Abstract
Test-and-treat programs are central to the global control of HIV, but transmitted drug resistance threatens the effectiveness of these programs. HIV mutations conferring resistance to antiretroviral drugs reduce replicative fitness in vitro, but their effect on propagation in vivo is less understood. Here, we estimate transmission fitness of these mutations in antiretroviral-naïve populations in the U.S. National HIV Surveillance System by comparing their frequency of clustering in a genetic transmission network relative with wild-type viruses. The large dataset (66,221 persons), comprising 30,196 antiretroviral-naïve persons, permitted the evaluation of sixty-nine resistance mutations. Decreased transmission fitness was demonstrated for twenty-three mutations, including M184V. In contrast, many high prevalence mutations (e.g. K103N, Y181C, and L90M) had transmission fitness that was indistinguishable from or exceeded wild-type fitness, permitting the establishment of large, self-sustaining drug resistance reservoirs. We highlight implications of these findings on strategies to preserve global treatment effectiveness.
Keywords: fitness, transmission network, HIV, drug resistance
1. Introduction
More than 10 million people worldwide are receiving antiretroviral therapy (ART) which, when effective at suppressing HIV replication, is of benefit both in reversing immunodeficiency disease and reducing transmissibility (Palella et al. 1998; Cohen et al. 2011; Joint United Nations Programme on HIV/AIDS (UNAIDS) 2013; Montaner et al. 2014). Recent evidence demonstrating that earlier use of ART results in better clinical outcomes than delayed treatment has prompted recommendations for early initiation of ART, accelerating further the implementation of a treatment as prevention strategy (World Health Organization 2015). Likewise the recommendation for the use of pre-exposure prophylaxis (PrEP) with tenofovir and FTC for prevention in persons who are at high risk of HIV acquisition has expanded the importance of antiretroviral medications in protecting uninfected persons (Centers for Disease Control and Prevention 2014; World Health Organization 2015).
The global scale-up of ART in resource-limited countries is an unprecedented public health accomplishment that was enabled by the availability of fixed-dose first-line and second-line regimens (World Health Organization 2015). The first-line regimen consists of a non-nucleoside reverse transcriptase inhibitor (NNRTI; efavirenz/nevirapine) plus two nucleoside analog reverse transcriptase inhibitors (NRTIs; tenofovir and 3TC/FTC), whereas the second-line regimen consists of a ritonavir-boosted protease inhibitor (PI; lopinavir or atazanavir) plus two NRTIs (most commonly zidovudine and 3TC/FTC) (World Health Organization 2015). Failure of first-line therapy can occur in up to 30% of patients per year and is frequently associated with the acquisition of viruses with NNRTI K103N and/or NRTI M184V mutations (Barth et al. 2010; McMahon et al. 2013). In a recent guidance, WHO maintained NNRTI-based ART as a preferred first-line regimen but provided the option of using the integrase inhibitor class of drugs (e.g. dolutegravir) in first-line ART. Updated second-and third-line ART regimens were also provided (World Health Organization 2015).
As ART gains a central role in both treatment and prevention worldwide, there are concerns that drug resistance might limit its effectiveness, particularly as lifelong ART will increasingly start earlier and target large populations with varying degrees of adherence and testing for viral suppression. Transmitted drug resistance (TDR), the acquisition of a virus containing drug resistance-associated mutations (DRAMs), can propagate DRAMs and potentially diminish the effectiveness of ART at a population level. The global scale-up of ART in resource-limited countries remains vulnerable to widespread drug resistance because of limited fixed-dose regimens in settings of sub-optimal clinical drug resistance monitoring. Surveillance systems that collect HIV sequence data to track the prevalence of DRAMs among ART-naïve persons have provided important epidemiologic information on TDR worldwide including concerning rise in TDR (Wheeler et al. 2010; Gupta et al. 2012; Rhee et al. 2015). However, a better understanding of the drivers of TDR is critical for optimal management of ART programs. New approaches for analyzing large collections of sequence data—beyond assessing DRAM prevalence—can investigate less-understood questions underlying TDR, such as transmission efficiency and the source of drug-resistant viruses.
DRAMs, which arise as a consequence of ART-driven selection provide an advantage to the virus in the presence of drug but can decrease viral replicative fitness in the absence of antiretroviral drugs. This process, which has been well studied in vitro, has demonstrated that some DRAMs have strongly deleterious effects on replicative fitness (e.g. M184V, K65R, and T215Y), whereas other DRAMs have weaker fitness effects (e.g. L90M, Y181C, D67N, L210W) (Mammano et al. 2000; Cong et al. 2007; Jain et al. 2011; Castro et al. 2013). However, relating the fitness effects of DRAMs that are observed in vitro to transmission fitness in vivo (i.e. the propensity of a mutation to propagate among hosts relative to wild-type virus at the population level) remains poorly understood. Persistence in vivo influences transmission fitness of DRAMs. Available data from untreated populations has shown differences in DRAM persistence, ranging from months to several years with shorter durations linked to viruses with low replicative capacity in vitro (Cong et al. 2007; Jain et al. 2011; Castro et al. 2013; Pingen et al. 2014). Another factor that influences transmission fitness is virus loads (Quinn et al. 2000; Cohen et al. 2011).
Previous work has compared DRAM frequencies in ART-naïve and ART-experienced patients to infer transmission fitness of viruses with DRAMs but found discordant results (Leigh Brown et al. 2003; de Mendoza et al. 2004; Corvasce et al. 2006; Poon et al. 2015; Winand et al. 2015), particularly with regard to K103N. A direct comparison of DRAM frequencies between ART-naïve and ART-experienced patients will not provide a clear picture of viral fitness, because the different transmission rates among these groups are confounded by variable DRAM persistence dynamics, difference in behavior due to awareness of HIV status, and the effect of ART on virus load and mutation persistence in ART-experienced persons. More recently, studies have used genetic clustering in viruses with TDR to infer whether TDR emanated from an ART-naïve or ART-experienced source (Mourad et al. 2015; Rhee, et al. 2015). However, a comprehensive understanding of the effect of DRAMs on transmission fitness underlying the propagation of TDR is still lacking.
Here, we use a novel molecular approach that compares transmissibility of wild-type viruses with those viruses containing DRAMs. We investigate the effect of DRAMs on transmission fitness using a genetic transmission network constructed from a large dataset from the U.S. National HIV Surveillance System. We use clustering in this network as a proxy for transmissibility to approximate the transmission fitness of DRAMs in ART-naïve persons relative to wild-type. We find evidence that certain DRAMS reduce transmission fitness but identify other DRAMs that are as fit or more fit than wild-type strains, resulting in self-sustaining drug resistance reservoirs. We discuss the implications of these findings on the global scale-up of test-and-treat programs currently underway and on strategies to preserve ART effectiveness.
2. Methods
2.1 HIV sequences
We analyzed HIV-1 protease (pro) and reverse transcriptase (RT) containing sequences of the polymerase (pol) gene reported to the U.S. National HIV Surveillance System from 2001 through 2014 from 66,236 persons. These sequences were collected from twenty-seven jurisdictions around the United States [see Oster et al. (2015) for a more detailed description of the population and transmission dynamics]. Subtyping was performed using a local installation of COMET (COntext-based Modeling for Expeditious Typing) (Struck et al. 2014). Only subtypes A, B, C, D, F, and G and CRFs 01_AE and 02_AG were included, because their drug resistance profile can be reliably characterized using Sierra (Liu and Shafer 2006). Sequences had a minimum length of 500 nucleotides, and when more than one sequence per person was available, the earliest collected sequence was used. Additional meta-data including transmission risk factor, demographics, and evidence of ART use was also analyzed.
Persons were classified as ART-naïve if they received their first genotype within 3 months of diagnosis and had no evidence of prior ART use, in accordance with CDC guidelines. Persons were classified as ART-experienced if they acknowledged using ARTs prior to the date of their initial genotype. The 108 DRAMs considered in this study comprise the CDC surveillance drug resistance mutation list applied to the data of the U.S. National HIV Surveillance System (Wheeler, et al. 2010). This list includes twenty-four NNRTI mutations, forty NRTI mutations, and forty-four PI mutations, all found in the pro and RT regions of pol.
2.2 Network construction
The transmission network was built using a local version of HIV-TRACE (www.hivtrace.org) following the protocol described in Wertheim et al. (2014) and Oster et al. (2015). Sequences were codon-aligned to an HXB2 reference sequence (positions 2253–3749). Forty-eight codons associated with drug resistance were excised from the alignment. Tamura-Nei 93 (TN93; Tamura and Nei 1993) pairwise genetic distance was estimated for all pairs of sequences to identify potential transmission partners, defined as persons whose viral sequences were ≤0.015 substitutions per site divergent. To prevent spurious clustering due to sequences with high levels of ambiguous nucleotides (e.g. R, Y, etc.), sequences with >1.5% ambiguities were penalized by averaging the genetic distance between the ambiguous and known nucleotides (e.g. Y is 0.5 substitutions from both C and T). For sequences with ≤1.5% ambiguities, these genetic distances were resolved (e.g. Y is zero substitutions from both C and T). Fifteen sequences were less than 0.015 substitutions per site divergent from the reference sequence (HXB2) and were excluded as possible contaminants. Transmission clusters were assembled by connecting viral sequences from potential transmission partners.
2.3 Frequency of clustering in strains containing DRAMs in ART-naïve persons
Relative transmission fitness was measured in the ART-naïve population as the frequency of strains with a specific DRAM that clustered with other strains with that DRAM divided by the frequency of wild-type strains that clustered with other wild-type strains.
Because transmission clusters comprise closely related viruses, mutations tend to be tightly shared among viruses within a cluster (i.e. founder effects). Therefore, statistical non-independence can lead to bias when assessing whether strains containing certain DRAMs clustered more or less frequently than strains lacking that DRAM. For example, if the appearance of a DRAM coincided with expansion of a larger cluster, it is difficult to disentangle the fitness effect of this DRAM with the growth of the cluster (Vega et al. 2015). Due to this network structure, a standard Χ2 test is inappropriate to establish whether strains containing a DRAM cluster more or less frequently than expected. A valid statistical test needs to correct for the network structure.
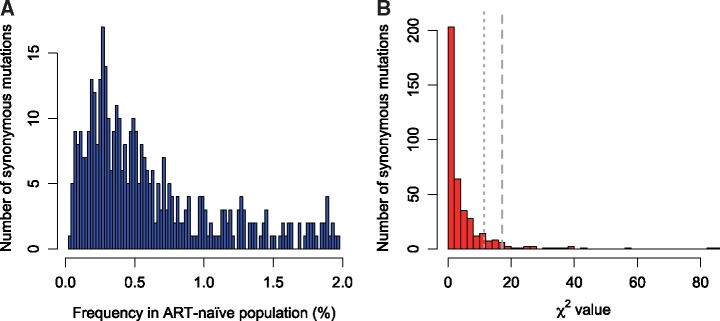
To ameliorate this problem, we constructed a null distribution of expected deviations from random clustering using low frequency synonymous variants in the ART-naïve population. These synonymous variants serve as a proxy for neutral markers across the transmission network. For all major alleles (frequency ≥ 95%), we identified majority and minority synonymous variants. We then calculated the deviation from expected clustering frequency using a Χ2 value for the clustering frequency of the minority synonymous variant, relative to the majority synonymous variant at that site. Using the distribution of Χ2 values from 396 low frequency synonymous variants, we constructed a test statistic for evaluating deviation from neutrality for specific DRAMs (Fig. 1). We considered synonymous variants with a frequency ≤2% in our cohort, because most DRAMs have a frequency of <2% in the ART-naïve population. Using this test statistic, we can explicitly test whether the clustering frequency of strains containing a particular DRAM is more extreme than would be expected for a synonymous variant.
Figure 1.
Synonymous mutations with a prevalence ≤2.0% in the ART-naïve population. (A) Frequency of synonymous mutations. (B) Distribution of Χ2 values for expected clustering frequency of 396 synonymous variants with prevalence ≤2.0% in ART-naïve persons in the U.S. National HIV Surveillance System genetic transmission network. This distribution served as a test statistic for determining statistical significance for clustering of drug resistance associated mutations (DRAMs). Long gray dashed line indicates corrected P = 0.05, and short gray dashed line indicates corrected P = 0.10.
For each DRAM, we calculated the deviation from expected clustering frequency (Χ2) and compared this value to the distribution of Χ2 values from synonymous sites. Only DRAMs for which we expected ≥5 clustered and non-clustered strains were included. We expect this test to be highly conservative, because it is likely that many of these synonymous variants also have fitness effects that can increase or decrease their clustering frequency. Therefore, we chose to report results from both the standard Χ2 test and network-informed test (highlighting results below an α of 0.1 for the network-informed test).
2.4 Large clusters
We characterized people in large clusters which included viruses from at least twenty people. Large clusters with evidence of TDR were defined as ≥33% of the viral sequences sharing at least one DRAM with a potential transmission partner in that cluster. The demographic and transmission risk factor composition of large clusters with and without TDR was compared using multivariable modeling that controlled for clustering of sequences.
2.5 Molecular dating analysis
Molecular dating analysis using a Bayesian Markov Chain Monte Carlo (MCMC) approach was performed for all large clusters with evidence of TDR using BEAST v.1.8.1 (Drummond and Rambaut 2007; Drummond et al. 2012). The rate of evolution was calibrated using date of genotyping. For each cluster, we ran chains of 10 million generations using a TN93 substitution model, a constant population size, and a strict molecular clock. More complicated evolutionary models [i.e. GTR+Γ4, exponential population growth, and uncorrelated lognormal relaxed molecular clock (Drummond et al. 2006)] were not supported by the data. For example, the 95% highest posterior density interval of the exponential growth rate parameter and the standard deviation of the lognormal rate distribution abutted against zero. The first 10% of generations were removed as burn-in, and mixing was assessed using Tracer v1.6. Trees were visualized using FigTree v1.4.2.
3. Results
3.1 ART-naïve transmission network
We used HIV-TRACE to construct an HIV genetic transmission network comprising 5,343 clusters. Of the 66,221 persons included in the analysis, 21,106 (31.9%) clustered with at least one other potential transmission partner [i.e. persons whose viruses were ≤0.015 substitution/site divergent, implying a direct or indirect epidemiological connection (Wertheim et al. 2017)]. People likely infected via sexual transmission constituted the majority of individuals in this dataset (81.8%) and were over-represented in the transmission network (89.7%; Table 1). We restricted our investigation into transmission fitness to ART-naïve individuals. Among the 30,196 ART-naïve persons in the network, 11,692 (38.7%) were potential transmission partners with another ART-naïve individual (Table 1). Individuals with a sexual transmission risk factor were also predominant among the ART-naïve persons (89.3%; Table 1). People who were ART-experienced or had unknown ART status were included in the network only for subsequent molecular dating analysis (see below), because ART-experienced people can easily confound fitness estimates. Due to strict inclusion criteria (see Section 2), 97.8% of people in this study were infected with subtype B (Table 1).
Table 1.
Transmission risk factor and subtype for HIV-infected people from twenty-seven U.S. jurisdictions included in this study.
| Attribute | Total |
Clustered in networka |
||
|---|---|---|---|---|
| Any ART-status n (%) | ART-naïve n (%)b | Any ART-status n (%) | ART-naïve n (%)b | |
| All | 66,221 (100%) | 30,196 (100%) | 21,106 (100%) | 11,692 (100%) |
| Risk factor | ||||
| MSM | 33,680 (50.9%) | 17,150 (56.8%) | 14,341 (67.9%) | 8,421 (72.0%) |
| Heterosexual | 20,473 (30.9%) | 9,946 (32.9%) | 4,484 (21.3%) | 2,336 (20.0%) |
| PWID | 10,844 (16.4%) | 2,810 (9.3%) | 2,133 (10.1%) | 869 (7.4%) |
| Other | 1,224 (1.8%) | 290 (1.0%) | 148 (0.7%) | 66 (0.6%) |
| Subtype | ||||
| B | 64,781 (97.8%) | 29,489 (97.7%) | 20,916 (99.1%) | 11,607 (99.3%) |
| Non-B | 1,440 (2.2%) | 707 (2.3%) | 190 (0.9%) | 85 (0.7%) |
aClustered with another ART-naïve individual in network.
bPersons were classified as ART-naïve if they received their first genotype within 3 months of diagnosis and had no evidence of prior ART use.
ART, antiretroviral therapy; MSM, men who have sex with men; PWID, people who inject drugs.
The relative transmission fitness of a DRAM in ART-naïve persons can be estimated as a proportion: the frequency of strains with that specific DRAM that cluster with other strains with that DRAM divided by the frequency of wild-type strains that cluster with other wild-type strains. A decrease or excess in the proportion of strains clustering is indicative of fewer or greater transmission events, respectively. To determine if this increase or decrease in DRAM clustering frequency is significant, we performed both a standard Χ2 test and constructed a more conservative test statistic, based on the observed clustering frequency of low prevalence neutral markers (i.e. synonymous mutations), to correct for the underlying network structure (Fig. 1; see Section 2 for details). We employed this more conservative test, because it has been suggested that network structure may explain the persistence of DRAMs (Brenner et al. 2013; Vega et al. 2015): DRAMs in expanding clusters increase in prevalence due to founder effects, rather than driving the cluster expansion themselves.
Of the 5,127 viruses from ART-naïve persons that contained at least one DRAM, 1,951 (38.0%) clustered with another ART-naïve person in the network, and 1,549 (30.2%) clustered with another ART-naïve person who shared that DRAM (Table 2). Differences in clustering frequency were also detected for viral strains containing different classes of DRAMs (e.g. NRTIs, NNRTIs, and PIs), with strains sharing NRTI mutations having the lowest frequency of clustering (Table 2).
Table 2.
Number of people with DRAMs of different classes included in study and their frequency of clustering with ART-naïve people with the same DRAM.
| DRAM class | Number clustering | Total | Clustering % | Population prevalence | P-valuea | Corrected P-valueb |
|---|---|---|---|---|---|---|
| DRAM or wild-type | 11,692 | 30,196 | 38.7% | – | – | – |
| Any DRAM | 1,549 | 5,127 | 30.2% | 16.98% | <0.001 | <0.001 |
| NNRTI | 924 | 2,651 | 34.9% | 8.78% | <0.001 | 0.086 |
| NRTI | 458 | 2,028 | 22.6% | 6.72% | <0.001 | <0.001 |
| PI | 421 | 1,443 | 29.2% | 4.78% | <0.001 | 0.010 |
aP-value from the unadjusted Χ2 test.
bProbability that the proportion of strains with DRAM clustering is non-random, after correction for network structure (see Section 2).
DRAM, drug resistance associated mutation; NNRTI, non-nucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; PI, protease inhibitor.
3.2 Fitness of HIV variants containing specific DRAMs
Of the 108 different DRAMs found in the ART-naïve population, sixty-nine mutations were present at a high enough frequency to permit further statistical analysis. Twenty-four DRAMs had clustering frequencies that deviated significantly from neutral expectation (Tables 3–5), most of which resulted in a significant reduction in transmission fitness. For example, strains containing M184V had a relative fitness of 0.09 (corrected P = 0.003; Table 3), indicating a substantial reduction in transmissibility. Most of the DRAMs associated with reduced relative fitness were NRTI mutations (e.g. T215Y, K219Q, and K70R). In contrast, NRTI mutations that had no significant impact on transmission fitness included M41L and the T215Y/F revertant mutations: T215D/S/C/V. Of the thirteen ART-naïve individuals with a K65R mutation, none clustered with another person with K65R (relative fitness = 0.00; Table 3). But due to the rarity of K65R, we were unable to detect evidence for a significant reduction in transmission fitness (corrected P = 0.205). We lacked power to characterize rare DRAMs with expected strong negative transmission fitness consequences (e.g. K65R and L74V; Table 3).
Table 3.
Number of people with NRTI (nucleoside reverse transcriptase inhibitor) DRAM containing strains, frequency of clustering with ART-naïve people with the same DRAM, and relative fitness in ART-naïve persons.
| Mutation | Number clustering | Total | Clustering % | Population prevalence | Relative fitnessa | P-valueb | Corrected P-valuec |
|---|---|---|---|---|---|---|---|
| M41L | 157 | 500 | 31.4% | 1.66% | 0.81 | 0.001 | 0.111 |
| T69N | 79 | 395 | 20.0% | 1.31% | 0.52 | <0.001 | 0.010 |
| T215D | 110 | 258 | 42.6% | 0.85% | 1.10 | 0.215 | 0.543 |
| T215S | 89 | 257 | 34.6% | 0.85% | 0.89 | 0.203 | 0.528 |
| D67N | 38 | 189 | 20.1% | 0.63% | 0.52 | <0.001 | 0.030 |
| M184V | 6 | 169 | 3.6% | 0.56% | 0.09 | <0.001 | 0.003 |
| K219Q | 27 | 151 | 17.9% | 0.50% | 0.46 | <0.001 | 0.030 |
| L210W | 41 | 147 | 27.9% | 0.49% | 0.72 | 0.009 | 0.202 |
| T69A | 13 | 144 | 9.0% | 0.48% | 0.23 | <0.001 | 0.010 |
| T215C | 39 | 128 | 30.5% | 0.42% | 0.79 | 0.068 | 0.376 |
| E44D | 20 | 118 | 16.9% | 0.39% | 0.44 | <0.001 | 0.038 |
| A62V | 19 | 113 | 16.8% | 0.37% | 0.43 | <0.001 | 0.040 |
| T215E | 23 | 90 | 25.6% | 0.30% | 0.66 | 0.014 | 0.235 |
| T69D | 8 | 86 | 9.3% | 0.28% | 0.24 | <0.001 | 0.028 |
| K70R | 6 | 61 | 9.8% | 0.20% | 0.25 | <0.001 | 0.040 |
| T215Y | 4 | 44 | 9.1% | 0.15% | 0.23 | <0.001 | 0.071 |
| K219E | 5 | 44 | 11.4% | 0.15% | 0.29 | <0.001 | 0.093 |
| K219R | 2 | 43 | 4.7% | 0.14% | 0.12 | <0.001 | 0.045 |
| V75I | 8 | 39 | 20.5% | 0.13% | 0.53 | 0.030 | 0.278 |
| T215I | 0 | 30 | 0.0% | 0.10% | 0.00 | <0.001 | 0.051 |
| L74I | 4 | 26 | 15.4% | 0.09% | 0.40 | 0.025 | 0.270 |
| F77L | 0 | 24 | 0.0% | 0.08% | 0.00 | <0.001 | 0.086 |
| D67G | 0 | 24 | 0.0% | 0.08% | 0.00 | <0.001 | 0.086 |
| M184I | 0 | 22 | 0.0% | 0.07% | 0.00 | <0.001 | 0.098 |
| K219N | 0 | 17 | 0.0% | 0.06% | 0.00 | 0.002 | 0.144 |
| V75M | 0 | 16 | 0.0% | 0.05% | 0.00 | 0.003 | 0.149 |
| T215F | 0 | 15 | 0.0% | 0.05% | 0.00 | 0.005 | 0.169 |
| K70E | 0 | 15 | 0.0% | 0.05% | 0.00 | 0.005 | 0.169 |
| T215V | 2 | 15 | 13.3% | 0.05% | 0.34 | 0.080 | 0.409 |
| L74V | 0 | 14 | 0.0% | 0.05% | 0.00 | 0.007 | 0.182 |
| K65R | 0 | 13 | 0.0% | 0.04% | 0.00 | 0.010 | 0.205 |
aRatio of frequency of strains with DRAM clustering to frequency of strains with wild-type clustering.
bP-value from the unadjusted Χ2 test.
cProbability that the proportion of strains with DRAM clustering is non-random, after correction for network structure (see Section 2); P-values < 0.10 are denoted in bold.
Table 4.
Number of people with PI (protease inhibitor) DRAM containing strains, frequency of clustering with ART-naïve people with the same DRAM and relative fitness in ART-naïve persons.
| Mutation | Number clustering | Total | Clustering % | Population prevalence | Relative fitnessa | P-valueb | Corrected P-valuec |
|---|---|---|---|---|---|---|---|
| L90M | 187 | 386 | 48.4% | 1.28% | 1.26 | <0.001 | 0.066 |
| T74S | 107 | 244 | 43.9% | 0.81% | 1.13 | 0.112 | 0.452 |
| V11I | 53 | 174 | 30.5% | 0.58% | 0.79 | 0.033 | 0.295 |
| Q58E | 29 | 173 | 16.8% | 0.57% | 0.43 | <0.001 | 0.023 |
| M46I | 13 | 170 | 7.6% | 0.56% | 0.20 | <0.001 | 0.008 |
| M46L | 26 | 137 | 19.0% | 0.45% | 0.49 | <0.001 | 0.040 |
| V82A | 28 | 113 | 24.8% | 0.37% | 0.64 | 0.003 | 0.149 |
| D30N | 41 | 102 | 40.2% | 0.34% | 1.04 | 0.835 | 0.879 |
| N88D | 41 | 97 | 42.3% | 0.32% | 1.09 | 0.539 | 0.735 |
| I85V | 25 | 89 | 28.1% | 0.29% | 0.73 | 0.052 | 0.341 |
| I54V | 19 | 69 | 27.5% | 0.23% | 0.71 | 0.074 | 0.389 |
| I84V | 20 | 65 | 30.8% | 0.22% | 0.79 | 0.235 | 0.563 |
| L24I | 10 | 33 | 30.3% | 0.11% | 0.78 | 0.415 | 0.694 |
| V32I | 2 | 32 | 6.3% | 0.11% | 0.16 | <0.001 | 0.093 |
| I47V | 2 | 19 | 10.5% | 0.06% | 0.27 | 0.022 | 0.263 |
| F53L | 2 | 16 | 12.5% | 0.05% | 0.32 | 0.058 | 0.351 |
| G73S | 0 | 15 | 0.0% | 0.05% | 0.00 | 0.005 | 0.169 |
| I54M | 4 | 15 | 26.7% | 0.05% | 0.69 | 0.488 | 0.725 |
| L76V | 6 | 14 | 42.9% | 0.05% | 1.11 | 0.965 | 0.972 |
aRatio of frequency of strains with DRAM clustering to frequency of strains with wild-type clustering.
bP-value from the unadjusted Χ2 test.
cProbability that the proportion of strains with DRAM clustering is non-random, after correction for network structure (see Section 2); P-values < 0.10 are denoted in bold.
Table 5.
Number of people with NNRTI (non-nucleoside reverse transcriptase inhibitor) DRAM containing strains, frequency of clustering with ART-naïve people with the same DRAM, and relative fitness in ART-naïve persons.
| Mutation | Number clustering | Total | Clustering % | Population prevalence | Relative fitnessa | P-valueb | Corrected P-valuec |
|---|---|---|---|---|---|---|---|
| K103N | 676 | 1821 | 37.1% | 6.03% | 0.97 | 0.332 | 0.629 |
| Y181C | 95 | 245 | 38.8% | 0.81% | 1.00 | 0.971 | 0.980 |
| G190A | 64 | 217 | 29.5% | 0.72% | 0.76 | 0.006 | 0.179 |
| K103S | 47 | 134 | 35.1% | 0.44% | 0.91 | 0.440 | 0.710 |
| P225H | 30 | 116 | 25.9% | 0.38% | 0.67 | 0.006 | 0.174 |
| A98G | 28 | 103 | 27.2% | 0.34% | 0.70 | 0.021 | 0.260 |
| K101E | 11 | 99 | 11.1% | 0.33% | 0.29 | <0.001 | 0.028 |
| Y188L | 28 | 96 | 29.2% | 0.32% | 0.75 | 0.069 | 0.381 |
| H221Y | 13 | 80 | 16.3% | 0.26% | 0.42 | <0.001 | 0.063 |
| L228R | 10 | 61 | 16.4% | 0.20% | 0.42 | 0.001 | 0.101 |
| G190S | 3 | 28 | 10.7% | 0.09% | 0.28 | 0.004 | 0.162 |
| E138Q | 2 | 27 | 7.4% | 0.09% | 0.19 | 0.002 | 0.136 |
| L100I | 0 | 23 | 0.0% | 0.08% | 0.00 | <0.001 | 0.093 |
| K101P | 4 | 19 | 21.1% | 0.06% | 0.54 | 0.178 | 0.510 |
| K101H | 9 | 18 | 50.0% | 0.06% | 1.29 | 0.459 | 0.715 |
| V106A | 0 | 17 | 0.0% | 0.06% | 0.00 | 0.002 | 0.144 |
| V106M | 0 | 14 | 0.0% | 0.05% | 0.00 | 0.007 | 0.182 |
| G190E | 0 | 14 | 0.0% | 0.05% | 0.00 | 0.007 | 0.182 |
| Y188H | 0 | 13 | 0.0% | 0.04% | 0.00 | 0.010 | 0.205 |
aRatio of frequency of strains with DRAM clustering to frequency of strains with wild-type clustering.
bP-value from the unadjusted Χ2 test.
cProbability that the proportion of strains with DRAM clustering is non-random, after correction for network structure (see Section 2); P-values < 0.10 are denoted in bold.
In contrast to these DRAMs with negative fitness consequences, L90M had a relative fitness significantly greater than 1.0 (Table 4), indicating that strains containing this DRAMs had a transmission fitness that exceeded wild-type strains in the ART-naïve population (corrected P = 0.066).
Other DRAMs (i.e. K103N, K103S, and Y181C) that have relatively high prevalence in the ART-naïve population, greater than 0.4%, were associated with relative fitness close to 1.0 (Table 5), indicating wild-type-like transmissibility
3.3 Clusters of transmitted drug resistance
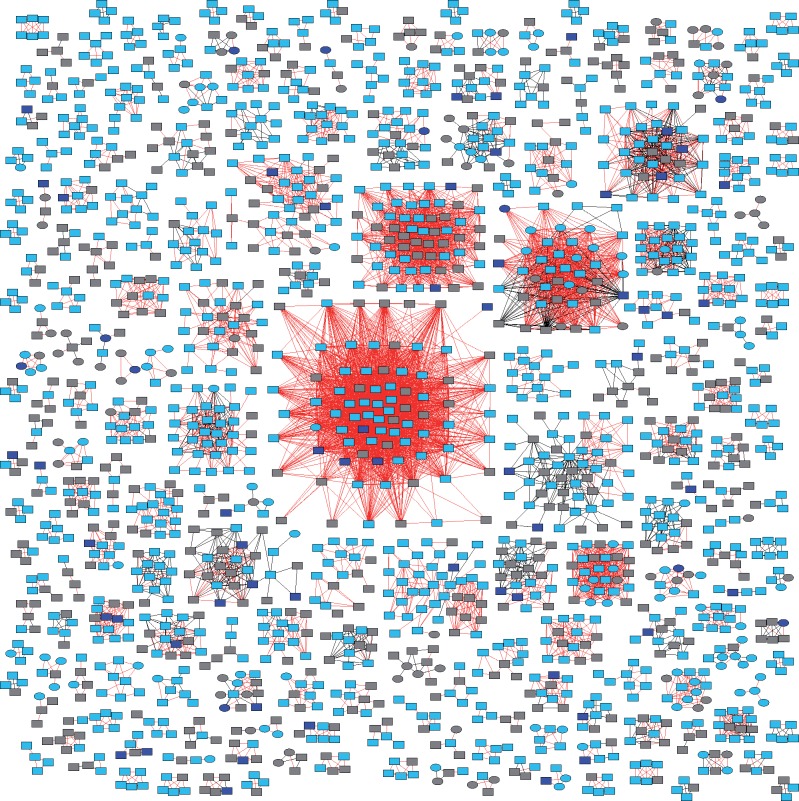
Across the entire U.S. National HIV Surveillance System transmission network, including viruses from ART-naïve, ART-experienced, and ART-unknown persons, we found 212 clusters comprising four or more people with evidence of TDR (i.e. ≥33% of cluster members sharing DRAMs with their potential transmission partners; Fig. 2). Fifteen of these clusters comprised at least twenty people, which we classified as large clusters with evidence of TDR (Table 6). We investigated whether external factors were driving the growth of large clusters with TDR by comparing the demographic and transmission risk factor composition of these fifteen large clusters with TDR with the sixty-five large clusters (i.e. ≥20 persons) without evidence of TDR, but found no significant differences in the composition of large clusters with and without evidence of TDR. These large clusters were all subtype B.
Figure 2.
HIV-1 clusters with evidence of transmitted drug resistance (TDR): ≥33% of nodes share a drug resistance associated mutation with a potential transmission partner. Two hundred and twelve clusters with four or more members are shown. Squares are men and circles are women. Red edges indicate a shared drug resistance associated mutation (i.e. TDR). ART-naïve nodes are light blue and ART-experienced nodes are dark blue. Gray nodes indicate that ART status was unknown.
Table 6.
Phylogenetic analysis of large clusters with evidence of TDR.
| Cluster | No. taxa | Predominant TDR Mutation(s) | Mutation TMRCA year: mean (95% HPD) | % members with TDR | No. ART- experienced |
|---|---|---|---|---|---|
| 194 | 80 | L90M | 2000 (1998–2002) | 100% | 5 |
| T74S | 2000 (1998–2002) | 100% | |||
| 192 | 58 | Y181C | 2005 (2004–6) | 100% | 2 |
| 48 | 51 | K103N | 2001 (1998–2004) | 39.2% | 2 |
| 1805 | 44 | K103N/S | 2004 (2003–6) | 88.6% | 3 |
| 374 | 42 | V11I | 2001 (1998–2002) | 92.9% | 1 |
| 385 | 40 | L90M | 1993 (1987–98) | 100% | 2 |
| 375 | 34 | K103N/S | 2000 (1999–2004) | 100% | 0 |
| 472 | 33 | K103N/S | 2003 (2000–5) | 100% | 4 |
| 84 | 28 | T74S | 2002 (2000–4) | 96.4% | 0 |
| 509 | 27 | K103N | –a | 37.0% | 4 |
| 609 | 27 | K103N | –a | 44.4% | 0 |
| Y181C | –a | 100% | |||
| T215D | –a | 100% | |||
| L210W | –a | 63.0% | |||
| M41L | –a | 88.9% | |||
| 786 | 23 | K103N | 2004 (2002–5) | 56.6% | 2 |
| 175 | 21 | T74S | 2002 (1997–2005) | 66.7% | 1 |
| 524 | 21 | K103N | 2005 (2000–8) | 81.0% | 0 |
| 1019 | 20 | Y181C/V | 1998 (1993–2003) | 65.0% | 1 |
| M46L | 2005 (2003–9) | 25.0% |
aMarkov chain Monte Carlo (MCMC) runs did not converge due to insufficient temporal signal.
TDR, transmitted drug resistance; TMRCA, time of most recent common ancestor, HPD highest posterior density.
Large clusters with TDR included DRAMs representative of resistance to all three drug classes (Table 6). A number of DRAMs conferring resistance to NNRTIs and PIs (K103N, L90M, Y181C, and T74S) are represented in multiple large clusters with TDR. Most of these clusters had high proportions of individuals with TDR (>90%); however, a few of the clusters with K103N did have lower frequencies of TDR ranging from 37% to 56.6% (e.g. clusters 48, 509, 609, 786 in Table 6). This pattern was sometimes due to the appearance of K103N within an already established cluster or reversion to wild-type in a predominantly K103N cluster.
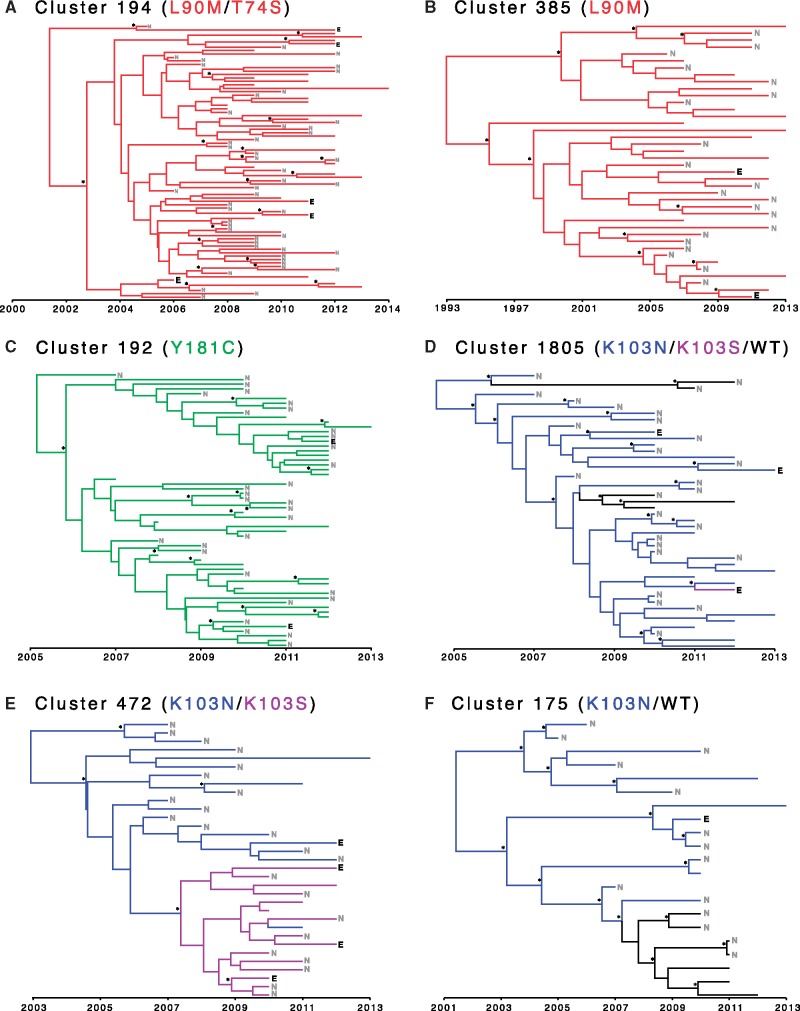
Molecular dating showed that large clusters with TDR typically have a time of most recent common ancestor (TMRCA) dating back to the early 2000s (Table 6). Two older mutations were found in clusters 385 and 1019 in which L90M and Y181C have been propagating since the 1990s. Several of these large clusters have not experienced reversion away from DRAM back towards wild-type, including the two largest clusters with TDR. Notably, viruses from ART-experienced people containing DRAMs were often nested in the phylogeny among viruses from ART-naïve people with the same DRAMs (Fig. 3; Table 6), demonstrating successive transmission of DRAMs from ART-naïve persons to individuals who later initiated ART.
Figure 3.
Molecular dating analysis of six large HIV-1 clusters with evidence of transmitted drug resistance. Phylogenies are colored according to predominant drug resistance associated mutations. Persons who were ART-experienced at the time of genotyping are denoted with a black “E.” Persons who were ART-naïve at the time of genotyping are denoted with a gray “N.” Persons whose ART status was unknown have unlabeled branch tips. Nodes with posterior support ≥0.90 are designated with an asterisk. See Table 6 for more details on clusters.
4. Discussion
Acquired drug resistance in ART-treated persons is the original source of TDR, but the ability of TDR to further spread at the population level among drug-naïve individuals heightens public health concerns as TDR threatens the effectiveness of ART. In this study, we assessed the relative transmission fitness of HIV strains containing DRAMs across a large U.S. National HIV Surveillance System genetic transmission network. This network reflects mostly sexual transmission, the main route of HIV spread globally, and subtype B, viruses which share similar DRAM profiles with non-B viruses, underscoring the relevance of these findings to the global HIV epidemic. After correcting for founder effects in the underlying network, we demonstrate that transmission fitness of strains containing certain major DRAMs were indistinguishable from or exceeded the transmission fitness of wild-type viruses.
Persons infected with virus containing L90M were involved in increased rates of transmission across the network, which may explain why this mutation is still prevalent among transmitted viruses long after replacement of nelfinavir/saquinavir. And viruses containing other prevalent DRAMs, such as K103N and Y181C, were not significantly different from wild-type virus. We also show that viruses with these DRAMs formed large transmission clusters and document long-standing transmission chains tracing back over a decade among ART-naïve persons. These findings are in accordance with the long intra-host persistence of transmitted K103N and L90M averaging 3.7 and 5.8 years, respectively (Jain et al. 2011; Castro et al. 2013), and document the establishment of endemic self-sustained drug resistance reservoirs that persist in ART-naïve populations.
The ease of propagation of K103N strains is of particular importance to resource-limited countries, where a standard first-line NNRTI-based regimen is widely used without routine drug resistance or virus load testing. In this setting, transmitted NNRTI resistance increases risk of ART failure and selection for DRAMs, further increasing the risk of resistance propagation and limiting second-line treatment success (Hamers et al. 2012; Lee et al. 2014; TenoRes Study Group 2016). A recent meta-analysis found that transmitted NNRTI resistance post ART scale-up continues to rise in different parts of the world with yearly odds increases ranging from 1.06- to 1.19-fold (Rhee et al. 2015). Thus, if NNRTI TDR continues to increase, fueled by higher transmission from ART-treated populations and efficient onward spread among ART-naïve persons, TDR will continue to weaken the efficacy of first-line NNRTI-based therapy. The point at which this regimen becomes ineffective is hard to predict, but mathematical models calibrated by new data from this and other studies will be important for inferring this threshold (Wagner et al. 2012; Phillips et al. 2014). Such modeling will assist public health policy to expedite a shift to a first-line therapy that contains an integrase inhibitor (dolutegravir) as a preferred regimen. Preceding a necessary shift to a dolutegravir-based regimen, strategies to safeguard the efficacy of the available NNRTI-based first-line therapy will include both virus load and drug resistance testing to better manage ART and identify which patients should start with the second-line PI-containing regimen. However, given that the resources and capacity to perform drug resistance testing in resource-limited countries are limited, expedited development and implementation of inexpensive and easy-to-use resistance screening approaches may be necessary (Johnson et al. 2008; Hoffmann et al. 2011).
The enhanced transmission of L90M strains is also noteworthy, as it has implications on second-line regimens containing lopinavir or atazanavir in resource-limited countries. L90M is associated with decreased susceptibility to both of these drugs as well as to early generation saquinavir, nelfinavir, and indinavir (Rhee et al. 2010). In the largest cluster with TDR, every virus had L90M and T74S, and the cluster had a TMRCA of 2000 (1998–2002), consistent with the first availability of PIs between 1995 and 1997 (see https://aidsinfo.nih.gov/education-materials/fact-sheets/21/58/fda-approved-hiv-medicines). L90M is one of the most commonly transmitted PI mutations globally, highlighting the need to better understand its clinical impact on the efficacy of second-line therapy (Rhee et al. 2015).
In contrast to resource-limited countries, the propagation of K103N and L90M will be more easily managed in resource-rich countries because of the availability of drug resistance testing to identify active drugs and multiple regimens, including recently recommended integrase inhibitor-based regimens for first-line therapy. Nonetheless, the clinical management of TDR will likely raise the cost and complexity of ART, underscoring the importance of reducing the size of these drug resistance reservoirs. Importantly, the high level of TDR propagation documented in the U.S. National HIV Surveillance System is concordant with that found in Switzerland and the United Kingdom, where 70–84% of TDR had a drug-naïve source (Drescher et al. 2014; Mourad et al. 2015). All these data point to the critical role of untreated or undiagnosed individuals in the spread of DRAMs and suggest that the waning of TDR will be limited unless a larger proportion of the HIV-infected population with DRAMs are diagnosed earlier and treated with appropriate ART regimens to achieve virus suppression and prevent the spread of drug resistance (Drescher et al. 2014; Mourad et al. 2015). Our findings in the USA show a higher TDR prevalence than in resource-limited countries. This observation reflects the longer history of ART in the USA, but also serves as a harbinger of the future trends of TDR in resource-limited countries, which will likely include the establishment of similar drug resistance reservoirs particularly with NNRTI-resistant viruses that will continue to grow possibly at a faster rate than that oberved in the USA fueled by the the lack of drug resistance testing and close management of treated populations (Rhee et al. 2015).
We also identified DRAMs that have low transmission fitness and propagate poorly compared with wild-type viruses. M184V and K65R, mutations that can potentially diminish the efficacy of ART and PrEP, were rarely detected and even less likely to be transmitted across clusters. These results are reassuring, because they suggest that these mutations are unlikely to propagate in the ART-naïve population and have little impact on the efficacy of PrEP. Their low transmission fitness reflects a high replicative impairment by these mutations leading to inefficient sexual transmission, and fast reversion dynamics, which reduces the chance of onward transmission (Jain et al. 2011; Castro et al. 2013). This finding is consistent with data in macaque models in which both M184V and K65R were found to reduce transmission efficiency requiring higher concentrations of virus to achieve infection (Cong et al. 2011, 2013). In addition, macaques infected with these mutant viruses exhibited lower virus loads compared with those infected with wild-type virus, suggesting lower potential for onward transmission (Cong et al. 2011).
Additional DRAMs with low propagation potential included the thymidine analog mutations (TAMs) T69N/D, D67N, K219Q, and F77L, which can be traced back to the predominant use of zidovudine in the early days of ART but remain particularly relevant to resource-limited countries where zidovudine is used in second-line therapy (Cong et al. 2007). The low transmission fitness of these DRAMs predicts waning TDR prevalence over time, but the rate of this decline can be lessened by new transmissions from ART-treated patients.
Prior to 2013, the U.S. National HIV Surveillance System database preferentially collected sequences from persons who were first genotyped within 3 months of diagnosis. Therefore, viruses from ART-experienced persons comprise only 7.7% of the sequences in this database. Nonetheless, we can still make inference about the origin of DRAMs in ART-experienced persons found in large clusters with TDR (Fig. 3). Because these DRAMs were typically present in the clusters dating back to the time of initial diversification, it is likely that the DRAMs in these ART-experienced people represent transmitted drug resistance, rather than de novo mutation selected by ART. For example, Cluster 472 has a TMRCA in 2003 (2000–2005), and all individuals in this cluster, including the presumed ancestor, has K103N (Fig. 3E;Table 6). Four ART-experienced people carrying K103N were documented in this cluster; the most parsimonious explanation for the presence of DRAMs in these ART-experienced people is that at least three of these ART-experienced people were infected by TDR.
Correcting for network structure by using the clustering frequency of synonymous variants represents a novel approach for estimating the fitness effects of heritable traits across a transmission network. However, this approach results in a severe loss of statistical power to detect fitness effects. For example, a Χ2 value of 3.84 is sufficient to detect a significant difference at P = 0.05 and 1 degree of freedom in a non-network adjusted test. For the novel test statistic used here, the comparable Χ2 value for significance with a corrected P = 0.05 is 17.18 with 1 degree of freedom. This test statistic is likely highly conservative, because synonymous mutations in HIV are unlikely to be universally selectively neutral due to RNA secondary structure and codon preferences. Both positive and negative selection on synonymous variants will push the test statistic towards more extreme values, resulting in more conservative estimates of significance. Therefore, it is probable that the deviations from neutrality reported here do reflect natural selection pressure and are not an artifact of network structure.
In conclusion, we use a novel approach to systematically assess the transmission fitness of DRAMs using a large and well-characterized surveillance dataset. The stringent network methods and the large dataset validate the conclusions of the analysis. We document the existence of self-sustaining drug resistance reservoirs among U.S. transmission networks that have important implications for the success of the global ART programs. The study shows how this type of sequence analysis is an important component of TDR surveillance and how it can aid in developing mitigation strategies to minimize the impact of drug resistance and inform strategic decisions on changing ART regimens to preserve effective ART programs.
Acknowledgements
We acknowledge the local and state health department staff instrumental in collecting and reporting HIV sequence data. Thanks to Sergei L. Kosakovsky Pond and Steven Weaver for their contributions to HIV-TRACE and to Ben Murrell for his statistical advice. JOW was funded in part by an NIH-NIAID K01 Career Development Award (K01AI110181) and the California HIV/AIDS Research Program (ID15-SD-052).
Disclaimer
The findings and conclusions of this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. The use of trade names and commercial sources is for identification only and does not imply endorsement by CDC.
Conflicts of interest: None declared.
References
- Barth R. E. et al. (2010) ‘Virological follow-up of adult patients in antiretroviral treatment programmes in sub-Saharan Africa: a systematic review’, Lancet Infectious Diseases, 10: 155–66. [DOI] [PubMed] [Google Scholar]
- Brenner B., Wainberg M. A., Roger M. (2013) ‘Phylogenetic inferences on HIV-1 transmission: implications for the design of prevention and treatment interventions’, AIDS, 27: 1045–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro H. et al. (2013) ‘Persistence of HIV-1 transmitted drug resistance mutations’, Journal of Infectious Diseases, 208: 1459–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. 2014. Pre-exposure Prophylaxis for the Prevention of HIV Infection in the United States – 2014. A Clinical Practical Guide. Atlanta, GA: Centers for Disease Control and Prevention.
- Cohen M. S. et al. (2011) ‘Prevention of HIV-1 infection with early antiretroviral therapy’, New England Journal of Medicine, 365: 493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong M. E., Heneine W., Garcia-Lerma J. G. (2007) ‘The fitness cost of mutations associated with human immunodeficiency virus type 1 drug resistance is modulated by mutational interactions’, Journal of Virology, 81: 3037–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong M. E. et al. (2013) ‘Prophylactic efficacy of oral emtricitabine and tenofovir disoproxil fumarate combination therapy against a tenofovir-resistant simian/human immunodeficiency virus containing the K65R mutation in macaques’, Journal of Infectious Diseases, 208: 463–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong M. E. et al. (2011) ‘Generation and mucosal transmissibility of emtricitabine- and tenofovir-resistant SHIV162P3 mutants in macaques’, Virology, 412: 435–40. [DOI] [PubMed] [Google Scholar]
- Corvasce S. et al. (2006) ‘Evidence of differential selection of HIV-1 variants carrying drug-resistant mutations in seroconverters’, Antiviral Therapy, 11: 329–34. [PubMed] [Google Scholar]
- de Mendoza C. et al. (2004) ‘Evidence for differences in the sexual transmission efficiency of HIV strains with distinct drug resistance genotypes’, Clinical Infectious Diseases, 39: 1231–8. [DOI] [PubMed] [Google Scholar]
- Drescher S. M. et al. (2014) ‘Treatment-naive individuals are the major source of transmitted HIV-1 drug resistance in men who have sex with men in the Swiss HIV Cohort Study’, Clinical Infectious Diseases, 58: 285–94. [DOI] [PubMed] [Google Scholar]
- Drummond A. J. et al. (2006) ‘Relaxed phylogenetics and dating with confidence’, PLoS Biology, 4: e88.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A. (2007) ‘BEAST: Bayesian evolutionary analysis by sampling trees’, BMC Evolution Biology, 7: 214.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J. et al. (2012) ‘Bayesian phylogenetics with BEAUti and the BEAST 1.7’, Molecular Biology and Evolution, 29: 1969–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. K. et al. (2012) ‘Global trends in antiretroviral resistance in treatment-naive individuals with HIV after rollout of antiretroviral treatment in resource-limited settings: a global collaborative study and meta-regression analysis’, Lancet, 380: 1250–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamers R. L. et al. (2012) ‘Effect of pretreatment HIV-1 drug resistance on immunological, virological, and drug-resistance outcomes of first-line antiretroviral treatment in sub-Saharan Africa: a multicentre cohort study’, The Lancet Infectious Diseases, 12: 307–17. [DOI] [PubMed] [Google Scholar]
- Hoffmann D. et al. (2011) ‘Measuring enzymatic HIV-1 susceptibility to two reverse transcriptase inhibitors as a rapid and simple approach to HIV-1 drug-resistance testing’, PLoS One, 6: e22019.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain V. et al. (2011) ‘Differential persistence of transmitted HIV-1 drug resistance mutation classes’, Journal of Infectious Diseases, 203: 1174–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. A. et al. (2008) ‘Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy’, PLoS Medicine, 5: e158.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joint United Nations Programme on HIV/AIDS (UNAIDS). 2013. Global report: UNAIDS report on the global AIDS epidemic 2013. Geneva, Switzerland: UNAIDS.
- Lee G. Q. et al. (2014) ‘Prevalence and virologic consequences of transmitted HIV-1 drug resistance in Uganda’, AIDS Research and Human Retroviruses, 30: 896–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh Brown A. J. et al. (2003) ‘Transmission fitness of drug-resistant human immunodeficiency virus and the prevalence of resistance in the antiretroviral-treated population’, Journal of Infectious Diseases, 187: 683–6. [DOI] [PubMed] [Google Scholar]
- Liu T. F., Shafer R. W. (2006) ‘Web resources for HIV type 1 genotypic-resistance test interpretation’, Clinical Infectious Diseases, 42: 1608–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammano F. et al. (2000) ‘Retracing the evolutionary pathways of human immunodeficiency virus type 1 resistance to protease inhibitors: virus fitness in the absence and in the presence of drug’, Journal of Virology, 74: 8524–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon J. H. et al. (2013) ‘Viral suppression after 12 months of antiretroviral therapy in low- and middle-income countries: a systematic review’, Bulletin of the World Health Organization, 91: 377–385E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaner J. S. et al. (2014) ‘Expansion of HAART coverage is associated with sustained decreases in HIV/AIDS morbidity, mortality and HIV transmission: the “HIV Treatment as Prevention” experience in a Canadian setting’, PLoS One, 9: e87872.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourad R. et al. (2015) ‘A phylotype-based analysis highlights the role of drug-naive HIV-positive individuals in the transmission of antiretroviral resistance in the UK’, AIDS, 29: 1917–25. [DOI] [PubMed] [Google Scholar]
- Oster A. M. et al. (2015) ‘Using Molecular HIV Surveillance Data to Understand Transmission Between Subpopulations in the United States’, Journal of Acquired Immune Deficiency Syndromes, 70: 444–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palella F. J., Jr et al. (1998) ‘Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection’, New England Journal of Medicine, 338: 853–60. [DOI] [PubMed] [Google Scholar]
- Phillips A. N. et al. (2014) ‘Effectiveness and cost-effectiveness of potential responses to future high levels of transmitted HIV drug resistance in antiretroviral drug-naive populations beginning treatment: modelling study and economic analysis’, Lancet HIV, 1: e85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingen M. et al. (2014) ‘Persistence of frequently transmitted drug-resistant HIV-1 variants can be explained by high viral replication capacity’, Retrovirology, 11: 105.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon A. F. et al. (2015) ‘The impact of clinical, demographic and risk factors on rates of HIV transmission: a population-based phylogenetic analysis in British Columbia, Canada’, Journal of Infectious Diseases, 211: 926–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn T. C. et al. (2000) ‘Viral load and heterosexual transmission of human immunodeficiency virus type 1’, Rakai Project Study Group. New England Journal of Medicine, 342: 921–9. [DOI] [PubMed] [Google Scholar]
- Rhee S. Y. et al. (2015) ‘Geographic and temporal trends in the molecular epidemiology and genetic mechanisms of transmitted HIV-1 drug resistance: an individual-patient- and sequence-level meta-analysis’, PLoS Medicine, 12: e1001810.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S. Y. et al. (2010) ‘HIV-1 protease mutations and protease inhibitor cross-resistance’, Antimicrobial Agents and Chemotherapy, 54: 4253–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struck D. et al. (2014) ‘COMET: adaptive context-based modeling for ultrafast HIV-1 subtype identification’, Nucleic Acids Research, 42: e144.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Nei M. (1993) ‘Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees’, Molecular Biology and Evolution, 10: 512–26. [DOI] [PubMed] [Google Scholar]
- TenoRes Study Group. (2016) ‘Global epidemiology of drug resistance after failure of WHO recommended first-line regimens for adult HIV-1 infection: a multicentre retrospective cohort study’, The Lancet Infectious Diseases, 16: 565–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega Y. et al. (2015) ‘Epidemiological surveillance of HIV-1 transmitted drug resistance in Spain in 2004-2012: relevance of transmission clusters in the propagation of resistance mutations’, PLoS One, 10: e0125699.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner B. G., Garcia-Lerma J. G., Blower S. (2012) ‘Factors limiting the transmission of HIV mutations conferring drug resistance: fitness costs and genetic bottlenecks’, Scientific Reports, 2: 320.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim J. O. et al. (2017) ‘Social and genetic networks of HIV-1 transmission in New York City’, PLoS Pathogens, 13: e1006000.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim J. O. et al. (2014) ‘The global transmission network of HIV-1’, Journal of Infectious Diseases, 209: 304–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler W. H. et al. (2010) ‘Prevalence of transmitted drug resistance associated mutations and HIV-1 subtypes in new HIV-1 diagnoses, U.S.-2006’, AIDS, 24: 1203–12. [DOI] [PubMed] [Google Scholar]
- Winand R. et al. (2015) ‘Assessing transmissibility of HIV-1 drug resistance mutations from treated and from drug-naive individuals’, AIDS, 29: 2045–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. 2015. Guideline on when to start antiretroviral therapy and on pre-exposure prophylaxis for HIV. Geneva: WHO. [PubMed]