascular plants demonstrated that type 3 occurs only in eudicots, type 2 in monocots, basal angiosperms, monilophytes, and lycophytes, and type 1 in any vascular plants.6 This indicates that the type 2 mechanism is more ancient than the type 3 mechanism, and the random distribution pattern in type 1 could result from degeneration of the type 2 or type 3 mechanism. Because lycophytes have the type 2 mechanism, the type 2 mechanism is likely to emerge first in early vascular plants with the root. The type 2 mechanism had lasted until the emergence of angiosperms and then of monocots. However, fundamental changes seem to have happened in root epidermal fate determination during the eudicot evolution. The common ancestor of eudicots discarded the type 2 mechanism and then replaced it with the position-dependent type 3 mechanism.
In contrast to the upstream fate-determining machinery, the hair cell-specific toolkits for tip growth, such as cell wall dynamics, secretory pathway, polarity cues, etc., are likely to be conserved in different taxa. A question during the evolutionary process of cell fate-determining mechanisms is how the root hair-specific toolkit genes found a regulatory connection to the newly adopted upstream fate-determining machinery. Different fate-determining mechanisms might have taken different regulatory modules or a common regulatory module for the hair-cell specificity of the toolkit gene expression.7 To assess this question, Kim et al. (2006) characterized the cis-element for root hair-specific gene expression (RHE for Root Hair specific Element).
Before the RHE was identified, only several genes were known to be expressed in a root hair-specific manner in Arabidopsis,8,9,10,11 although a later study screened more RHE-containing root hair-specific (RHS) genes.12 Elaborate promoter analyses were first conducted with 2 root hair-specific EXPANSIN A (EXPA) genes (EXPA7 and EXPA18). Functional analyses of the promoters by sequential deletion and 1-bp-level substitution focused down to at least 7 core nucleotides that are required for root hair-specific gene expression.6 Phylogenetic analyses of root hair-specific EXPAs and other genes from diverse angiosperms revealed their likely orthologs, and the promoters of these orthologs really contain the RHE-like motifs.6 Further functional analyses of these RHE-like motifs by deletion and substitution showed that they really function as RHE for root hair-specific expression of the orthologs.6 The functional analysis and multiple alignment analysis of RHE-like motifs from those root hair-specific genes revealed a conserved 16- or 17-bp-long RHE sequence.6 The RHE includes a more conserved right part (RP) with highly conserved ‘CACG’, a less conserved left part (LP) with highly conserved ‘T’, and a linker between LP and RP which varies in length and thus generates 16- or 17-bp-long functional RHE in final length. The alignment analysis of 27 RHEs showed a RHE consensus sequence with a palindromic relationship between LP and RP, although individual RHEs mostly are partially palindromic. This partial palindromic structure of RHE led to a hypothesis that a group of structurally related transcription factors (Root Hair-specific Factor, RHFs) might combinatorically bind to the RHE, in which RP accommodates a single RHF species and LP recruits more diverse sister RHF species.
In a long search for RHF, our recent study demonstrated that an Arabidopsis basic helix-loop-helix (bHLH) transcription factor, ROOT HAIR DEFECTIV SIX-LIKE 4 (RSL4), directly binds to the RHE in in vitro and in vivo binding assays and upregulates the RHE-containing RHS genes (Fig. 1).13 RSL4 is one of RSL members that form a small subgroup of related bHLHs and have been implicated in root hair formation.14,15 Targeting RSL4 as a RHF candidate is attributable to a transcriptome analysis where RSL4 target genes were identified using the rsl4 mutant and the RSL4 overexpression transformant.14 These RSL4 target genes turn out to include many known RHS genes,12 which prompted to test if RSL4 modulates RHS genes by directly binding to the RHE.
Hwang et al.'s study also identified RSL4 orthologs from another eudicot (poplar), a monocot (rice), and a lycophyte (Selaginella moellendorffii) and demonstrated that these orthologs also are able to bind to the RHE in vitro and in vivo and enhance root hair growth of Arabidopsis, suggesting that the RSL4-RHE regulatory module for root hair specificity is conserved in vascular plants. RHE-containing EXPA7-orthologous genes were identified in Selaginella, and this Selaginella RHE was shown to be functional to direct root hair-specific gene expression in Arabidopsis.13 Rice and Selaginella roots show the type 2 hair cell-distribution pattern.1,2,6 This indicates that the RSL4-RHE module is operational for root hair specificity even under the type 2 fate-determining mechanism and had emerged with vascular plants or even before them. Some orthologous genes of RSL4 are found in bryophytes such as Marchantia polymorpha (a liverwort) and Physcomitrella patens (a moss), and the promoter regions of these orthologous genes contain RHE-like motifs as does the RSL4 gene promoter.13 Rhizoids of bryophytes are similar with root hairs of vascular plants in that both elongate via tip growth. Therefore, it is conceivable that the RSL4-RHE module evolved together with early land plants for their adaptation to dry environments.
So far, the developmental pathway for root hair development seems to be fully characterized in type 3 species, which includes the receptor for the positional cue, transcriptional interaction for fate determination, the major root hair suppressor in the non-hair cell position, upper morphogenetic modulators (class I RSLs) and lower morphogenetic modulators (class II RSLs including RSL4) in the hair cell position, and root hair-morphogenetic worker genes (RHSs) and their adaptor RHE to recruit the RSL4 modulator. However, RSL4 is the target not only of the developmental pathway via the class I RSL but also of the hormonal (auxin) pathway. Auxin is likely to play to mediate environmental factors that stimulate root hair growth.16 Further studies of the auxin signaling to RSL4 will provide a wider spectrum of regulatory network for root hair growth.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This study was supported by the grants from the Next-Generation BioGreen 21 program (The Agricultural Genome Center PJ011195) of the Rural Development Administration and the Mid-career Researcher Program (2015002633) of the National Research Foundation.
References
- 1.Dolan L. Pattern in the root epidermis: An interplay of diffusible signals and cellular geometry. Ann Bot 1996; 77:547-53; http://dx.doi.org/ 10.1093/aob/77.6.547 [DOI] [Google Scholar]
- 2.Clowes FAL. Pattern in root meristem development in angiosperms. New Phytol 2000; 146:83-94; http://dx.doi.org/ 10.1046/j.1469-8137.2000.00614.x [DOI] [Google Scholar]
- 3.Schiefelbein JW. Constructing a plant cell. The genetic control of root hair development. Plant Physiol 2000; 124:1525-31; PMID:11115870; http://dx.doi.org/ 10.1104/pp.124.4.1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim CM, Dolan L. Root hair development involves asymmetric cell division in Brachypodium distachyon and symmetric division in Oryza sativa. New Phytol 2011; 192:601-10; PMID:21848982; http://dx.doi.org/ 10.1111/j.1469-8137.2011.03839.x [DOI] [PubMed] [Google Scholar]
- 5.Grierson C, Schiefelbein J. Genetics of Root Hair Formation In Root Hairs (ed Emons AMC, Ketelaar T), 2008; pp. 1-25. Springer, Heidelberg. [Google Scholar]
- 6.Kim DW, Lee SH, Choi SB, Won SK, Heo YK, Cho M, Park YI, Cho H-T. Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 2006; 18:2958-70; PMID:17098810; http://dx.doi.org/ 10.1105/tpc.106.045229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho HT. A cis-element for root hair specificity has been co-opted repeatedly through the divergence of upstream fate-determining machineries. Plant Signal Behav 2007; 2:117-8; PMID:19704754; http://dx.doi.org/ 10.4161/psb.2.2.3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernhardt C, Tierney ML. Expression of AtPRP3, a proline-rich structural cell wall protein from Arabidopsis, is regulated by cell-type-specific developmental pathways involved in root hair formation. Plant Physiol 2000; 122:705-14; PMID:10712533; http://dx.doi.org/ 10.1104/pp.122.3.705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baumberger N, Ringli C, Keller B. The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev 2001; 15:1128-39; PMID:11331608; http://dx.doi.org/ 10.1101/gad.200201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumberger N, Steiner M, Ryser U, Keller B, Ringli C. Synergistic interaction of the two paralogous Arabidopsis genes LRX1 and LRX2 in cell wall formation during root hair development. Plant J 2003; 35:71-81; PMID:12834403; http://dx.doi.org/ 10.1046/j.1365-313X.2003.01784.x [DOI] [PubMed] [Google Scholar]
- 11.Cho H-T, Cosgrove DJ. Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 2002; 14:3237-53; PMID:12468740; http://dx.doi.org/ 10.1105/tpc.006437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Won SK, Lee YJ, Lee HY, Heo YK, Cho M, Cho H-T. Cis-element- and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiol 2009; 150:1459-73; PMID:19448035; http://dx.doi.org/ 10.1104/pp.109.140905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang Y, Choi H-S, Cho H-M, Cho H-T. Tracheophytes contain conserved orthologs a basic helix-loop-helix transcription factor to modulate ROOT HAIR SPECIFIC genes. Plant Cell 2017; 29:39-53; PMID:28087829; http://dx.doi.org/ 10.1105/tpc.16.00732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi K, Menand B, Bell E, Dolan L. A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nature Genetics 2010; 42:264-7; PMID:20139979; http://dx.doi.org/ 10.1038/ng.529 [DOI] [PubMed] [Google Scholar]
- 15.Pires ND, Yi K, Breuninger H, Catarino B, Menand B, Dolan L. Recruitment and remodeling of an ancient gene regulatory network during land plant evolution. Proc Natl Acad Sci USA 2013; 110:9571-6; PMID:23690618; http://dx.doi.org/ 10.1073/pnas.1305457110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee RD-W, Cho H-T. Auxin, the organizer of the hormonal/environmental signals for root hair growth. Front Plant Sci 2013; 4:448; PMID:24273547; http://dx.doi.org/ 10.3389/fpls.2013.00448 [DOI] [PMC free article] [PubMed] [Google Scholar]