Abstract
FlyBase (http://flybase.org) is the primary repository of genetic and molecular data of the insect family Drosophilidae. For the most extensively studied species, Drosophila melanogaster, a wide range of data are presented in integrated formats. Data types include mutant phenotypes, molecular characterization of mutant alleles and aberrations, cytological maps, wild-type expression patterns, anatomical images, transgenic constructs and insertions, sequence-level gene models and molecular classification of gene product functions. There is a growing body of data for other Drosophila species; this is expected to increase dramatically over the next year, with the completion of draft-quality genomic sequences of an additional 11 Drosphila species.
SCOPE OF FLYBASE
FlyBase includes information about the structure and function of genes and gene products of the Drosophila genome (1). Although the primary species represented is that workhorse of classic genetics, Drosophila melanogaster, the database currently includes records for genes of more than 400 other Drosophila species, and will house genomic information for the 11 additional species included in the Drosophila comparative genomics sequencing effort. Phenotypic and genetic interaction information about mutants, and wild-type gene and enhancer-trap expression patterns are linked to strains in the Drosophila Stock Centers, from which extensive collections of mutant and wild-type strains are available. Mutant phenotypes (2) and gene expression patterns are described using controlled vocabularies, including anatomical terms linked to illustrations in the Anatomy section of FlyBase. Data concerning chromosome aberrations, natural transposons, genetically engineered constructs and transgene insertions are presented with hyperlinks to affected genes and resulting mutant alleles.
An overview of the classes of data found in FlyBase may be seen on the homepage (http://flybase.org; for further description see Supplementary Figure 1). Features recently added to FlyBase include an External Database Links section in Gene reports, expanded Batch query options and an extensive Drosophila Resources compilation (http://flybase.bio.indiana.edu/allied-data/resources.html), which provides a comprehensive list of links to both network resources (e.g. sequence analysis tools) and material resources (e.g. clone and microarray suppliers) external to the FlyBase project.
Data are compiled by curators and annotators from sources including the scientific literature, large-scale genome sequencing projects and online resources such as the GenBank (NCBI)/EMBL/DDBJ nucleotide sequence databases and the UniProt (3) protein database. FlyBase curators work with curators of other databases, such as the Gene Ontology (GO) consortium (4) to ensure consistency of annotation across databases. The D.melanogaster genome annotation, Release 4.0 at the time of writing (5–7), has been enhanced by hand curation of all gene models (8,9), including integration of error reports submitted by the user community.
Table 1 shows a snapshot of FlyBase content as of September 2004. The remainder of this paper will focus on genes and gene models in FlyBase.
Table 1. Number of data records/statements in FlyBase: September 13, 2004.
| Gene records (D.melanogaster) | 28 015 |
| Genes records (species other than D.melanogaster) | 10 766 |
| Genes with genome annotations (D.melanogaster) | 14 872 |
| Genes with genome annotations, protein coding (D.melanogaster) | 14 367 |
| Genes with GO annotations (D.melanogaster) | 9643 |
| Genes with GO annotations (other species) | 143 |
| Mutant alleles (D.melanogaster) | 66 279 |
| Mutant alleles (other species) | 5058 |
| Phenotypic data controlled vocabulary statements (D.melanogaster) | 142 756 |
| Genetic interaction controlled vocabulary statements (D.melanogaster) | 76 580 |
| Natural transposons (D.melanogaster) | 145 |
| Natural transposon insertions mapped to euchromatic genome (D.melanogaster) | 1571 |
| Genetically engineered transposons (D.melanogaster) | 14 075 |
| DNA sequence accession records curated to genes (D.melanogaster) | 77 629 |
| DNA sequence accession records curated to genes (other species) | 17 912 |
| References | 128 602 |
| Stock Center stocks | 27 200 |
THE GENE REPORT
FlyBase provides several formats of gene report which differ by degree of completeness of data reported within the initial web page, the default being the Synopsis format. The Synopsis report for the maleless (mle) gene is shown in Figure 1. The Synopsis report displays commonly accessed gene information fields, an Available reports side panel to allow easy access to other report formats, and a text Summary generated automatically from the underlying data. The Abridged report format displays a wider range of information in the initial display than the Synopsis format, but collapses many of the details, such as individual Allele reports, into links in tables. The Full report format is the most comprehensive initial display.
Figure 1.
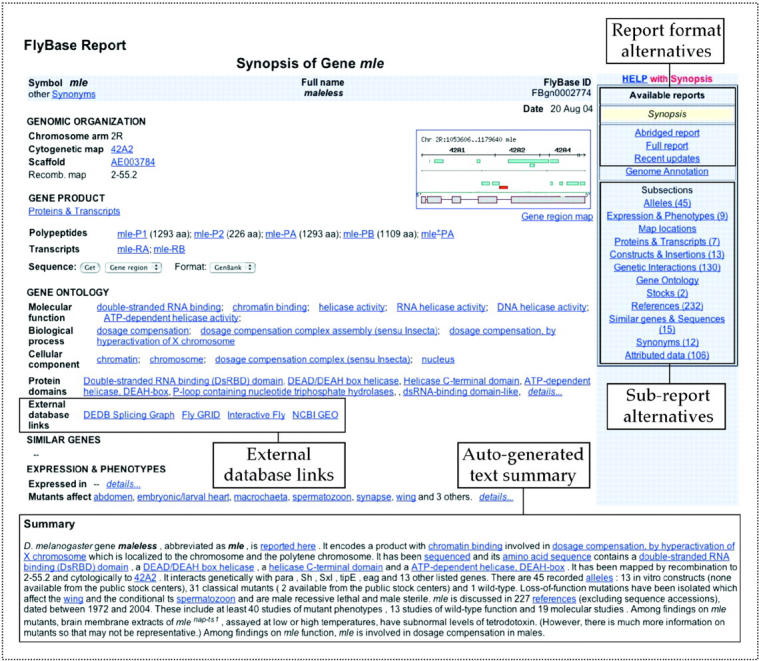
FlyBase gene report, highlighting different format and subsection report options, automated gene summaries and the recently added External Database section.
FlyBase also offers Subsection reports selected by data type, for example, alleles of that gene, references that discuss the gene and sequences in the DNA and protein data banks that correspond to the gene. Links to these and other subreports are listed in the Subsections panel of the Synopsis report. Recent additions include the Gene Ontology subreport, the Genetic Interactions subreport and the Constructs & Insertions subreport.
Gene reports now include an External Database Links section (http://flybase.bio.indiana.edu/allied-data/extdb/ExternalLinks.htm). This section houses links to databases external to FlyBase, to ease access to information about the gene that falls outside the scope of FlyBase data curation. The databases currently listed in this section include; the BDGP In Situ Gene Expression Database (10), Drosophila melanogaster Exon Database (http://proline.bic.nus.edu.sg/dedb), PANTHER Protein Classification (11,12), Fly GRID Interaction Data (13), Hybrigenics PIMRider interactions (14), Interactive Fly (15), Yale Developmental Gene Expression (16) and NCBI's Gene Expression Omnibus (17). Not all genes have an entry in all these databases. The number of external links in place via this facility exceeds 76 500.
THE GENE ANNOTATION REPORT
Detailed information about the annotated transcripts and other sequence-level data for a particular gene are to be found in the Annotation Report. This may be accessed from the Gene Report page from the link ‘Genome Annotation’ or by a direct query using the ‘Gene Annotations’ option in the homepage search box. The Annotation Query Form (http://flybase.bio.indiana.edu/annot/fbannquery.hform) allows queries based on location, gene class, peptide length, mapped expressed sequenced tags (ESTs) or cDNAs, GO terms, or terms within annotation comments.
An example of an Annotation Report is shown in Figure 2. Notable features include a graphic representation of the transcript structures aligned with supporting evidence, information about each transcript and protein product, links to sequence data and information about other data mapped experimentally to the genomic sequence, such as point mutations, aberration breakpoints, rescue fragments and experimentally defined regulatory regions. Accompanying comments describe any unusual characteristics of the gene model, such as atypical splice donor or acceptor, non-AUG translation start, or dicistronic transcript. At the top of the report is a link to the peptide analysis that includes a graphic display of homologous proteins and known InterPro (3) protein motifs.
Figure 2.

FlyBase annotation report. The panels show sequential extracts from the annotation report. At the top there are links to a Cytogenetic map, the GenBank scaffold sequence accession, and a Peptides ‘view analysis’ page showing alignments to related proteins and protein domain predictions. The ‘Sequence’ option allows the user to retrieve sequence for the gene region, transcripts, UTRs or proteins in a choice of formats. The Gene Annotation and Evidence panel shows two alternative transcripts and supporting EST, cDNA and protein (blastx) data. Note that the mle-RB transcript is based on data curated from the literature (not represented graphically), and that cDNA data supports an additional alternative transcript (to be added in the next annotation update). Details about the annotated transcripts and protein products are presented, and an ‘Other Features’ section describes mutational lesions, rescue fragments and other entities mapped onto the sequence level. These features appear on the GBrowse map, which may be accessed from a link at the top of the page.
GENE REGION MAPS: GBROWSE AND APOLLO
A molecular map of the region surrounding a gene may be accessed through the Gene Region Map (GBrowse) link on either the Gene Report page or the Gene Annotation Report. GBrowse (18) is a configurable genome viewer that allows the presentation of both molecularly mapped and cytologically mapped data (http://www.gmod.org/ggb/gbrowse.shtml; see Supplementary Figure 2). Annotations or larger genomic regions may also be viewed using the interactive viewing and editing tool, Apollo (19). Apollo is available for Windows, MacOSX or Unix systems and may be downloaded from the Apollo site (http://www.fruitfly.org/annot/apollo).
BULK DATA DOWNLOADS
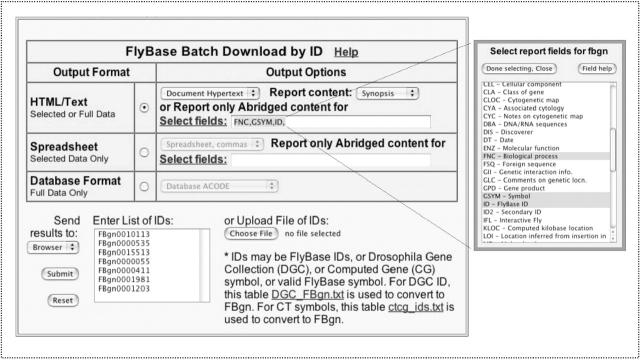
FlyBase offers a variety of routes for bulk data retrieval; a recent addition is the Batch Download Reports by ID facility shown in Figure 3. This tool allows the user to query the genes dataset for many records at once, by valid symbol or by FlyBase identification number. The users can select the output type they wish to retrieve (HTML/Text, Spreadsheet or Database format). For HTML/Text outputs, the user can choose Report Content (from Synopsis, Abridged, Full, Summary, Alleles, Sequences, Reviews, References). For HTML/Text or Spreadsheet outputs, it is possible to filter output by field, using the ‘Select fields’ function. A related tool, Batch Download Sequences by ID, allows querying for sequences for many genes simultaneously. Options for sequence retrieved are Gene Region, Transcript, Translation, 3′-untranslated region (3′-UTR) and 5′-UTR. Both Batch Download forms can be accessed from the Genes data directory or from the Genome Annotation and Sequences page.
Figure 3.
The FlyBase ‘Batch Download Reports by ID’ tool. In this example, seven genes are the subject of the query (listed in the ‘Enter List of Ids’ box), the user has selected ‘Document hypertext’ as the output format, and is in the process of selecting which data fields to retrieve.
In addition to bulk queries performed over the web interface, FlyBase data files are available for download by ftp from several of our mirror sites, in a text, acode or XML format. Protocols are described in the FlyBase Reference Manual section D (http://flybase.org/docs/lk/refman/refman-D.html).
D.MELANOGASTER GENOME RELEASES
The genomic sequence of D.melanogaster continues to be refined and expanded (http://flybase.bio.indiana.edu/annot/release3.html); the Berkeley Drosophila Genome Project has made public Release 4.0 of the genome sequence (http://www.fruitfly.org/annot/release4.html), and is currently finishing Release 5.0. FlyBase makes regular corrections and additions to the gene model annotations based on new data submissions to the sequence databases, user error reports and literature curation. We anticipate that comparative genomic analyses will play an increasing role in annotation assessment and improvement. Annotation updates are indicated by decimal numbers appended to the release number: e.g. Release 4.0 and Release 4.1. The heterochromatic portion of the genome is being analyzed by members of the Drosophila Heterochromatin Genome Project (http://www.dhgp.org); the heterochromatin annotations are accessible through FlyBase.
ADDITIONAL DROSOPHILA GENOMES
The National Human Genome Research Institute (NHGRI) has recognized the importance of comparative genomic analysis for the annotation of D.melanogaster and for understanding how genomes evolved. Towards this end, the major NHGRI-funded sequencing centers are sequencing 11 additional species of Drosophila (pseudoobscura, yakuba, simulans, virilis, ananassae, erecta, willistoni, grimshawi, mojavensis, persimilis and sechellia; status of projects reported at http://genome.gov/page.cfm?pageID=10002154). The genome sequences, annotations, syntenic relationships and other data from these genome projects will be incorporated into FlyBase, consistent with FlyBase's long-term commitment to maintaining genomic and genetic data on the family Drosophilidae.
THE CHADO DATABASE SCHEMA
FlyBase has been operating since 1992 and is now in the process of developing and populating a new database structure, an integrated implementation of the chado generic genome database schema (http://www.gmod.org/schema/). The initial design of the chado schema was undertaken by FlyBase developers at Harvard and Berkeley to fully integrate the finished D.melanogaster genome sequence and annotation with the vast body of Drosophila genetic and phenotypic data produced over the last 100 years. The chado schema is an open software project and is being developed in cooperation with the GMOD initiative (http://www.gmod.org).
REFERENCING FLYBASE
We suggest FlyBase be referenced in publications by citing this publication and the FlyBase web address (http://flybase.org).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
FlyBase is supported by grant P41 HG00739 from the National Human Genome Research Institute, National Institutes of Health, with additional support from the Medical Research Council (London).
NOTE ADDED IN PROOF
The initial analysis of the genome sequence of a second Drosophila species, D.pseudoobscura [Richards,S. et al. (2004) Genome Res., in press] can now be accessed at GenBank and Flybase (http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nucleotide&val=40362459 and http://flybase.bio.indiana.edu/cgi-bin/gbrowse_fb/dpse, respectively). It includes 12 197 gene annotations of D.pseudoobscura and their inferred orthology/synteny relationships with their D.melanogaster counterparts.
REFERENCES
- 1.The FlyBase Consortium (2003) The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res., 31, 172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drysdale R. (2001) Phenotypic data in FlyBase. Brief Bioinformatics, 2, 68–80. [DOI] [PubMed] [Google Scholar]
- 3.Apweiler R., Bairoch,A., Wu,C.H., Barker,W.C., Boeckmann,B., Ferro,S., Gasteiger,E., Huang,H., Lopez,R., Magrane,M. et al. (2004) UniProt: the Universal Protein knowledgebase. Nucleic Acids Res., 32 (Database issue), D115–D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris M.A., Clark,J., Ireland,A., Lomax,J., Ashburner,M., Foulger,R., Eilbeck,K., Lewis,S., Marshall,B., Mungall,C. et al. (2004) The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res., 32, D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams M.D., Celniker,S.E., Holt,R.A., Evans,C.A., Gocayne,J.D., Amanatides,P.G., Scherer,S.E., Li,P.W., Hoskins,R.A., Galle,R.F. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 6.Celniker S.E., Wheeler,D.A., Kronmiller,B., Carlson,J.W., Halpern,A., Patel,S., Adams,M., Champe,M., Dugan,S.P., Frise,E. et al. (2002) Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol., 3, R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoskins R.A., Smith,C.D., Carlson,J.W., de Carvalho,A.B., Halpern,A., Kaminker,J.S., Kennedy,C., Mungall,C.J., Sullivan,B.A., Sutton,G.G. et al. (2002) Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol., 3, R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Misra S., Crosby,M.A., Mungall,C.J., Matthews,B.B., Campbell,K.S., Hradecky,P., Huang,Y., Kaminker,J.S., Millburn,G.H., Prochnik,S.E., Smith,C.D., Tupy,J.L. et al. (2002) Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol., 3, R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaminker J.S., Bergman,C.M., Kronmiller,B., Carlson,J., Svirskas,R., Patel,S., Frise,E., Wheeler,D.A., Lewis,S.E., Rubin,G.M. et al. (2002) The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol., 3, R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomancak P., Beaton,A., Weiszmann,R., Kwan,E., Shu,S., Lewis,S.E., Richards,S., Ashburner,M., Hartenstein,V., Celniker,S.E. et al. (2002) Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol., 3, R88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mi H., Vandergriff,J., Campbell,M., Narechania,A., Majoros,W., Lewis,S., Thomas,P.D. and Ashburner,M. (2003) Assessment of genome-wide protein function classification for Drosophila melanogaster. Genome Res., 13, 2118–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas P.D., Campbell,M.J., Kejariwal,A., Mi,H., Karlak,B., Daverman,R., Diemer,K., Muruganujan,A. and Narechania,A. (2003) PANTHER: a library of protein families and subfamilies indexed by function. Genome Res., 13, 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breitkreutz B.J., Stark,C. and Tyers,M. (2003) The GRID: the General Repository for Interaction Datasets. Genome Biol., 4, R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colland F., Jacq,X., Trouplin,V., Mougin,C., Groizeleau,C., Hamburger,A., Meil,A., Wojcik,J., Legrain,P. and Gauthier,J.M. (2004) Functional proteomics mapping of a human signaling pathway. Genome Res., 14, 1324–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brody T. (1999) The Interactive Fly: gene networks, development and the Internet. Trends Genet., 15, 333–334. [DOI] [PubMed] [Google Scholar]
- 16.Arbeitman M.N., Furlong,E.E., Imam,F., Johnson,E., Null,B.H., Baker,B.S., Krasnow,M.A., Scott,M.P., Davis,R.W. and White,K.P. (2002) Gene expression during the life cycle of Drosophila melanogaster. Science, 297, 2270–2275. [DOI] [PubMed] [Google Scholar]
- 17.Edgar R., Domrachev,M. and Lash,A.E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res., 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stein L.D., Mungall,C., Shu,S., Caudy,M., Mangone,M., Day,A., Nickerson,E., Stajich,J.E., Harris,T.W., Arva,A. et al. (2002) The generic genome browser: a building block for a model organism system database. Genome Res., 12, 1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis S.E., Searle,S.M., Harris,N., Gibson,M., Lyer,V., Richter,J., Wiel,C., Bayraktaroglu,L., Birney,E., Crosby,M.A. et al. (2002) Apollo: a sequence annotation editor. Genome Biol., 3, R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.