Abstract
Nucleosome Positioning Region Database (NPRD), which is compiling the available experimental data on locations and characteristics of nucleosome formation sites (NFSs), is the first curated NFS-oriented database. The object of the database is a single NFS described in an individual entry. When annotating results of NFS experimental mapping, we pay special attention to several important functional characteristics, such as the relationship between type of gene activity and nucleosome positioning, the influence of non-histone proteins on nucleosome formation, type of the variant of nucleosome positioning (translational or rotational), indication of tissue types and states of cell activity, description of experimental methods used and accuracy of nucleosome position determination, and the results of applying theoretical and computer methods to the analysis of contextual and conformational DNA properties. At present, the NPRD database contains 438 entries and integrates the data described in 124 original papers. The database URL: http://srs6.bionet.nsc.ru/srs6/. Then click the button ‘Databank’ and open the link NUCLEOSOME.
INTRODUCTION
Nucleosomes are the major structural elements of chromatin. Each nucleosome is formed by a 147 bp DNA fragment wrapped around an octamer composed of pairs of histone molecules of four types. In addition to DNA compacting, the most important function of nucleosomes is their interaction with the molecular components of the nucleus machineries involved in DNA replication, repair and recombination. The key role in gene transcription is also assigned to nucleosomes (1,2).
Chromatin is, as a rule, represented by regular arrays of nucleosomes. The factors determining the regularity of nucleosome location in vivo and in vitro are, first, the sequence-directed nucleosome positioning, determined by the interaction of nucleosome formation sites (NFSs) sequences with the histone octamer (3) and, second, their interaction with various non-histone proteins (4,5).
So far, the relative role of these factors in nucleosome positioning is vague. Presumably, this is first and foremost connected with insufficient volume of experimental data, preventing their systematization and formalization.
The database NUCLEOSOMAL DNA by Ioshikhes and Trifonov (6), comprising 143 entries, compiled the information about only the nucleosomal center positions and techniques used for the nucleosomal mapping; moreover, the information on types of nucleosome positioning and their relationship between transcription regulation and the state of gene activity was absent. In addition, the volume of this database and the degree of representation of the data on nucleosome organization in genomes of higher eukaryotes were evidently insufficient for large-scale research on nucleosome organization of the genomes and, in particular, estimation of the abilities of individual genomic regions to position nucleosomes.
To solve these problems, we are developing a curated NFS-oriented Nucleosome Positioning Region Database (NPRD). Along with a detailed description of NFS localization, including their mapping relative to the borders of genes and their structural elements, the database contains important functional characteristics: the relationship between types of gene activity and nucleosome positioning, the influence of non-histone proteins on nucleosome formation, occurrence of translational or rotational nucleosome positioning, indication of tissue types and states of cell activity, description of experimental methods used and the accuracy of nucleosome position determination, and the results of applying theoretical and computer methods to the analysis of contextual and conformational DNA properties. The database in question provides the possibility of formalized description and assessment of the contributions of the factors listed above to nucleosome positioning taking into account the available information about biologically significant characteristics of the regions considered.
We think that our database is important for developers of new computer methods of nucleosomal DNA analysis and recognition; and experimenters involved in transcription regulation and chromatin structure investigation.
FORMAT OF THE NPRD DATABASE
The NPRD format developed allows for accumulating, integrating and systematizing miscellaneous experimental data on locations and characteristics of NFSs and detailed information about the other factors influencing nucleosome positioning, extracted from the published sources. Each NPRD entry corresponds to one annotated NFS. Below, we give a description of the fields in the order of their appearance (Table 1).
Table 1. Description of the NPRD fields.
| Field name | Description |
|---|---|
| AccessionNumber | Identifier of an entry. This is the only unique identifier of an entry. It follows the pattern NXXXXX, where N is a letter and X is a number |
| Number | The tag used in original paper |
| Annotator | Annotator name and date of the last editing |
| Taxon | Organism classification |
| Method | Type of experiment (in vivo, in vitro) |
| Species | Organism species |
| DNABankLink | Link to EMBL/GenBank |
| Description | List of names of genes and their products |
| Gene | Source of the sequence: ‘gene’ if sequence is located within a gene, ‘genomic’ if otherwise |
| Region | Gene region. Possible values: ‘5′region’, ‘3′region’, ‘exon’, ‘intron’, ‘5′UTR’, ‘3′UTR’ and ‘CDS’ |
| Function | Gene region function. The format is: ‘Function’; ‘ TRRD link’ (if is available); Possible ‘Function’ values are: ‘promoter’ and ‘enhancer’ |
| MainBounds | Main position of nucleosome center. The format is: ‘AC (EMBL/GenBank accession number)’; ‘start point name (e.g. ST, transcription start; SS, beginning of the sequence; and SR, translation start)’: ‘start point position in EMBL/GenBank entry’; ‘main position of nucleosome center relative to start point’; and [‘probability of main position of nucleosome center (if this position is referenced)’; ‘error in determining the main position of nucleosome center in base pair]. Examples: Z46939; ST: 737; –298;(80%,10);’ and ‘Z46939; ST: 737; –298;();’—if information on probability and error is absent |
| VarBounds | Variable positions of nucleosome center. The format is similar to that of MainBounds field |
| MainBounds | Main position of nucleosome borders. The format is similar to that of MainBounds field |
| VarBounds | Variable positions of nucleosome borders. The format is similar to that of MainBounds field |
| Comments | Comments on nucleosome site |
| PositioningFactora | The name(s) of transcription factor(s) or structural non-histone protein(s) influencing nucleosome formation. The result of the factor action (positive effect; negative effect; and no effect) |
| Commentsa | Comments on positioning factor |
| Sequence | Nucleotide sequence in the simple format |
| Disorder_Positiona | Discrepancies between the data on nucleotide sequence in the paper annotated and the corresponding sequence in EMBL/GenBank |
| PosEvidence | Experimental evidence of nucleosome positioning: cell type or source, DNA/histone source and experiment type |
| NegEvidencea | Experimental evidence of the absence of nucleosome positioning: cell type or source, DNA/histone source and experiment type |
| ConditionEffecta | Effects of various physical and chemical factors (salts, temperature, etc.) on nucleosome formation |
| KeyWords | Key words |
| Reference | Complete bibliographic reference to the annotated paper with a link to PubMed |
aDenotes an optional field.
An example of an entry representing HNF-4-alpha gene proximal promoter nucleosome organization is given in Table 2. In active cells, the promoter and enhancer of this gene were occupied by positioned nucleosomes unlike in non-expressing cell lines, where positioning of nucleosomes was random. According to Hatzis and Talianidis model (7), formation of an active pre-initiation complex occurs in a step-by-step fashion: (i) poised or committed state (enhancer and promoter were occupied by the cognate DNA-binding proteins); (ii) recruitment of CBP and P/CAF (histone acetyltransferases), Brg-1 (chromatin remodeling protein) to the enhancer region and assembly of the RNA pol-II holoenzyme at the proximal promoter region; (iii) unidirectional movement of the DNA–protein complex formed on the enhancer along the intervening sequences and spreading of histone hyperacetylation and (iv) formation of a stable enhancer–promoter complex, hyperacetylation of nucleosomes located at the promoter, remodeling of the nucleosome located at the transcription start site and release of RNA pol-II from the promoter. Information on nucleosome positioning and its correlation with the gene activity is presented through the fields (Table 2): ‘Function’, ‘MainBounds’, ‘Comments’, ‘PosEvidence’, ‘NegEvidence’ and ‘KeyWords’.
Table 2. An example entry in the NPRD database.
| AccessionNumber | N00955 |
| Number | 1 |
| Annotator | V. G. Levitsky June 9, 2004 |
| Method | in vivo |
| Species | Homo sapiens (human) |
| Taxon | Eukaryota; Metazoa; Chordata; Vertebrata; Mammalia; Eutheria; Primates; Catarrhini; Hominidae; Homo |
| DNABankLink | GenBank; HS1013A22; AL132772; |
| Description | Hepatocyte nuclear factor 4-alpha gene, HNF-4-alpha gene, transcription factor HNF-4 gene, transcription factor 14 gene, HNF4A, NR2A1, TCF14, HNF4 |
| Gene | Gene |
| Region | 5′-region; |
| Function | promoter, 5′-UTR, CDS, TRRDUNITS4:P02102; |
| MainBounds | AL132772; SN:1997; −180 to −1; (); |
| Comments | The analysis revealed that the proximal promoter region was occupied by an array of positioned nucleosomes, evidenced by the regularly spaced—about 150 bp ladder of bands…efficient cross-linking of the HNF-4 enhancer and promoter DNAs suggests that the two regions must be in close proximity at the time of transcription initiation and onward |
| Sequence | cagaggctaggccaagactcccagcagatcttcccagaggacggtttgaaaggaaggcagagagggcactgggaggaggcagtgggagggcggagggcgggggccttcggggtgggcgcccagggtagggcaggtggccgcggcgtggaggcagggagaatgcgactctccaaaaccctc |
| PosEvidence | Human differentiating CaCo-2 cells (gene active): MNase digestion, restriction enzyme hypersensitivity assay; |
| NegEvidance | ovarian A2780 carcinoma cells (gene repressed): MNase digestion; |
| Keywords | enhancer–promoter communication, histone hyperacetylation; |
| Authors | Hatzis, P. and Talianidis, I. |
| Title | Dynamics of enhancer–promoter communication during differentiation-induced gene activation |
| Source | Mol. Cell. |
| Year | 2002 |
| Volume | 10 |
| Issue | 6 |
| Pages | 1467–1477 |
| PubMed | 12504020 |
An NFS in the human hepatocyte nuclear factor 4-alpha gene promoter is crucial for determining expression status.
ACCESS TO THE NPRD AND DATA RETRIEVAL TOOLS
The database URL: http://srs6.bionet.nsc.ru/srs6/. Then click the button ‘Databank’ and open the link NUCLEOSOME. Sequence Retrieval System (SRS) is used as a basic software tool for accessing the NPRD via the Internet; this provides efficient linking to the nucleotide sequences (EMBL/GenBank) and to the literature sources (PubMed). A system of hyperlinks integrates the NPRD with the informational system TRRD (8) (e.g. Table 2, field ‘Function’), allowing for a quick access to both the experimental information on the regulation of expression of a particular gene, whereto an NFS is localized, and the programs for computer analyses of regulatory sequences of the gene, providing users the possibility of additional data mining.
DATABASE STATISTICS
The database contains 438 entries integrating the data of 124 original papers. Now, the Internet-accessible version contains 122 entries, constituting about one-third of the database volume.
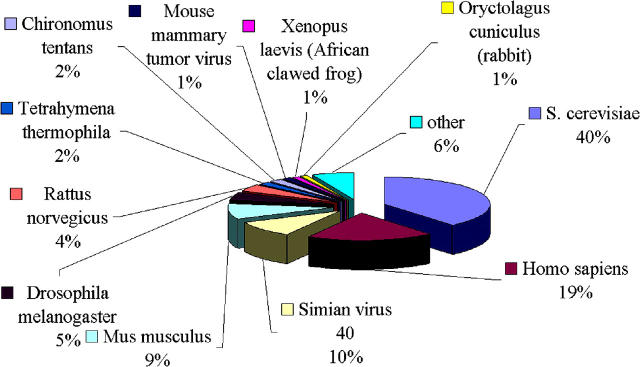
Representation of species is shown in Figure 1: sequences of higher eukaryotes constitute over 40% of the database; of them, 33% are mammalian genomes.
Figure 1.
Representation of species in the NPRD database.
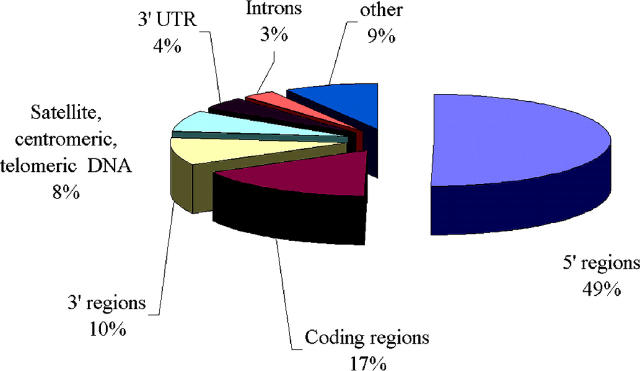
Figure 2 illustrates the representation in the NPRD regions of genes and other functional components of the genomes. Note that the promoter regions constitute about a half of these regions, presumably indicating an increased interest of researchers to the relationship between nucleosome organization and transcription regulation.
Figure 2.
Representation of gene and genomic regions in the NPRD database.
RESEARCH BASED ON DATA STORED IN THE NPRD
We have earlier designed an integrated information system Nucleosomal DNA Organization (9) (http://wwwmgs.bionet.nsc.ru/mgs/gnw/nucleosom/), comprising, along with NPRD, RECON program for nucleosome formation potential prediction (10) and NFS recognition by DNA conformational and physicochemical properties (9). Both programs were trained on NFS data later included in the NPRD dataset. Using the software package RECON, we have carried out the computer analysis of several classes of genome sequences: promoters of various classes, enhancers, coding and non-coding parts of genes, areas of splicing sites and sites of an insert of mobile genetic elements (10–15).
The volume of information, compiled in the NPRD, gives the possibility to study contextual characteristics of NFSs, increase the efficiency of their identification in genomic sequences and perform a more detailed research into nucleosome organization of genomes.
FUTURE DEVELOPMENTS
The database NPRD is being constantly developed and supplemented with new experimental data from the available literature sources. We are planning to update the database annually. Concurrently, its integration with the existing databases and its search capabilities are being constantly improved.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are grateful to Drs T. M. Khlebodarova and O. G. Smirnova for participating in the annotation, and to Dr N. L. Podkolodny for interface design. The work was supported by the RFBR (grant nos 03-04-48555, 01-07-90376, 02-07-90355, 02-07-90359, 03-07-96833, 03-04-48469 and 03-07-90181); Ministry of Industry, Science, and Technologies of the Russian Federation (grant no. 43.073.1.1.1501); Russian Federal Research Development Program ‘Research and Development in Priority Directions of Science and Technology’ (contract no. 28/2003); grant MCB RAS (no. 10.4); SB RAS (integration project no. 119); NATO (grant no. LST.CLG.979816); the CRDF and the Ministry of Education of Russian Federation within the Basic Research and Higher Education Program (Y1-B-08-02).
REFERENCES
- 1.Kornberg R.D. and Lorch,Y. (1999) Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell, 98, 285–294. [DOI] [PubMed] [Google Scholar]
- 2.Aalfs J.D. and Kingston,R.E. (2000) What does ‘chromatin remodeling’ mean? Trends Biochem. Sci., 25, 548–555. [DOI] [PubMed] [Google Scholar]
- 3.Trifonov E.N. (1997) Genetic level of DNA sequences is determined by superposition of many codes. Mol. Biol. (Moskow), 31, 759–767. [PubMed] [Google Scholar]
- 4.Widom J. (1998) Structure, dynamics, and function of chromatin in vitro. Annu. Rev. Biophys., 27, 285–327. [DOI] [PubMed] [Google Scholar]
- 5.Becker P.B. (2002) Nucleosome sliding: facts and fiction. EMBO J., 21, 4749–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ioshikhes I. and Trifonov,E.N. (1993) Nucleosomal DNA sequence database. Nucleic Acids Res., 21, 4857–4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatzis P. and Talianidis,I. (2002) Dynamics of enhancer–promoter communication during differentiation-induced gene activation. Mol. Cell, 10, 1467–1477. [DOI] [PubMed] [Google Scholar]
- 8.Kolchanov N.A., Ignatieva,E.V., Ananko,E.A., Podkolodnaya,O.A., Stepanenko,I.L., Merkulova,T.I., Pozdnyakov,M.A., Podkolodny,N.L., Naumochkin,A.N. and Romashchenko,A.G. (2002) Transcription Regulatory Regions Database (TRRD): its status in 2002. Nucleic Acids Res., 30, 312–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levitsky V.G., Ponomarenko,M.P., Ponomarenko,J.V., Frolov,A.S. and Kolchanov,N.A. (1999) Nucleosomal DNA property database. Bioinformatics, 15, 582–592. [DOI] [PubMed] [Google Scholar]
- 10.Levitsky V.G., Podkolodnaya,O.A., Kolchanov,N.A. and Podkolodny,N.L. (2001) Nucleosome formation potential of eukaryotic DNA: tools for calculation and promoters analysis. Bioinformatics, 17, 998–1010. [DOI] [PubMed] [Google Scholar]
- 11.Levitsky V.G., Katokhin,A.V., Podkolodnaya,O.A. and Furman,D.P. (2004) Nucleosomal DNA organization: an integrated information system. In Kolchanov,N. and Hofestaedt,R. (eds), Bioinformatics of Genome Regulation and Structure. Kluwer Academic Publishers, Boston/Dordrecht/London, pp. 3–12. [Google Scholar]
- 12.Levitsky V.G., Podkolodnaya,O.A., Kolchanov,N.A. and Podkolodny,N.L. (2001) Nucleosome formation potential of exons, introns, and Alu repeats. Bioinformatics, 17, 1062–1064. [DOI] [PubMed] [Google Scholar]
- 13.Kutsenko A.S., Gizatullin,R.Z., Al-Amin,A.N., Wang,F., Kvasha,S.M., Podowski,R.M., Matushkin,Y.G., Gyanchandani,A., Muravenko,O.V., Levitsky,V.G., Kolchanov,N.A., Protopopov,A.I., Kashuba,V.I., Kisselev,L.L., Wasserman,W., Wahlestedt,C. and Zabarovsky,E.R. (2002) NotI flanking sequences: a tool for gene discovery and verification of the human genome. Nucleic Acids Res., 30, 3163–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Podkolodnaia O.A., Levitskii,V.G. and Podkolodnyi,N.L. (2001) Locus-controlling regions: description in the LCR-TRRD database. Mol. Biol. (Moskow), 35, 943–951. [PubMed] [Google Scholar]
- 15.Levitsky V.G. (2004) RECON: a program for prediction of nucleosome formation potential. Nucleic Acids Res., 32, W346–W349. [DOI] [PMC free article] [PubMed] [Google Scholar]