Abstract
Potato virus Y (PVY) is a major pathogen of potatoes and other solanaceous crops worldwide. It is most closely related to potyviruses first or only found in the Americas, and it almost certainly originated in the Andes, where its hosts were domesticated. We have inferred the phylogeny of the published genomic sequences of 240 PVY isolates collected since 1938 worldwide, but not the Andes. All fall into five groupings, which mostly, but not exclusively, correspond with groupings already devised using biological and taxonomic data. Only 42 percent of the sequences are not recombinant, and all these fall into one or other of three phylogroups; the previously named C (common), O (ordinary), and N (necrotic) groups. There are also two other distinct groups of isolates all of which are recombinant; the R-1 isolates have N (5′ terminal minor) and O (major) parents, and the R-2 isolates have R-1 (major) and N (3′ terminal minor) parents. Many isolates also have additional minor intra- and inter-group recombinant genomic regions. The complex interrelationships between the genomes were resolved by progressively identifying and removing recombinants using partitioned sequences of synonymous codons. Least squared dating and BEAST analyses of two datasets of gene sequences from non-recombinant heterochronously-sampled isolates (seventy-three non-recombinant major ORFs and 166 partial ORFs) found the 95% confidence intervals of the TMRCA estimates overlap around 1,000 CE (Common Era; AD). We attempted to identify the most accurate datings by comparing the estimated phylogenetic dates with historical events in the worldwide adoption of potato and other PVY hosts as crops, but found that more evidence from gene sequences of non-potato isolates, especially from South America, was required.
Keywords: Potato virus Y, phylogenetics, recombination, least squares dating, probabilistic dating.
1. Introduction
Potato virus Y (PVY) is the type species of the genus Potyvirus, one of the largest, most widespread, and economically important genera of plant viruses. It is a major world pathogen of potatoes, and other solanaceous crops, such as tobacco, tomato, and pepper, and is probably the most damaging virus of the world’s potato crop (Loebenstein et al. 2001; Stevenson et al. 2001; Kerlan 2006; Kerlan and Moury 2008; Gray et al. 2010; Ogawa et al. 2012; Karasev and Gray 2013; Jones 2014). PVY was first described by Smith (1931), its C strain by Bawden (1936) who considered it to be a distinct but related virus (potato virus C; PVC), and its N strain by Nobrega and Silberschmidt (1944). PVC was subsequently recognized as the common strain of PVY by Bawden and Sheffield (1944) and PVY’s original or “ordinary” strain then became its O strain. The O and C strains from potato were distinguished by their reactions with the potatoes Ny and Nc hypersensitivity genes, and the third strain, the N strain, caused systemic veinal necrosis in tobacco. The biological strains named O, C, and N were later adopted (De Bokx and van der Want 1987; Jones 1990) and were mostly congruent with their phylogenetics (Boonham et al. 2002; Moury et al. 2002). Isolates from pepper and tomato mostly belong to the C phylogenetic group (Moury 2010), whereas those from tobacco were mostly placed in the N and O phylogenetic groups (Tian et al. 2011). However, as more genomes have been sequenced and more potato hypersensitivity genes found (Singh et al. 2008; Moury 2010; Karasev et al. 2011; Karasev and Gray 2013; Jones 2014; Rowley, Gray, and Karasev 2015; Kehoe and Jones 2016), continued use of the same names for biological and phylogenetic groups has proved to be increasingly confusing due to the lack of complete coincidence between them. A new strain nomenclature system for sub-dividing the phylogenetic groups using Latinised numerals has therefore been proposed (Jones 2014; Jones and Kehoe 2016; Kehoe and Jones 2016). Their groupings correlate with the five broad ‘phylogroups’ we discuss in this article; the traditional C, O, and N groups plus two groups of recombinants, R1 and R2.
The genomic sequences of more than 200 isolates of PVY are now publicly available in the international databases. They come from all continents except Antarctica but none have been collected from crops in the potato’s main center of domestication in the Andean region of Bolivia and Peru, where nine potato species are cultivated in contrast to only one (Solanum tuberosum) outside the Andean region (Hawkes 1978; Brown 1993; Brown and Henfling 2014). The phylogenetic history of PVY is complex. It has undoubtedly involved spread within and between wild and domesticated potato species, wild ancestors of tomato, pepper, tobacco, and other solanceous crops in South or Central America, and also within plantings of potato and other cultivated solanaceous plants throughout the world (Klinkowski and Schmelzer 1960; Silberschmidt 1960; Brücher 1969; Jones 1981; Spetz et al. 2003). It has also involved recombination between lineages (Glais, Tribodet, and Kerlan 2002; Moury et al. 2002; Lorenzen et al. 2006; Schubert, Fomitcheva, and Sztangret-Wisniewska 2007; Ogawa et al. 2008; Singh et al. 2008; Hu et al. 2009a, b; Visser and Bellstedt 2009; Moury and Simon 2011; Cuevas et al. 2012; Karasev and Gray 2013). Many of these recombinants cause tuber necrosis (Beczner et al. 1984; Boonham et al. 2002; Karasev and Gray 2013).
Here, we report an analysis updating the phylogenetic history of PVY. It is based on the genome sequences available in the international databases in January 2016. They were from isolates collected from around the world, but none were from the potato crop’s domestication center in the Andes, where greater diversity would be expected. The majority of the isolates were from potatoes, but several also came from pepper, tomato, or tobacco. As recombination confounds phylogenetic analysis, our strategy was to separate the genomic sequences that are mostly non-recombinant (n-rec) from those that had major recombinant (rec) regions. To simplify the separation of rec from n-rec sequences, the PVY ORF sequences were partitioned into alignments of codons that only varied synonymously (syn codons), and those that had also varied non-synonymously (n-syn codons), and these were then analyzed separately. This strategy identified two sets of n-rec sequences, one of full-length ORFs and the other partial ORFs, for dating the PVY phylogeny. Our dating analyses, like those of Visser, Bellstedt, and Pirie (2012), find that the PVY population outside the Andean region mostly diverged over the past few centuries. This was after the Spanish colonization of South America following the defeat of the Inca empire in 1,532, and after the introduction of one species of potato (S. tuberosum) to Europe in the second half of the 16th century and later to other continents (Salaman and Hawkes 1949; Salaman 1954; Brown 1993; Hawkes and Fransisco-Ortega 1993; Brown and Henfling 2014). We found, like Visser, Bellstedt, and Pirie (2012), that the damaging rec variants emerged and spread only during the past century.
2. Methods and data sources
Two hundred and forty complete genomic sequences of PVY (Supplementary Table S1) were obtained from Genbank in January 2016. Each was edited using BioEdit (Hall 1999) to extract its main ORF. These were aligned, using the encoded amino acids as guide, by the TranslatorX online server (Abascal, Zardoya, and Telford 2010; http://translatorx.co.uk) with its MAFFT option (Katoh and Standley 2013) to give an alignment of 9,201 nucleotides (available at http://192.55.98.146/_resources/e-texts/blobs/240PVYORFs.zip). BlastN and BlastP (Altschul et al. 1990) online facilities of Genbank were used with the Chile 3 sequence, and representative PVY sequences from the N and C phylogroups, to search for related sequences. A simple pairwise sliding-window method DnDscan (Gibbs et al. 2006), available at http://192.55.98.146/_resources/e-texts/blobs/DnDscan1.ZIP, was used to identify codons in the alignments that had only evolved synonymously or had also evolved non-synonymously. These were partitioned using SEQSPLIT v1.0, a Fortran program written by, the late John Armstrong (available at http://192.55.98.146/_resources/e-texts/blobs/SeqSplit.ZIP). Sequences were tested for the presence of phylogenetic anomalies using the full suite of options in RDP4 (Maynard Smith 1992; Holmes, Worobey and Rambaut 1999; Padidam, Sawyer and Fauquet 1999; Gibbs et al. 2000; Martin and Rybicki 2000; McGuire and Wright 2000; Posada and Crandall 2001; Martin et al. 2005; Boni, Posada and Feldman 2007; Lemey et al. 2009; Martin et al. 2015); regions found to be anomalous by three or fewer methods and < 10−6 random probability were ignored. For one test, codons in the aligned sequences with gaps were removed using POSORT (available at http://192.55.98.146/_resources/e-texts/README-POSORT.pdf).
Models for ML analysis were tested using TOPALi (Milne et al. 2009) and the ProtTest 3 server at http://darwin.uvigo.es (Darriba et al. 2011); the best fit models were found to be GTR+Г4 + I (Tavaré 1986) for nucleotide sequences and LG+ Г4 + I (Le and Gascuel 2008) for amino acid sequences. Phylogenetic trees were inferred using the neighbor-joining (NJ) facility in ClustalX (Jeanmougin et al. 1998), the SplitsTree method (Huson and Bryant 2006) and PhyML 3.0 (ML) (Guindon and Gascuel 2003), and the support for their topologies assessed using the log-likelihood support for the trees and the SH-support (Shimodaira and Hasegawa 1999) for their nodes. Nucleotide diversities (Nei and Li 1979) were computed using DAMBE5 online (Xia 2013). Trees were drawn using Figtree Version 1.3 (http://tree.bio.ed.ac.uk/software/figtree/), and pairs of trees were compared using PATRISTIC (Fourment and Gibbs 2006) to test for mutational saturation and were confirmed by the method of Xia (2013). Most virus isolate collection dates (Supplementary Table S1) were obtained from Genbank files, or from Visser, Bellstedt, and Charleston (2012), and dates for some N and NTN isolates were not previously published (Ohshima K—unpublished data); the dating of sequence EU563512 is mentioned in the Section 4. The temporal signal in sets of aligned sequences, and the dates of the most common recent ancestor (TMRCA) and other nodes of inferred phylogenies were estimated by TempEst (Rambaut et al. 2016), the ‘Least Squares Dating’ method of To et al (2015) using Version lsd-0.3beta, and by the probabilistic methods of BEAST v1.8.2 (Drummond et al. 2012). In BEAST analyses Bayes factors were used to select the best-fitting molecular-clock model and coalescent priors for the tree topology and node times, and we compared strict and relaxed (uncorrelated exponential and uncorrelated lognormal) molecular clocks, as well as five demographic models (constant population size, expansion growth, exponential growth, logistic growth, and the Bayesian skyline plot). Posterior distributions of parameters, including the tree, were estimated from Markov Chain Monte Carlo samples taken every 104 from 108 steps after discarding the first 10 percent, and checked using Tracer v1.6 (http://tree.bio.ed.ac.uk/software/tracer/). These provided the TMRCAs, and other dates were obtained from the ‘maximum clade credibility tree’, namely the tree that was commonest among those observed. The adequacy of the temporal signal in our data was also checked by using ten independently date-randomized replicates in both the least squared dating (LSD) and BEAST analyses (Ramsden, Holmes and Charleston, 2009; Duchêne 2015).
3. Results
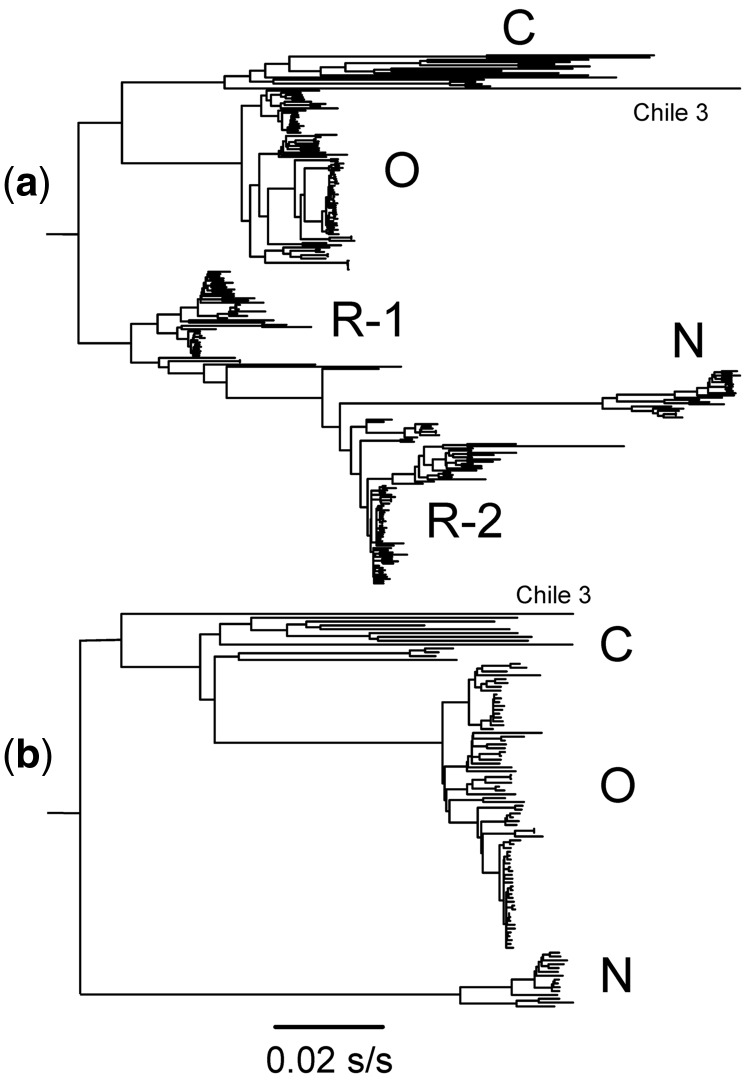
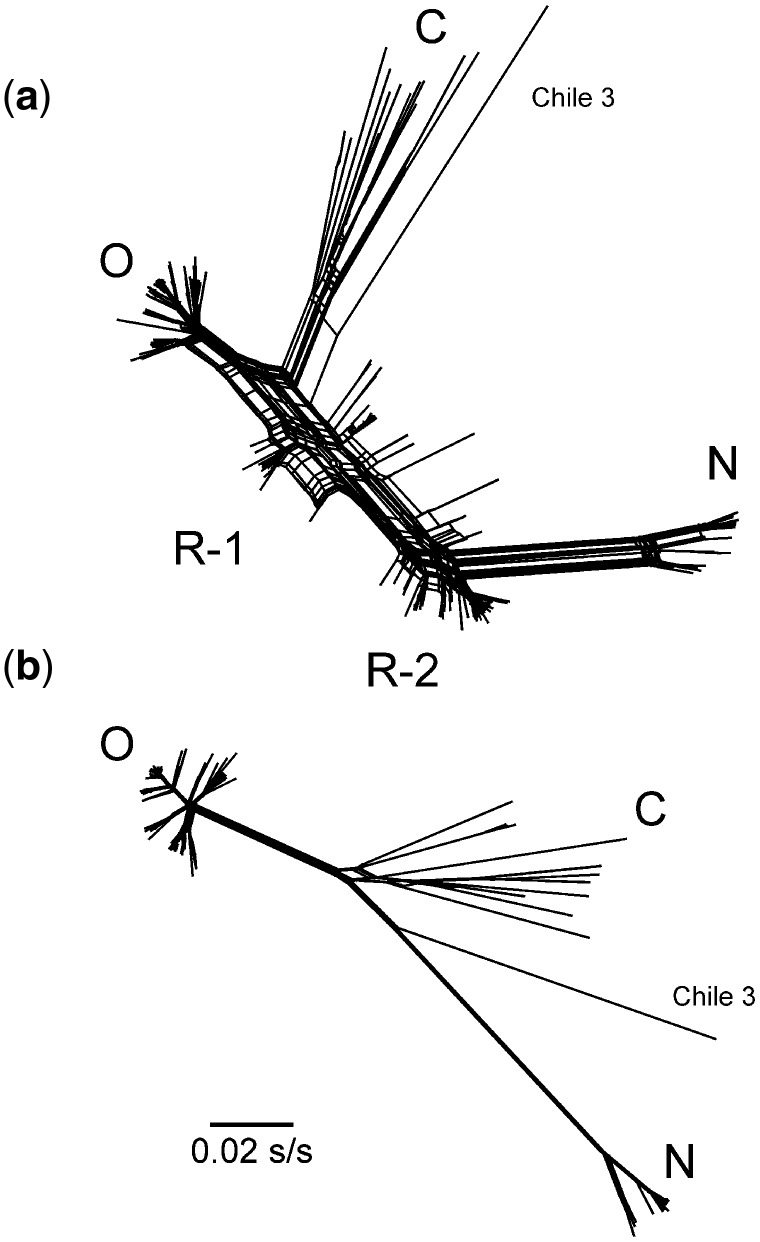
The principal ORFs of 240 genomic sequences of PVY gave an alignment 9,201 nucleotides long with 6.7 percent of the 3,067 codons invariate. The ORFs formed five major groupings in a mid-point rooted NJ tree (Fig. 1A). Their relationships closely resembled those found in a maximum likelihood phylogeny reported by Kehoe and Jones (2016) who examined seventy-three isolates from among the 240 that we examined. In both our NJ phylogeny and the published ML phylogeny, the C phylogroup had the longest branches and formed the basal tree. One of its lineages diverges to give the O group and three others, N, R-1, and R-2; see Fig. 1 legend for the correspondences between our phylogroups and those of Kehoe and Jones (2016). In both phylogenies, many sister sequences have very asymmetric relationships; when summed to the midpoint root, the shortest branches are less than 25 percent of the longest. This may indicate that evolutionary rates have varied or, more likely, that some of the sequences are recombinant. The same arrangement of the same phylogroups was inferred by SplitsTree analysis (Fig. 2A) with the C and O phylogroups linked to others by a complex web of interrelationships indicating that many of the ORFs are recombinants, some of which are closely similar as shown by clustering of the links between the groups.
Figure 1.
The branches of NJ phylogenies calculated from the main genomic ORFs of (A) 240 isolates of PVY and (B) 103 of the isolates that showed no significant evidence of recombination (see text). The marked clusters are of groups named in Kehoe and Jones (2016); cluster C is of groups C1(II) and C2(III); O is of O(I) and O5(X); N is of N(IV), XIII and NA-N(IX); NTN-1 is of NTN-NW, SYR-I(XII), NTN-B(VI), NTN-NW, SYR-II(XI), N-Wi(VII), and N:O(VIII); and NTN-2 of NTN-A(V). Chile 3 is isolate Accession Code FJ214726.
Figure 2.
The branches of SplitsTree phylogenies of the main genomic ORFs of (A) 240 isolates of PVY and (B) 103 of those isolates that showed no significant evidence of recombination. The marked clusters are the same as those in Fig. 1.
The ORFs were also directly searched for phylogenetic anomalies using the RDP suite of programs, and more than half gave significant evidence of recombination but with the assignment of ‘parental’ sequences often uncertain as many alternative combinations of ORFs were identified as possible ‘parents’.
3.1 The search for the n-rec PVY ORFs
The search for the n-rec ORFs was simplified by using alignments obtained by partitioning the variable codons into sequences of the syn codons (50.4 percent) and the n-syn codons (42.9 percent) they contained. NJ trees of these two alignments were closely similar topologically to those of the complete sequences (Fig. 1A) and, when compared in a patristic-distance graph (Fig. S1), they showed no evidence of mutational saturation along the main axis of the tree, and this was confirmed using the binary test for saturation (Xia 2013). However, there were discrete clusters of points off the main diagonal, as would be expected for rec sequences, and the main diagonal was very broad with the spread being greater in the n-syn axis (X) than in the syn axis (Y), again indicating the presence of recombinants.
ML trees obtained from the complete ORF sequences, and from their syn and n-syn codons, were also closely similar however the syn codon tree had a greater statistical support than those calculated from the n-syn codons or the complete sequences; log-likelihoods of −45390.6 and −60825.8 and −109855.39165, respectively. Furthermore, more nodes in the syn sequence tree had SH-support >0.9 than the same nodes in the other trees.
A preliminary search of the syn codon sequences using RDP, as well as NJ trees and patristic distance graphs, showed that all members of the R-1 and R-2 phylogroups had some large rec regions with the same ‘parents’. A representative selection of 48 sequences (Supplementary Table S1) was used to resolve these groupings. The selection included all sixteen sequences of the C phylogroup together with eight sequences from each of the other phylogroups, chosen to be as representative as possible of the basal divergences of those phylogroups (i.e. those with the shortest branch lengths, and therefore least changed after the phylogroup had been established). Sequences with the same pattern of recombination and parentage were progressively identified and removed. The most strongly supported phylogenetic anomaly was shared by all eight R-2 sequences. They had an R-1 phylogroup sequence (EF026076) as the major parent and an N phylogroup minor parent (AJ890346); the latter region was from around nt 5707 to the 3′ terminus of the complete ORFs (38 percent of the ORF). It was identified by seven out of nine RDP methods with probabilities ranging from 10−27 to 10−112. All R-2 sequences were therefore removed, and the remaining forty sequences again analyzed by RDP, which then found the most strongly supported and shared anomaly to be in all R-1 sequences. These had an O phylogroup major parent (EF026074) and an N phylogroup minor parent (X97895), that had, in most (see below), been provided from the 5′ terminus to nt 2205 (24 percent of the ORF); this region was identified by seven out of nine RDP methods with probabilities ranging from 10−23 to 10−84. The syn and n-syn sequences of the remaining thirty-two C, O, and N phylogroup ORFs gave almost identical ML and NJ phylogenetic trees, and a patristic distance graph of those trees had most points close to a single diagonal; the statistical support for the syn ML tree was again greater than for the n-syn ML tree (log-likelihood −24723.4 and −29299.9, respectively).
Finally, the syn codon sequences from C and O phylogroup isolates, that were not among the forty-eight representative sequences, were added to the remaining thirty-two C and O syn sequences to produce an alignment of 120 sequences. This was then searched using RDP for other more specific rec regions (i.e. sequence specific, not phylogroup specific), and seventeen sequences were found to have significant inter- and intra-phylogroup rec regions (i.e. three C phylogroup sequences, seven from the O phylogroup, and seven from the N phylogroup). One of those N phylogroup sequences was X97895 from isolate N605, which has been used in several studies as a reference sequence, but was found by us to be a recombinant between AJ890346 and DQ157180, representing the two main lineages of the N phylogroup. Figure 1B shows the NJ tree of the complete ORFs of the remaining 103 sequences. This phylogeny is much more symmetrical than that of the original 240 sequences (Fig. 1A) and its shortest branches (tip to root) are around 70 percent of the length of the longest. Figure 2B shows their SplitsTree graph, which again has a much simpler linkage structure in its main branches than the 240 sequence tree (Fig. 2A). Figure S1 also shows the patristic distance graph comparing the NJ trees of the 103 syn and n-syn sequences, and this also confirms that these n-rec sequences have evolved in a biologically coherent manner with most points aligned with, and close to, a diagonal that has a slope around 0.8, and shows no evidence of mutational saturation.
3.2 The R-1 and R-2 phylogroups
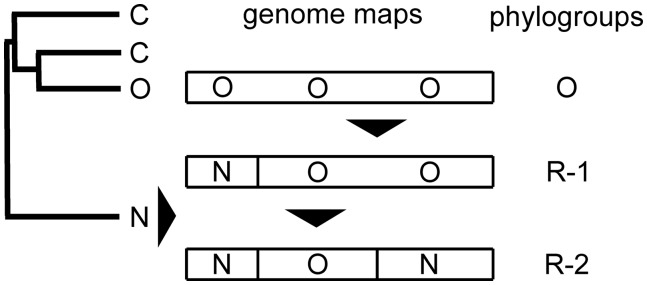
The genome maps of the three rec phylogroups are shown in Fig. 3. Phylogroup N is the sister lineage to the C and O phylogroups and most genomes of these three phylogroups are non-recombinant, whereas all the R-1 and R-2 phylogroup sequences are recombinants and have parents in the O and N phylogroups.
Figure 3.
A cartoon summarizing the relationships and genomic maps of the five major phylogroups of the PVY isolates shown in Figs 1 and 2.
The ORFs of all forty-three R-1 phylogroup sequences are most closely related to ORFs of the O phylogroup, especially that of sequence EF026074. However all have a 5′ terminal region (24 percent of the complete ORF) that is closely related to the homologous region of N phylogroup ORFs, especially that of X97895. In around half the ORFs (e.g. DQ157179, HQ912870, JF927762) the N phylogroup region is from its 5′ end to around nt 2205, and in others (e.g. AJ890349, HQ912863, JN935419, and KJ801915) it is from nts 301 to 2,205, and is preceded by a short region closest to O sequences. In two of the sequences (HM991454 and JQ969040) the N phylogroup region is most closely related to AB331517, not X97896. These differences indicate that more than one event and O parent was involved in establishing the R-1 phylogroup. Some of the R-1 sequences (e.g. AJ889868 and KJ634023) also have a short rec region around nts 5,600–6,300 that is most closely related to that region of R-2 isolates.
The sequences of all seventy-six R-2 ORFs are closest to that of an R-1 phylogroup sequence, EF026076, but all have a 3′ region (nts 5707-end; 38 percent of the ORF) that is most closely related to the homologous region of AJ890346, an N phylogroup sequence. In addition around one-fifth of the R-1 and R-2 sequences also have smaller individual regions that in RDP analyses are recorded as significant inter- and intra-phylogroup recombinations.
The C phylogroup ORFs sequences, including that of the Chile 3 isolate, are the most variable; nucleotide diversity π = 0.0974 ± 0.0460. Those of the other four phylogroups are much less variable, indicating that they have probably diverged more recently; the N phylogroup population is the most variable, π = 0.0256 ± 0.0121, compared with the O phylogroup, 0.0211 ± 0.0100, R-1 phylogroup, 0.0196 ± 0.0093, and R-2 the least variable, 0.0188 ±0.0089.
3.3 Rooting the PVY phylogeny
BlastN and BlastP searches using representative sequences from all of the n-rec PVY phylogroups found the most closely related genomic sequences to be those of sunflower chlorotic mottle virus (JN863233), pepper severe mosaic virus (AM181350) and bidens mosaic virus (KF649336). In both ML and NJ trees of seventy-three dated n-rec complete ORFs (see below), including these three viruses as an outgroup, the N phylogroup was placed as sister to all other PVY phylogroups, as in Fig. 1B. However, when ML or NJ trees were calculated from the encoded amino acid sequences, or from the 166 core nucleotide sequences (see below) together with the homologous regions of the three outgroup sequences, the Chile 3 sequence (FJ214726) was placed as sister to all other PVY lineages. This difference was found not only when complete aligned sequences were used, but also when all indels (9.5 percent of the sequences) had been removed. Nonetheless all the major nodes in trees found from complete and degapped sequences of both ORF and amino acid trees, had >0.97 SH-type statistical support. Thus the exact position of the basal node of the present PVY phylogeny is unresolved but is either side of a single robustly supported node linking the Chile 3 sequence and the N phylogroup to all other PVYs. The reason for this rooting uncertainty is unknown; the individual mutational differences between the Chile 3, N phylogroup and all other sequences are spread throughout the full length of the sequences. Furthermore the points representing the pairwise syn and n-syn comparisons of the Chile 3 sequence and other PVY isolates in a patristic distance graph (Fig. S1) were close to the main diagonal trend, indicating that the Dn/Ds ratios of the Chile 3 isolate are not unusual, but similar to those of other PVY isolates.
3.4 Dating the PVY phylogeny
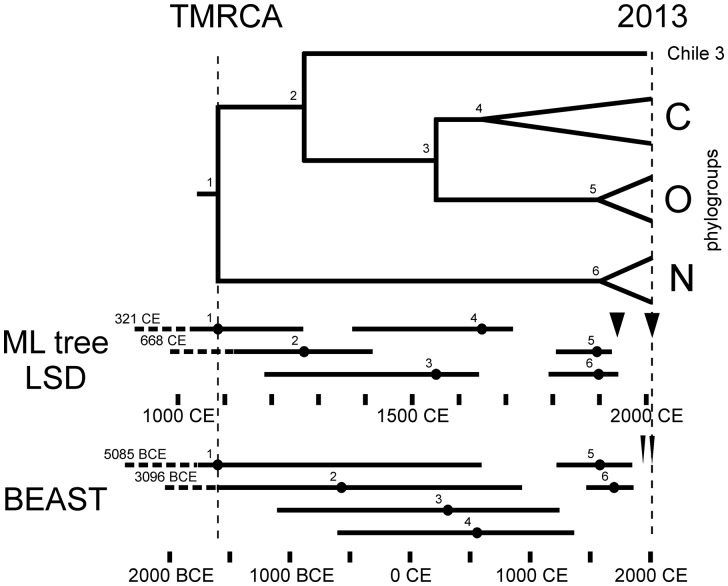
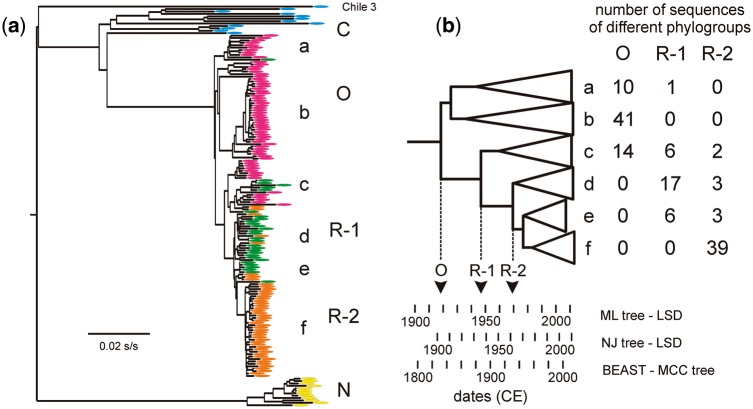
The 240 PVY genomic sequences we analyzed provided two sets of heterochronously dated sequences (Supplementary Table S1). Seventy-three non-recombinant isolates from the C, O, and N phylogroups provided a set of full length ORFs; the earliest sample was collected in 1938 CE (Common Era or AD), the most recent in 2013 CE and their mean sample collection date was 1999.4 CE. As described above the R-1 and R-2 genomes are recombinants that share the central core region of their genomes (nts 2,206–5,706) with those of the O phylogroup. Sample collection dates are known for ninety-eight R-1 and R-2 isolates (Supplementary Table S1). Thus, together with the core regions of the seventy-three dated recombinant sequences, their shared central region provides another dataset of 171 sequences for estimating PVY phylogenetic divergence dates; it covers the same range of sampling dates, but with a mean of 2004.1 CE. Although there are twice as many sequences, they are only 38 percent of the length of the complete ORFs. The alignment of the 171 core regions was re-examined by RDP, and five found to have a short 5′ terminal region of N phylogroup sequence (closest to JQ969036), so these were removed to leave 166 sequences in the dataset. ML and NJ trees inferred from these core regions differed only in minor details and resembled closely those of the seventy-three n-rec ORFs (Fig. 1), except that they placed all the O, R-1, and R-2 sequences in single tight cluster that did not cleanly resolve into O, R-1, and R-2 lineages (Fig. 4). The poor resolution within this cluster of O, R-1, R-2 sequences probably results from the small diversity within the cluster; nucleotide diversity π = 0.018 ± 0.008 compared with 0.048 ± 0.023 in all 166 core sequences, and 0.075 ± 0.035 in the seventy-three n-rec complete ORFs. Nonetheless the distribution of O, R-1 and R-2 sequences within the lineages (Fig. 4) indicate clearly that O isolates are ancestral to R-1 isolates, and R-2 isolates are the most recent.
Figure 4.
A cartoon summarizing the divergence dates of a ML phylogeny of seventy-three dated n-rec ORF sequences (Table 1 - line 1) estimated by the LSD method, and the dates from the MCC tree of a BEAST analysis of the same data. Bars indicate the 95% CI ranges for both analyses. Arrows indicate the period, 1935 CE–2016 CE, during which the isolates were collected.
TempEst analyses of the two datasets (73 n-rec ORFs and 166 cores) found that both had a strong temporal signal; correlation coefficients 0.436 and 0.228, P = 0.00012 and 0.0032, for the complete and core sequences, respectively. They had small residuals with no trend, and these were mostly associated with the C phylogroup sequences and that of Chile 3. The estimated TMRCAs were 1411.6 CE and 1076.8 CE, respectively (Fig. 5). Separate TempEst analyses of the seventy-three n-rec cores and ninety-three rec cores in the 166 core dataset found that the seventy-three n-rec core sequences had a temporal correlation of 0.448 (P = 0.00007) with a TMRCA of 1459.8 CE, whereas the regression for the ninety-three rec core sequences was 0.0356 (P = 0.73). Thus the temporal signal detected by TempEst in the 166 core sequences seems to be mostly, if not exclusively, in its seventy-three n-rec core sequences
Figure 5.
The NJ phylogeny of the central core regions of the genomes of 166 dated C, O, N, R-1, and R-2 isolates. (A) the Accession Codes of the C isolates are blue, O red, R-1 green, R-2 orange, and N yellow. (B) Summary of the phylogroup composition of the collapsed clusters a–f. The node dates are from LSD estimates of MJ and NJ phylogenies and from the MCC phylogeny of a BEAST analysis. These node dates were used to calculate the date scales assuming linearity. Labeled arrows indicate the dates of the likely origins of the O, R-1, and R-2 populations.
The LSD method was used in its ‘constrained’ mode (i.e. an ancestral node must be older than its daughter nodes) to calculate the dates, evolutionary rates, and confidence intervals, for the TMRCAs of the ML and NJ trees of the seventy-three ORF sequences, and also their encoded amino acid sequences (Table 1). Estimates of the TMRCA dates obtained from complete sequences were close to the mean of those from partitioned syn and n-syn codon sequences (Table 1). However, the TMRCA date estimates from ML trees were 80–120 years earlier than those obtained from NJ trees, and there were even larger differences between the TMRCA date estimates from ORF versus amino acid sequences; the former were 250–290 years more ancient than those from amino acid sequences. Table 1 also records the estimated dates of the five principal nodes in the phylogenies. Randomizing the collection dates confirmed that the data contained a clear temporal signal as the ML phylogeny of seventy-three n-rec complete sequences had a TMRCA date of 1085 CE, but gave a mean TMRCA date from twenty collection date randomizations of 12,555 BCE (standard deviation 7,076 years; range 10 BCE–17,036 BCE). Thus the TMRCA date calculated with the true sampling dates is outside the range of dates obtained with randomized collection dates, and 1.9 standard deviations from their mean.
Table 1.
LSD dates of the TMRCA and major nodes in phylogenies of 73 n-rec PVY sequences.
| Datasetb | Datac | Algorithmd | Sites used | Node datesa |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Node 1e | Node 2 | Node 3 | Node 4 | Node 5 | Node 6 | Ratef (×10−4s/s/y) | ||||
| ORF | Nucs | ML | All | 1085.4 (1251–123) | 1269.3 | 1551.2 | 1651.4 | 1898.2 | 1901.8 | 2.07 (0.98–2.55) |
| n-syn codons | 1158.3 (1265–792) | 1320.8 | 1575.3 | 1680.5 | 1906.6 | 1903.4 | 2.78 (1.94–3.23) | |||
| syn codons | 1017.6 (1234–151) | 1243.7 | 1538.3 | 1629.3 | 1881.3 | 1904 | 1.77 (0.92–2.21) | |||
| NJ | All | 1204.9 (1496–-17272) | 1280.8 | 1515.2 | 1443.1 | 1890 | 1834.9 | 1.11 ((0.05–1.71) | ||
| n-syn codons | 1243.5 (1513 to − 230) | 1309.1 | 1543 | 1462.8 | 1901.9 | 1839.6 | 1.53 (0.55–2.28) | |||
| syn codons | 1152.5 (1467 to − 7.2 × 108) | 1242 | 1438.7 | 1459.8 | 1846.6 | 1828.4 | 0.91 (1 × 10−6–1.46) | |||
| AAs | ML | All | 1363.6 (1501–238) | 1427.1 | 1603 | 1702.1 | 1943 | 1933.6 | 1.05 (0.41–1.26) | |
| NJ | All | 1459.9 (1642–70) | 1513 | 1615 | 1702.3 | 1933.1 | 1910.4 | 0.86 (0.23–1.19) | ||
| Core | Nucs | ML | All | 1151.2 (1296–703) | 1259.9 | 1626.6 | 1622.8 | 1918.2 | 1925.7 | 2.02 (1.34–2.43) |
| NJ | All | 1307.7 (1521 to − 71) | 1307.7 | 1624.8 | 1544.6 | 1906.2 | 1897.5 | 1.19 (0.40–1.57) | ||
Node dates: positive dates are CE (Common Era = AD), negative are BCE (=BC).
Dataset: ORF—major open reading frame; Core, nucleotides 2206 − 5706.
Data: Nucs, nucleotides; AAs, encoded amino acids.
Algorithm: ML, maximum likelihood (PhyML); NJ, neighbor-joining (ClustalX).
Nodes numbered as in Fig. 4; 95% confidence intervals for Node 1 only.
Rate: evolutionary rate, substitutions/site/year; 95% confidence intervals for Node 1.
To et al. (2015) stated that although LSD can analyze trees obtained by any method, ‘more accurate results are expected from trees obtained using maximum-likelihood methods’, which, with our data, gave the earliest TMRCA estimates, and these were most precise in that their 95 percent confidence limits were much smaller (Table 1). They estimate that the present PVY population diverged from a common ancestor around 1085 CE with commensurately more recent dates for the divergences within the phylogeny (Fig. 6).
Figure 6.
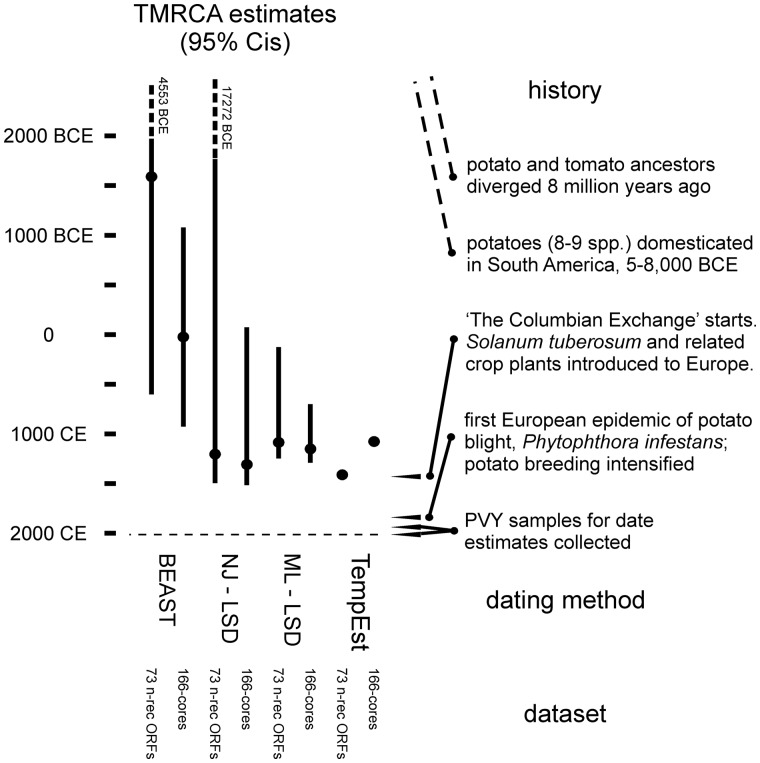
A cartoon summarizing the TMRCA dates and 95% CIs (vertical scale) of PVY estimated by TempEst, LSD (ML and NJ trees) and BEAST (MCC tree) analyses of the seventy-three n-rec ORF and 161-core sequences, together with historical events that may have influenced the evolution of the virus.
LSD analysis of the 166 core sequences gave TMRCA dates (Table 1) for the ML and NJ trees close to those estimated for the seventy-three complete sequences, and all major nodes had >0.9 SH-type statistical support including that of the combined cluster of O, R-1, and R-2 phylogroup sequences and its major nodes. The mean root date from twenty collection date randomizations of the ML data was 11,491 BCE (standard deviation 5,716 years; range 1292 BCE–15,269 BCE), again confirming the strong temporal signal.
The lineages within the cluster of O, R-1, and R-2 sequences in the ML and NJ trees of core sequences, when collapsed, had identical groupings (Fig. 4), although the dates estimated for these lineages differed (Table 1 and Fig. 4). For example the ML tree estimated the origins of O isolates, R-1 isolates and R-2 isolates to be 1918.2 CE, 1946.7 CE, and 1969.4 CE, respectively (Fig. 4), whereas the NJ tree estimates were all earlier and were, respectively, 1906.2 CE, 1933.3 CE, and 1960.3 CE.
In BEAST analyses of the two datasets the best-supported model for mutational substitution was GTR + I + Γ4, while the best model for the rate of substitution was the ‘relaxed uncorrelated lognormal’ clock, and a population of constant size. All datasets passed date-randomization tests. The phylogenies inferred by BEAST had essentially the same topology as those inferred by ML and NJ, however the dates (Table 2) were significantly earlier than those estimated by the regression methods. The TMRCA of the seventy-three n-rec ORF sequences was 1590 BCE, and was twenty-four CE for the 166 cores. The topology and dates of the other nodes were obtained from the maximum clade credibility tree, were commensurately earlier and showed exactly the same pattern of slightly unresolved clustering of the cluster of O, R-1, R-2 core sequences as in the ML and NJ trees (Fig. 4A) with their origins estimated to be 1832.9 CE, 1880.6, and 1932, respectively (Fig. 4B). The 166 core sequences were partitioned into those from the seventy-three from n-rec sequences and those from the ninety-three from rec sequences, they gave TMRCA estimates of 214 BCE and 7 CE, respectively, and passed the date randomization test, indicating that they contained significant temporal signals, in contrast to the results of the TempEst tests.
Table 2.
BEAST dates of TMRCA nodes in phylogenies of PVY sequences.
| Dataset | 73 n-rec complete ORFs | All 166 core regions | 73 n-rec core regions | 93 rec core regions |
|---|---|---|---|---|
| Sequence length (nt) | 9201 | 3501 | 3501 | 3501 |
| No. of sequences | 73 | 166 | 73 | 93 |
| Sampling date range | 1938–2013 | 1938–2013 | 1938–2012 | 1970–2013 |
| TMRCAa (years) | 3603 (1411–6566) | 1989 (1084–3096) | 2227 (951–3952) | 2006 (688–3755) |
| TMRCAb (dates) | −1590 (602 to − 4553) | 24 (929 to − 1083) | −214 (1062 to − 1939) | 7 (1325 to − 1742) |
| Substitution rate (nt/site/year) b | 5.97 × 10−5 (2.50 × 10−5–9.56 × 10−5) | 9.99 × 10−5 (6.88 × 10−5–1.32 × 10−4) | 9.24 × 10−5 (4.33 × 10−5–1.39 × 10−4) | 8.66 × 10−5 (3.71 × 10−5–1.37 × 10−4) |
TMRCA, ‘time to the most recent common ancestor’; years before 2013. 95% credibility intervals (CI) in parentheses.
TMRCA, dates; positive dates are CE (Common Era = AD), negative are BCE (=BC). 95% credibility intervals (CI) in parentheses.
Part of Fig. 5 summarizes and compares the TMRCAs and confidence intervals estimated by the different methods, and shows that despite the TMRCA estimates covering a six-fold range, the 95% confidence limits of most overlap between 500 CE and 1500 CE.
Finally, a simple comparison was made of the basal branch length of the PVY cluster as a proportion of the basal branch length of the potyviruses. A ML phylogeny was calculated from four representative PVY ORF sequences [AB331515 (N phylogroup), EU563512 (C), FJ214726 (C), HM367075 (O)] aligned with those of the other 103 representative potyviruses and rymoviruses used for Fig. 1 of Gibbs, Nguyen, and Ohshima (2015). The mean patristic distances of the sequences connected through the root of the phylogeny (i.e. from Narcissus degeneration virus, NC_008824; Onion yellow dwarf virus, NC_005029; Shallot yellow stripe virus, NC_007433; Vallota speciosa virus isolate Marijiniup 7, JQ723475 to all the other potyviruses) was 2.258 substitutions/site, and through the root of the PVY sequences (i.e. from AB331515 (N) to all the other PVYs) was 0.227 substitutions/site, a ratio of 9.94:1.
4. Discussion
This article reports that using syn codon sequences simplified the resolution of relationships in a complex population of PVY genomes. The phylogenies and RDP analyses calculated from the syn codon sequences were simpler to interpret than those calculated from the comparable n-syn codon or complete sequences, and had greater statistical support. Syn codon changes reflect the temporal component of phylogenetic change of gene populations better than n-syn codons, as changes of the latter are also linked to changes of the encoded protein, and incoherent changes of syn and n-syn codons in complete sequences may confound analysis of either, especially when rec sequences are present. Significantly Ohshima et al (2016) also found that Bayesian estimates of the ages of cucumber mosaic cucumovirus (CMV) populations were more precise if made using syn codon sequences rather than complete sequences; CMV has three genomic segments, which encode five ORFs and they found that the age estimates for different ORFs using syn codon sequences had a coefficient of variation around half that of complete sequences, and the 95% CI ranges of those estimates was around 25–50 percent smaller than those of the original complete sequences.
The strategy of progressively removing the most strongly supported cluster specific recombinants resolved the complexity of the relationships of the rec and n-rec ORFs. In Fig. 1A (i.e. a NJ tree), and in Fig. 2 of Kehoe and Jones (2016) (i.e. an ML tree), the N phylogroup forms an outlier of the R-2 cluster. We have shown that this topology is false, as the dominance of the recombinant linkages between the phylogroups seriously compromise agglomerative methods of tree-building in the following way. When sequences of all PVY phylogroups are present in an analysis, the R-1 sequences are more closely related to their O parent than their N parent, as the former provided a greater percentage (76 percent) of their sequence, and therefore the R-1 and O phylogroups link. Likewise the R-2 sequences are more closely related to their R-1 parent than their N parent, as the former provided 62 percent of their sequence, and so again the R-2 phylogroup links to the R-1 phylogroup rather than the N parent. Finally the N phylogroup clusters with the R-2 ORFs as they share two-thirds of their sequence, whereas the true links of the N phylogroup as the sister lineage to all the other phylogroups are more distant. Thus the inferred inter-phylogroup relationships, when all are present, are false (Figs 1A and 2A), and the true topology of the phylogroups is only revealed when the recombinants are removed.
In the second part of this project we attempted to date the major features of the PVY phylogeny using different heterochronous tip-dating methods. The resulting estimates, although overlapping as judged by their 95% CIs, differed significantly in scale, so that the TMRCAs estimated, especially for the basal nodes, by the probabilistic method were between two to four times earlier than those estimated by the regression methods. So here we discuss whether events of the history of PVY and its hosts are congruent with those of the PVY phylogeny, and provide, in essence, independent node dating that supports some estimates more than others.
All our analyses supported a single PVY phylogeny that has several distinctive features (Fig. 6). PVY is a recent member of the ‘PVY lineage’ of potyviruses, most of whose members were isolated first, or only, from plants in the Americas (Gibbs and Ohshima 2010), and although PVY was first identified in the UK, it almost certainly came from the Americas. Furthermore the basal divergences of the PVY phylogeny produce three lineages, and one is the Chile 3 isolate found in Capsicum baccatum only in South America (Moury 2010). Thus the basal (TMRCA) node in the PVY phylogeny most likely represents an event that occurred in South America. The other two basal PVY lineages have been found throughout the world, but not yet in South America. The flora and fauna of South America was biologically isolated from Europe until the start of the ‘Columbian Exchange’ in the late 15th century CE (Crosby 1972) when the early trans-Atlantic maritime traders first took American animals and plants, including potato tubers, to Europe and vice versa. Thus the basal nodes of the PVY phylogeny are likely to represent events that predate the late 15th century, and this conclusion agrees with all our date estimates of those nodes; the most recent is 1411 CE with estimates from regression analyses ranging from 1085 CE to 1411 CE, and those from probabilistic analyses from 1590 BCE to 24 BCE.
The second basal lineage, the C phylogroup, probably diverged in Europe, and consists mostly (7/12 isolates) of isolates found in tobacco, tomato and pepper crops. Potato, pepper, tomato and tobacco were first introduced to Europe during the Columbian Exchange from their domestication centres in the Andean regions of Peru and Bolivia (potato), more widely in the Andes (tomato, tobacco) and further north in the Americas (pepper) (Bai and Lindhout 2007; Pickersgill 2007; Brown and Henfling 2014; Kraft et al. 2014). There is no evidence that PVY is transmitted by sexually produced seed, so it was most likely introduced to Europe in the small numbers of S. tuberosum tubers first taken from South America to the Canary Islands in 1562 CE, to Spain in 1570 CE and the UK in 1588 CE, if so then all the initial PVY population of Europe probably originated from infected S. tuberosum tubers in those cargoes. The divergence to several non-potato hosts probably occurred outside South America, although it is unknown to what extent different C phylogroup isolates are ecologically adapted to particular hosts. The dates estimated by the regression methods are congruent with this interpretation whereas those estimated by the probabilistic methods are not (Fig. 6; nodes 3 and 4).
The third basal lineage, the N phylogroup, is known only from a cluster of closely related isolates, so it probably radiated recently, and at present we have no idea of when and how it first migrated from the Americas. It has been isolated worldwide, mostly from potatoes (13/15 isolates).
The earliest potato breeding programs of any size began in 1810, but did not become widespread until the second half of the 19th century. Initially there were very few potato introductions to Europe, and for the first two centuries the crops had little genetic diversity. Virus diseases that seriously debilitated established cultivars, but did not pass through sexually produced potato seed, probably encouraged selection and, as a result, more cultivars were grown in the late 18th and early 19th centuries. These were taken from natural berries produced by self pollination. Most were selfed derivatives so selection reduced the gene pool (Glenndinning 1983). Potato blight disease caused by the oomycete Phytophthora infestans appeared in the mid 19th century and eliminated almost all cultivars as they were so inbred, further reducing the gene pool. Breeding among blight survivors and new introductions from the Andean region of South America led to many new cultivars being grown by early 20th century (Glenndinning 1983). This introduction of new potato germplasm led to genetic diversity in the crop, which apparently introduced the selection pressure in the form of PVY resistance genes that diversified the PVY population in the early 20th century. Our analyses indicate that the parents of the recombinant R-1 and R-2 strains were members of existing O and N populations, therefore probably European. As the Nc resistance gene, which C strains ellicit, was widely distributed in early potato cultivars (Bawden 1936; Cockerham 1943; Bawden and Sheffield 1944), selection for avirulence to this gene was likely to be an important factor in the diversification of the O phylogroup population. Similarly, N strains, but not O strains, infect plants with both Nc and the less common Ny and Nz genes, so selection for avirulence would again have favored diversity in the N phylogroup population (Cockerham 1970; Jones 1990;Chikh-Ali et al. 2014; Kehoe and Jones 2016).
PVY strains causing veinal necrosis symptoms in tobacco were first reported in 1935 in the UK (Smith and Dennis 1940) and soon afterwards in Brazil, Europe and North America (Nobrega and Silberschmidt 1944; Bawden and Kassanis 1947, 1951; Richardson 1958; Klinkowski and Schmelzer 1960; Silberschmidt 1960; Todd 1961; Kahn and Monroe 1963; Brücher 1969). However, whether these strains included ones in both the N and R1 phylogroups is unknown. Strains inducing this symptom in tobacco produce mild symptoms in potato plants so they soon became widely distributed because infected plants were often missed when seed potato crops were rogued (Todd 1961; Jones 1990). Possibly, this increased spread can be attributed to emergence of the R1 population at that time, but evidence for that is lacking, and none of the sequences we analyzed came from early isolates. Although the R-1 population is not very damaging to potato, it is difficult to control as its symptoms are so mild and it overcomes resistance genes Nc, No, and Nz. By contrast the R-2 population is more damaging as it often causes tuber necrosis, and still overcomes these three resistance genes.
The tuber necrosis isolates, R-2 phylogroup, were first sighted in the early 1980s in Europe and shown to be recombinants (Beczner et al. 1984; Le Romancer, Kerlan, and Nedellec 1994; Boonham et al. 2002; Lorenzen et al. 2006). These reports are likely to be accurate as R-2 infections are so noticeable and, by the 1980s, potato crops in Europe were intensively monitored for disease. Our estimates of the origin of R-2 isolates were 1969.4 CE, 1960.3 CE, and 1932.4 CE for the ML-LSD, NJ-LSD and BEAST analyses, respectively. Thus the ML-LSD estimate agrees most closely with the crop record, and the BEAST estimate agrees least well, but all are possible. Similarly, PVY infections that may have been caused by R-1 isolates were first reported in 1935, and soon confirmed to be widespread. The ML-LSD, NJ-LSD and BEAST methods placed the origin of R-1 as 1946.7 CE, 1933.3 CE and 1880.6 CE respectively. Thus the NJ-LSD estimate is the most likely, the BEAST estimate less likely, and the ML-LSD impossible.
All our date estimates agree that the near simultaneous radiation of the O and N populations of PVY (Figs 4 and 6; nodes 5 and 6) coincided with the earliest potato breeding programs of any size. These programs increased the genetic diversity of available potato cultivars and this would have included the Ny and Nz resistance genes. They also coincided with cultural and agronomic changes to seed and ware potato crop production (Singh et al. 2008; Gray et al. 2010; Jones 2014). A combination of these factors is likely to have led to conditions favoring recombinants. This scenario fits better with the LSD dates, 1918.2 CE and 1906.2 CE, respectively, than with the earlier BEAST date of 1832.9 CE.
Most of our PVY date estimates are congruent with those of the recent history of the international potato crop, but this is not surprising as the estimated dates have very broad overlapping 95% CI estimates. One crucial event might however inform us whether some TMRCA estimates are more accurate than others, and that is whether the divergence of the C phylogroup (Fig. 6; nodes 3 and 4) occurred before or after PVY was introduced to Europe in the late 16th century; the TMRCAs estimated by the regression methods are congruent with a European divergence of the C phylogroup, whereas those estimated by the probabilistic methods indicate a much earlier divergence of the C phylogroup. This dilemma will only be resolved when the genomic sequences of more Chile 3-like isolates or of the C phylogroup are known.
Our estimates of the dates for the PVY phylogeny are somewhat earlier than those reported by Visser, Bellstedt, and Pirie (2012) who analyzed rec, n-rec, and partitioned PVY genomic sequences by probabilistic methods, although our 95% CI estimates overlap. The difference may merely be the result of population sampling differences as a much larger set of sequences was available to us, and it included the early dated isolate KP691327 (1943 CE). A further difference between our analyses is the date given to sequence EU563512, which as Visser, Bellstedt, and Pirie (2012) reported, was obtained ‘from a potato plant vegetatively propagated since 1938’ and sequenced in 2007 (Dullemans et al. 2011). We dated this sequence as 1938, the earliest of our sequences, whereas Visser, Bellstedt, and Pirie (2012) dated it as 2007. In making this choice we checked how this difference affected the date estimates of the seventy-three n-rec dataset, and found that using the 1938 date gave an ML-LSD TMRCA of 1085.4 CE (95% CI 1267 CE–321 CE), whereas using the 2007 date gave a TMRCA of 363 CE, and very much broader 95% CIs (918 CE to 1.8 × 109 BCE) and, when the sequence was omitted, an intermediate TMRCA of 687 CE (1016 CE–9503 BCE). Thus the 95% CI range was smallest using the 1938 date, and justified our choice.
Finally, we explored the possibility of extrapolating dates over an even larger timescale. Gibbs et al. (2008) suggested that the Neolithic invention of agriculture, which in Eurasia occurred in the northern Levantine/Mesopotamian area around 12,000 BCE (Bellwood 2005; Pinhasi, Fort, and Ammerman 2005), produced the conditions for the starburst potyvirus diversification, and this provides another possible dating point for potyviruses. We estimated that in an ML phylogeny of the major ORFs of a large representative set of potyviruses and four PVY isolates (one N, one O, and two C phylogroup isolates) the ratio of the mean patristic distances of the sequences connected through the root of the phylogeny and through the root of the PVY sequences was 9.94:1 indicating that a TMRCA of all potyviruses of around 10,000 BCE would give a TMRCA of PVY around 1000 CE.
Supplementary data
Supplementary data are available at Virus Evolution online.
Supplementary Material
Acknowledgements
We are very grateful to Hien To and Olivier Gascuel for great help with use of their LSD software.
Conflict of interest: None declared.
References
- Abascal F., Zardoya R., Telford M. J. (2010) ‘TranslatorX: Multiple Alignment of Nucleotide Sequences Guided by Amino Acid Translations’, Nucleic Acids Research, 38: W7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., et al. (1990) ‘Basic Local Alignment Search Tool’, Journal of Molecular Biology, 215: 403–10. [DOI] [PubMed] [Google Scholar]
- Bai Y., Lindhout P. (2007) ‘Domestication and Breeding of Tomatoes: What Have We Gained and What Can We Gain in the Future?’, Annals of Botany, 100: 1085–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bawden F. C. (1936) ‘The Viruses Causing Top Necrosis (Acronecrosis) of the Potato’, Annals of Applied Biology, 23: 487–97. [Google Scholar]
- Bawden F. C., Kassanis B. (1947) ‘The Behaviour of Some Naturally Occurring Strains of Potato Virus Y’, Annals of Applied Biology, 31: 503–16. [DOI] [PubMed] [Google Scholar]
- Bawden F. C., Kassanis B. (1951) ‘Serologically Related Strains of Potato Virus Y That Are Mutually Antagonistic in Plants’, Annals of Applied Biology, 38: 402–10. [CrossRef]Mismatch] [Google Scholar]
- Bawden F. C., Sheffield F. M. L. (1944) ‘The Relationships of Some Viruses Causing Necrotic Diseases of the Potato’, Annals of Applied Biology, 31: 33–40. [Google Scholar]
- Beczner L., et al. (1984) ‘Studies on the Etiology of Tuber Necrotic Ringspot Disease in Potato’, Potato Research, 27: 339–52. [Google Scholar]
- Bellwood P. (2005) First Farmers: The Origins of Agricultural Societies. Malden, Oxford and Carlton: Blackwell Publishing. [Google Scholar]
- Boni M. F., Posada D., Feldman M. W. (2007) ‘An Exact Nonparametric Method for Inferring Mosaic Structure in Sequence Triplets’, Genetics, 176: 1035–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonham N., et al. (2002) ‘Biological and Sequence Comparisons of Potato virus Y Isolates Associated with Potato Tuber Necrotic Ringspot Disease’, Plant Pathology, 51: 117–26. [Google Scholar]
- Brown C. R. (1993) ‘Origin and History of the Potato’, American Journal of Potato Research, 70: 363–73. [Google Scholar]
- Brown C. R., Henfling J. W. (2014) ′A History of the Potato′, in Navarre R., Pavek M.J. (eds.) The Potato: Botany, Production and Use, pp. 1–11.Wallingford, UK: CABI. [Google Scholar]
- Brücher H. (1969) ‘Observations on Origin and Spread of Y-N Virus in South America’, Angewandte Botanik, 43: 241–9. [Google Scholar]
- Chikh-Ali M., et al. (2014) ‘Evidence of a Monogenic Nature of the Nz Gene Against Potato Virus Y Strain Z (PVYZ) in Potato’, American Journal of Potato Research, 91: 649–54. [Google Scholar]
- Cockerham G. (1943) ‘The Reactions of Potato Varieties to Viruses X, A, B and C’, Annals of Applied Biology, 30: 338–44. [Google Scholar]
- Cockerham G. (1970) ‘Genetical Studies on Resistance to Potato Viruses X and Y’, Heredity, 25: 309–48. [Google Scholar]
- Crosby A. E. (1972). ′The Columbian Exchange: Biological and Cultural Consequences of 1494′, 268 pp. Greenwood Publishing Group.
- Cuevas J. M., et al. (2012) ‘Phylogeography and Molecular Evolution of Potato Virus Y’, PLoS One, 7: e37853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D., et al. (2011) ‘ProtTest 3: Fast Selection of Best-Fit Models of Protein Evolution’, Bioinformatics, 27: 1164–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bokx J. A., van der Want J. P. H. (eds.) (1987) ′Viruses of Potatoes and Seed-Potato Production′, 2nd edn. Wageningen: Centre for Agricultural Publishing and Documentation. [Google Scholar]
- Drummond A. J., et al. (2012) ‘Bayesian Phylogenetics with BEAUti and the BEAST 1.7’, Molecular Biology and Evolution, 29: 1969–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchêne S. (2015) ‘The Performance of the Date-Randomization Test in Phylogenetic Analyses of Time-Structured Virus Data’, Molecular Biology and Evolution, 32: 1895–906. [DOI] [PubMed] [Google Scholar]
- Dullemans A. M., et al. (2011) ‘Complete Nucleotide Sequence of a Potato Isolate of Strain Group C of Potato Virus Y from 1938’, Archives of Virology, 156: 473–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourment M., Gibbs M. J. (2006) ‘PATRISTIC: a Program for Calculating Patristic Distances and Graphically Comparing the Components of Genetic Change’, BMC Evolutionary Biology, 6: 1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs A. J., Ohshima K. (2010) ‘Potyviruses in the Digital Age’, Annual Review of Phytopathology, 48: 205–23. [DOI] [PubMed] [Google Scholar]
- Gibbs A. J., et al. (2008) ‘The Prehistory of Potyviruses: Their Initial Radiation Was during the Dawn of Agriculture’, PLoS One, 3: e2523.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs A. J., Nguyen H. D., Ohshima K. (2015) ‘The ‘Emergence’ of Turnip Mosaic Virus Was Probably a ‘Gene-for-Quasi-Gene’ Event’, Current Opinion in Virology, 10: 20–5. [DOI] [PubMed] [Google Scholar]
- Gibbs A. J., Armstrong J. S., Gibbs A. J. (2000) ‘Sister-Scanning: A Monte Carlo Procedure for Assessing Signals in Recombinant Sequences’, Bioinformatics, 16: 573–82. [DOI] [PubMed] [Google Scholar]
- Gibbs A. J., et al. (2006) ‘The Variable Codons of H3 Influenza A Virus Haemagglutinin Genes’, Archives of Virology, 52: 11–24. [DOI] [PubMed] [Google Scholar]
- Glais L., Tribodet M., Kerlan C. (2002) ‘Genomic Variability in Potato Potyvirus Y (PVY): Evidence that PVYNW and PVYNTN Variants Are Single to Multiple Recombinants Between PVYO and PVYN Isolates’, Archives of Virology, 147: 363–78. [DOI] [PubMed] [Google Scholar]
- Glenndinning D. R. (1983) ‘Potato Introductions and Breeding up to the 20th Century’, New Phytologist, 94: 479–505. [Google Scholar]
- Gray S., et al. (2010) ‘Potato Virus Y: An Evolving Concern for Potato Crops in the United States and Canada’, Plant Disease, 94: 1384–97. [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003) ‘A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood’, Systematic Biology, 52: 696–704. [DOI] [PubMed] [Google Scholar]
- Hall T. A. (1999) ‘BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT', Nucleic Acids Symposium Series, 41: 95–8. [Google Scholar]
- Hawkes J. G. (1978) ′History of the Potato′, in Harris P. M. (ed.) The Potato Crop, pp. 1–14.New York: Springer. [Google Scholar]
- Hawkes J. G., Fransisco-Ortega J. (1993) ‘The Early History of the Potato in Europe’, Euphytica, 70: 1–7. [Google Scholar]
- Holmes E. C., Worobey M., Rambaut A. (1999) ‘Phylogenetic Evidence for Recombination in Dengue Virus’, Molecular Biology and Evolution, 16: 405–9. [DOI] [PubMed] [Google Scholar]
- Hu X., et al. (2009a) ‘Sequence Characteristics of Potato Virus Y Recombinants’, Journal of General Virology, 90: 3033–41. [DOI] [PubMed] [Google Scholar]
- Hu X., et al. (2009b) ‘A Novel Recombinant Strain of Potato Virus Y Suggests a New Viral Genetic Determinant of Vein Necrosis in Tobacco’, Virus Research, 143: 68–76. [DOI] [PubMed] [Google Scholar]
- Huson D. H., Bryant D. (2006) ‘Application of Phylogenetic Networks in Evolutionary Studies’, Molecular Biology and Evolution, 23: 254–67. [DOI] [PubMed] [Google Scholar]
- Jeanmougin F., et al. (1998) ‘Multiple Sequence Alignment with Clustal X’, Trends in Biochemical Sciences, 23: 403–5. [DOI] [PubMed] [Google Scholar]
- Jones R. A. C. (1981) ′The Ecology of Viruses Infecting Wild and Cultivated Potatoes in the Andean Region of South America′, in Thresh J. M. (ed.) Pests, Pathogens and Vegetation, pp 89–107. London: Pitman. [Google Scholar]
- Jones R. A. C. (1990) ‘Strain Group Specific and Virus Specific Hypersensitive Reactions to Infection with Potyviruses in Potato Cultivars’, Annals of Applied Biology, 117: 93–105. [CrossRef]Mismatch] [Google Scholar]
- Jones R. A. C. (2014) ′Virus Disease Problems Facing Potato Industries Worldwide: Viruses Found, Climate Change Implications, Rationalising Virus Strain Nomenclature and Addressing the Potato Virus Y issue′, in Navarre R., Pavek M. J. (eds.) The Potato: Botany, Production and Uses,Wallingford, UK: CABI. [Google Scholar]
- Jones R. A. C., Kehoe M. A. (2016) ‘A Proposal to Rationalize Within-Species Plant Virus Nomenclature: Benefits and Implications of Inaction’, Archives of Virology, 161: 2051–7. [DOI] [PubMed] [Google Scholar]
- Kahn R. P., Monroe R. L. (1963) ‘Detection of the Tobacco Veinal Necrosis Strain of Potato Virus in Solanum cardenasii and S. andigenum Introduced into the United States’, Phytopathology, 53: 1356–9. [Google Scholar]
- Karasev A. V., Gray S. M. (2013) ‘Genetic Diversity of Potato virus Y Complex’, American Journal of Potato Research, 90: 7–13. [Google Scholar]
- Karasev A. V., et al. (2011) ‘Genetic Diversity and Origin of the Ordinary Strain of Potato virus Y (PVY) and Origin of Recombinant PVY Strains’, Phytopathology, 101: 778–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013) ‘MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability’, Molecular Biology and Evolution, 30: 772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehoe M. A., Jones R. A. C. (2016) ‘Potato virus Y Strain Nomenclature: Lessons From Comparing Isolates Obtained Over a 73 Year Period’, Plant Pathology, 65: 322–33. [Google Scholar]
- Kerlan C. (2006) ′Potato Virus Y′ in Descriptions of Plant Viruses, No. 414. Adams J., Antoniw J. (eds.), Rothamsted Research: UK. [Google Scholar]
- Kerlan C., Moury B. (2008) ′Potato Virus Y′ in Granoff A., Webster R. G. (eds.) Encyclopedia of Virology, 3rd edn, pp. 287–96. New York: Academic Press. [Google Scholar]
- Klinkowski M., Schmelzer K. (1960) ‘A Necrotic Type of Potato Virus Y’, American Potato Journal, 37: 221–8. [Google Scholar]
- Kraft K. H., et al. (2014) ‘Multiple Lines of Evidence for the Origin of Domesticated Chili Pepper, Capsicum annuum, in Mexico’, Proceedings of the National Academy of Science USA, 111: 6165–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Romancer M., Kerlan C., Nedellec M. (1994) ‘Biological Characterization of Various Geographical Isolates of Potato Virus Y Inducing Superficial Necrosis on Potato Tubers’, Plant Pathology, 43: 138–44. [Google Scholar]
- Le S. Q., Gascuel G. (2008) ‘An Improved General Amino Acid Replacement Matrix’, Molecular Biology and Evolution, 25: 1307–20. [DOI] [PubMed] [Google Scholar]
- Lemey P., et al. (2009) ‘Identifying Recombinants in Human and Primate Immunodeficiency Virus Sequence Alignments Using Quartet Scanning’, BMC Bioinformatics, 10: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebenstein G., et al. (eds.) (2001) Virus and Virus-Like Diseases of Potatoes and Production of Seed-Potatoes. Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- Lorenzen J. H., et al. (2006) ‘Whole Genome Characterization of Potato virus Y Isolates Collected in the Western USA and Their Comparison to Isolates From Europe and Canada’, Archives of Virology, 151: 1055–74. [DOI] [PubMed] [Google Scholar]
- Martin D., Rybicki E. (2000) ‘RDP: Detection of Recombination Amongst Aligned Sequences’, Bioinformatics, 16: 562–3. [DOI] [PubMed] [Google Scholar]
- Martin D. P., et al. (2005) ‘A Modified BOOTSCAN Algorithm for Automated Identification of Recombinant Sequences and Recombination Breakpoints’, AIDS Research and Human Retroviruses, 21: 98–102. [DOI] [PubMed] [Google Scholar]
- Martin D. P., et al. (2015) ‘RDP4: Detection and Analysis of Recombination Patterns in Virus Genomes’, Virus Evolution, 1: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne I., et al. (2009) ‘TOPALi v2: a Rich Graphical Interface for Evolutionary Analyses of Multiple Alignments on HPC Clusters and Multi-Core Desktops’, Bioinformatics, 25: 126–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J. (1992) ‘Analyzing the Mosaic Structure of Genes’, Journal of Molecular Evolution, 34: 126–9. [DOI] [PubMed] [Google Scholar]
- McGuire G., Wright F. (2000) ‘TOPAL 2.0: Improved Detection of Mosaic Sequences Within Multiple Alignments’, Bioinformatics, 16: 130–4. [DOI] [PubMed] [Google Scholar]
- Moury B. (2010) ‘A New Lineage Sheds Light on the Evolutionary History of Potato Virus Y’, Molecular Plant Pathology, 11: 161–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moury B., et al. (2002) ‘Evidence for Diversifying Selection in Potato virusy and in the Coat Protein of Other Potyviruses’, Journal of General Virology, 83: 2563–73. [DOI] [PubMed] [Google Scholar]
- Moury B., Simon V. (2011) ‘dN/dS-Based Methods Detect Positive Selection Linked to Trade-Offs Between Different Fitness Traits in the Coat Protein of Potato Virus Y’, Molecular Biology and Evolution, 28: 2707–17. [DOI] [PubMed] [Google Scholar]
- Nei M., Li W. H. (1979) ‘Mathematical Model for Studying Genetic Variation in Terms of Restriction Endonucleases’, Proceedings of the National Academy of Science USA, 76: 5269–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega N. R., Silberschmidt K. (1944) ‘Sobre uma provavel variante do virus “Y” da batatinha (Solanum virus 2, Orton) que tern a peculiaridade de provocar necroses em plantas de fumo’, Arquivos Do Instituto Biológico (Sao Paulo), 15: 307–30. [Google Scholar]
- Ogawa T., et al. (2008) ‘Genetic Structure of a Population of Potato Virus Y Inducing Potato Tuber Necrotic Ringspot Disease in Japan; Comparison with North American and European Populations’, Virus Research, 131: 199–212. [DOI] [PubMed] [Google Scholar]
- Ogawa T., et al. (2012) ‘The Genetic Structure of Populations of Potato Virus Y in Japan; Based on the Analysis of 20 Full Genomic Sequences’, Journal of Phytopathology, 160: 661–73. [Google Scholar]
- Ohshima K., et al. (2016) ‘Temporal Analysis of Reassortment and Molecular Evolution of Cucumber Mosaic Virus: Extra Clues From Its Segmented Genome’, Virology, 487: 188–97. [DOI] [PubMed] [Google Scholar]
- Padidam M., Sawyer S., Fauquet C. M. (1999) ‘Possible Emergence of New Geminiviruses by Frequent Recombination’, Virology, 265: 218–25. [DOI] [PubMed] [Google Scholar]
- Pickersgill B. (2007) ‘Domestication of Plants in the Americas: Insights From Mendelian and Molecular Genetics’, Annals of Botany, 100: 925–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinhasi R., Fort J., Ammerman A. J. (2005) ‘Tracing the Origin and Spread of Agriculture in Europe’, PLoS Biology, 3: e410.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D., Crandall K. A. (2001) ‘Evaluation of Methods for Detecting Recombination From DNA Sequences: Computer Simulations’, Proceedings of the National Academy of Science USA, 98: 13757–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., et al. (2016) ‘Exploring the Temporal Structure of Heterochronous Sequences Using TempEst’, Virus Evolution, 2: vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsden C., Holmes E. C., Charleston M. A. (2009) ‘Hantavirus Evolution in Relation to Its Rodent and Insectivore Hosts: No Evidence for Codivergence’, Molecular Biology Evolution, 26: 143–53. [DOI] [PubMed] [Google Scholar]
- Richardson D. E. (1958) ‘Some Observations on the Tobacco Veinal Necrosis Strain of Potato Virus Y’, Plant Pathology, 7: 133–5. [Google Scholar]
- Rowley J. S., Gray S. M., Karasev A. V. (2015) ‘Screening Potato Cultivars for New Sources of Resistance to Potato Virus Y’, American Journal of Potato Research, 92: 38–48. [Google Scholar]
- Salaman R. N. (1954) ‘The Early European Potato: Its Character and Place of Origin’, Journal of the Linnean Society. Botany, 53: 1–27. [Google Scholar]
- Salaman R. N., Hawkes J. G. (1949) ‘The Character of the Early European Potato’, Proceedings of the Linnean Society, London, 161: 71–84. [Google Scholar]
- Schubert J., Fomitcheva V., Sztangret-Wisniewska J. (2007) ‘Differentiation of Potato Virus Y Strains Using Improved Sets of Diagnostic PCR-Primers’, Journal of Virological Methods, 140: 66–74. [DOI] [PubMed] [Google Scholar]
- Shimodaira H., Hasegawa M. (1999) ‘Multiple Comparisons of Log-Likelihoods with Applications to Phylogenetic Inference’, Molecular Biology and Evolution, 16: 1114–6. [Google Scholar]
- Silberschmidt K. M. (1960) ‘Types of Potato Virus Y Necrotic to Tobacco: History and Recent Observation’, American Potato Journal, 37: 151. [Google Scholar]
- Singh R. P., et al. (2008) ‘Discussion Paper: The Naming of Potato Virus Y Strains Infecting Potato’, Archives of Virology, 153: 1–13. [DOI] [PubMed] [Google Scholar]
- Smith K. M. (1931) ‘On the Composite Nature of Certain Potato Virus Diseases of the Mosaic Group as Revealed by the Use of Plant Indicators and Selective Methods of Transmission’, Proceedings of the Royal Society B, 109: 251–66. [Google Scholar]
- Smith K. M., Dennis R. W. G. (1940) ‘Some Notes on a Suggested Variant of Solanum Virus 2 (Potato Virus Y)’, Annals of Applied Biology, 27: 65–70. [Google Scholar]
- Spetz C., et al. (2003) ‘Molecular Resolution of Complexes of Potyviruses Infecting Solanaceous Crops at the Centre of Origin in Peru’, Journal of General Virology, 84: 2565–78. [DOI] [PubMed] [Google Scholar]
- Stevenson W. R. et al. (eds.) (2001) Compendium of Potato Diseases, 2nd edn St. Paul, MN: APS Press. [Google Scholar]
- Tavaré S. (1986) ‘Some Probabilistic and Statistical Problems in the Analysis of DNA Sequences’, Lectures on Mathematics in the Life Sciences, 17: 57–86. [Google Scholar]
- Tian Y. P., et al. (2011) ‘Genetic Diversity of Potato Virus Y Infecting Tobacco Crops in China’, Phytopathology, 10: 377–87. [DOI] [PubMed] [Google Scholar]
- To T. H., et al. (2015) ‘Fast Dating Using Least-Squares Criteria and Algorithms’, Systematic Biology, 65: 82–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd J. M. (1961) ‘Tobacco Veinal Necrosis on Potato in Scotland: Control of the Outbreak and Some Characters of the Virus’, in Proceedings of the Fourth Conference on Potato Virus Diseases, Braunschweig, 1960, pp. 82–92. [Google Scholar]
- Visser J. C., Bellstedt D. U. (2009) ‘An Assessment of Molecular Variability and Recombination Patterns in South African Isolates of Potato Virus Y’, Archives of Virology, 154: 1891–900. [DOI] [PubMed] [Google Scholar]
- Visser J. C., Bellstedt D. U., Pirie M. D. (2012) ‘The Recent Recombinant Evolution of a Major Crop Pathogen, Potato Virus Y’, PLoS One, 7: e50631.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X. (2013) ‘DAMBE5. A Comprehensive Software Package for Data Analysis in Molecular Biology and Evolution’, Molecular Biology and Evolution, 30: 1720–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.