Abstract
Embryonic mosaicism, defined as the presence of karyotypically distinct cell lines within an embryo, has been frequently reported with a high incidence in preimplantation embryos derived from IVF and is thought to be one of the major biological limitations for the routine application of PGD for aneuploidies (PGD-A). The incidence of mosaicism in preimplantation embryos is in fact reported to be between 4 and 90%. However, these data are in sharp contrast with what is known from clinical pregnancies, where true foetal mosaicism is observed in less than 0.5% of cases. Here, we challenge these previous observations in preimplantation embryos, presenting an alternative perspective, which also considers the impact of technical variation to diagnose mosaicism as one possible cause contributing to overestimation of the incidence of mosaicism in embryos. Although euploid/aneuploid mosaicism may be present in blastocysts, the possibility of detecting this phenomenon within a single trophectoderm biopsy represents a contemporary challenge to bring about improvement to the practice of PGD-A. The purpose of this opinion paper is to provide a critical review of the literature, provide a possible alternative interpretation of the data, and discuss future challenges with diagnosing mosaicism in PGD-A cycles.
Keywords: chromosomal abnormalities, mosaicism, embryo selection, PGD PGS, IVF, ICSI outcome
Introduction
Embryonic mosaicism is defined as the presence of karyotypically different cell lines within the same individual, such as a preimplantation embryo (Delhanty et al., 1997; Youssoufian and Pyeritz, 2002). The primary origin of embryonic mosaicism is post-zygotic chromosome segregation errors as a consequence of mitotic non-disjunction. While other mechanisms have been considered, such as anaphase lag, and endoduplication or deletion, these events may be extremely rare (Gueye et al. 2014). Although meiotic aneuploidies are uniformly present in all cells and present with a well-defined clinical penetrance in reproductive health (Hassold and Hunt, 2001; Cohen, 2002), the embryonic fate and the clinical consequences of mosaic aneuploidies may depend on many variables, including which chromosome is involved, when the error occurs and thus what percentage of the embryo is aneuploid, and where it is located within the embryo (Johnson et al., 1990; Wapner et al., 1992; Wilkins-Haug et al., 1995). As a consequence, the clinical penetrance of a mosaic aneuploidy can be seen as unique for each event and difficult to be predicted in the absence of a well-defined phenotype.
Despite the fact that chromosomal mosaicism is diagnosed in <2% of prenatal specimens and only a small proportion of them (≈10%) is then confirmed in the foetus (Malvestiti et al., 2015), estimates of preimplantation stage mosaicism frequency range from 4% to as high as 90% (Taylor et al., 2014). Indeed, these estimates are believed to be a major biological limitation to the success of preimplantation aneuploidy screening (Mastenbroek and Repping, 2014). Moreover, the development of tools that might provide better sensitivity to the detection of low levels of aneuploidy in a mosaic trophectoderm (TE) biopsy has been proposed (Greco et al., 2015). In turn, different laboratories have begun to report the diagnosis of mosaicism in clinical cases of blastocyst PGD for aneuploidies (PGD-A).
This review aims to provide a critical evaluation of existing data on mosaicism in preimplantation embryos, to shed light on upcoming possibilities for detecting mosaicism in TE biopsies, and to propose how these data may be appropriately managed in a clinical setting.
Prevalence
Cleavage stage
One of the challenges facing PGD-A is the development of accurate estimates of the frequency of chromosomal mosaicism in the preimplantation stage of development. Access to all the cells from and individual embryo often requires specific patient consent for research and the approval of regulatory and ethical oversight committees. In many cases, the subsequent availability of embryos for dissociation and analysis may introduce significant selection bias, such as abnormal development or previous diagnosis of aneuploidy, making extrapolation to the general IVF population inappropriate.
Probably one of the most important limitations of prior estimates of embryonic mosaicism has to do with the inaccuracy of single cell PGD-A methods. In fact, all of the reported estimates of mosaicism are potentially impacted by the technical accuracy of methods used to predict aneuploidy. When analysing multiple single cells from an embryo it is close to impossible to distinguish between technical artefacts, and a genuine biological variation due to mosaicism. This is especially relevant for cleavage stage embryos where multiple single cells from the same embryos are analysed in separate reactions with a defined diagnostic error rates. Indeed, one single false positive aneuploidy observation is sufficient to result in a false positive diagnosis of mosaicism in the embryo. For example, suppose that a specific chromosome testing method is used on single cells and has a 10% false positive error rate. The analysis of six normal blastomeres is expected to result in a false positive mosaicism diagnosis in 50% of cases and 70% when 10 blastomeres are analysed. Most of the studies on this matter have used FISH to assess the incidence of mosaicism in preimplantation embryos, a method that was showed to have an even higher false positive error rate when used on interphase nuclei (Scriven and Bossuyt 2010; Treff et al., 2010; Northrop et al., 2010). It is not surprising then that FISH-based studies on single blastomeres have reported as high as 50–90% of human cleavage stage embryos being mosaic since statistically the analysis of eight normal blastomeres with 15% false positive error rate is expected to result in a false positive mosaicism diagnosis rate of nearly 75% (probability of obtaining at least one event with a false positive discovery rate of 15%: 1–0.858). It is thus extremely crucial to take some rectification measures for this phenomenon and correct for the expected false positive rate of each method during the data analysis in order to obtain a more reliable estimation of the actual incidence of mosaicism.
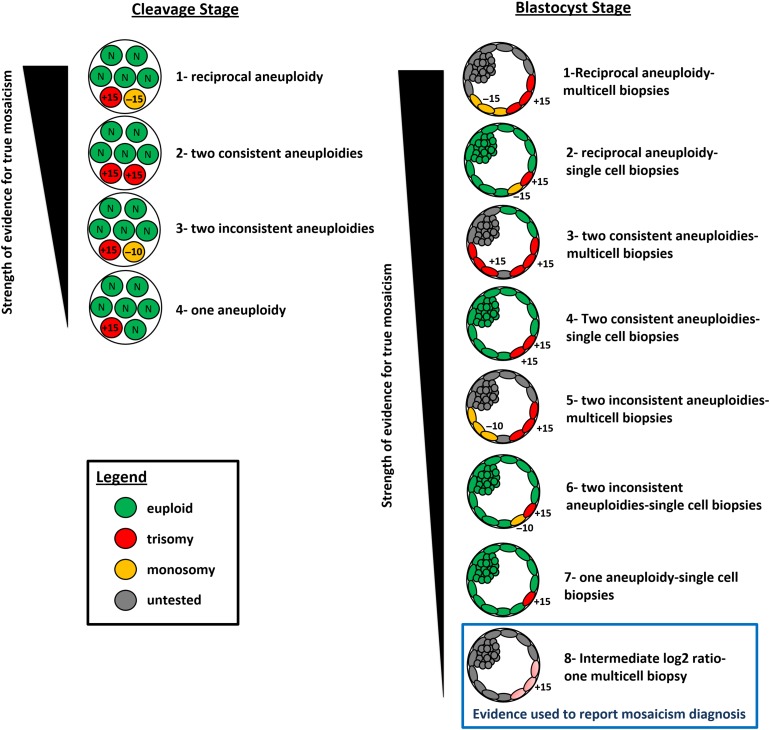
Another important consideration is the stringency of methods used to classify embryos as mosaic, which can vary significantly. For example, some studies only require one abnormal cell to be present in order to classify the embryo as mosaic. In contrast, the most rigorous criteria for classification would be the presence of reciprocal aneuploidy in two different cells or samples from the same embryo (Fig. 1). That is, one biopsy displaying a monosomy of a specific chromosome and another biopsy from the same embryo displaying trisomy for the same chromosome. This level of evidence for a true mitotic non-disjunction event would minimize the impact of technical artefacts on estimates of mosaicism, and perhaps with a minimal reduction in the sensitivity of detection. In our opinion the degree of mosaicism is therefore likely to have been extremely overestimated due to the lack of correction for the expected false positives and to the non-standardized criteria used for classification of embryos as mosaic.
Figure 1.
Proposed model for the strength of evidence when studying mosaicism in cleavage and blastocyst stage embryos. Even though one aneuploidy or two inconsistent aneuploidies provides a lower level of reliability it has been the most common criteria for defining mosaicism in preimplantation embryos. The highest level of evidence for identifying genuine mosaicism is a double biopsy and blinded analysis showing reciprocal aneuploidies. In this case the influence of technical errors is expected to be marginal. Despite being the primary method for clinical diagnosis by many laboratories, intermediate log2 ratios represents the lowest strength of evidence for true mosaicism.
More recently comprehensive chromosome screening (CCS) methods have been used to evaluate mosaicism in cleavage stage embryos (Wells and Delhanty 2000). Array comparative genomic hybridization (aCGH) was the most commonly used method for this purpose (Vanneste et al., 2009; Capalbo et al., 2013a, b; Mertzanidou et al., 2013a, b). Even though advanced genetic technologies have been well adapted to work on single cells, they do not provide 100% accuracy and are still biased by amplification artefacts (Capalbo et al., 2015). Depending on different procedural aspects, reagent batch and parameters for quality control and data analysis, a varying false positive error rate can be expected from the application of a specific aCGH protocol on a single cell. For instance, one can be more stringent in the analysis and consider only arrays results with an SD ≤ 0.15, >80% included clones, signal-to-background ratio >3 and derivative log-ratio <0.2 and get a higher accuracy but a smaller sample size (Jacobs et al., 2014), or less stringent and get more results but with lower reliability. Different versions of the analysis software and different protocols (dual channel array vs single channel array) may also impact performance. Recently, Illumina released a statement that indicated many aCGH PGS kits which had been released and in clinical use had compromised performance, resulting in a significantly increased risk of false positives. Unfortunately, it is possible that some published studies may have been impacted by this error. However, most studies do not report the specific lot numbers of reagents used making it difficult to assess which experiments may have been compromised.
Ideally, all studies investigating the presence of mosaicism with single cell analysis would first provide data on the methods accuracy on large dataset of single cells with previously established aneuploidies (positive controls). All aCGH studies performed so far on blastomeres failed to report a primary validation on single cells, with the use of each specific protocol , to estimate the false positive error rate, or do not report such data on a sufficient number of samples (Jacobs et al., 2014).
An example is a recent publication of Mertzanidou and colleagues using aCGH on all dissected blastomeres from 14 cleavage stage embryos donated for research from fertile patients undergoing monogenic PGD cycles. In that study, 20% of blastomere failed to produce a result due to amplification failure or low profile quality and 70% of embryos were classified as mosaic (Mertzanidou et al., 2013a, b). This study also provides an opportunity to indirectly evaluate the accuracy of detecting aneuploidy in general. For example, given that it is well established that the majority of human aneuploidy is derived from maternal meiotic errors, at least one of the embryos in this study should display uniform aneuploidy for at least one chromosome. Surprisingly, no embryo contained the same aneuploidy in all of its cells. Furthermore, given that a substantial contribution to mosaicism derives from mitotic non-disjunction, it can be expected that at least one embryo should have displayed reciprocal errors for at least one chromosome (Daphnis et al., 2005; Munné et al., 2005; Mantikou et al., 2012; Capalbo et al., 2013b). Surprisingly, even though the high failure rate of analysis could have lowered the detection of non-disjunction, not a single reciprocal aneuploid chromosome was observed. While the authors suggested that mosaicism in embryos is predominant, this data can be alternatively interpreted as a display of the poor reliability of that specific aCGH protocol and scoring criteria used when applied to single cells.
In contrast, another study involving characterization of mosaicism at the cleavage stage demonstrated a smaller prevalence by using a single nucleotide polymorphism (SNP) cell array based method of CCS (Treff et al., 2010). Before applying the method to blastomeres, this SNP array method was specifically evaluated for single cell accuracy using positive control cell lines and demonstrated 98.6% concordance with the expected karyotypes (Treff et al., 2010). In the analysis of mosaicism, blastomeres were randomized to either FISH or SNP array analysis from arrested cleavage stage embryos. While FISH evaluated fewer chromosomes and fewer cells per embryo (because of lower reliability of obtaining a result compared to SNP array), it still estimated a rate of mosaicism of 100%. In contrast, SNP array predicted a significantly lower mosaicism rate of only 31%, while also observing uniform aneuploidy as expected (meiotic errors). This study demonstrated that the use of inaccurate methods of aneuploidy screening can significantly overestimate the prevalence of mosaicism and further illustrates the caution necessary when interpreting results from poorly validated methods of PGD-A.
In addition to methodological artefacts, inaccurate predictions of mosaicism may also originate from differences in the cell cycle phase and DNA replication stage of the blastomeres being studied. Current methodology does not take into account the phase of the cell cycle, despite the variable copy number (CN) status of different genomic regions in the S phase. It is known that the DNA of the S-phase cell is progressively replicated from multiple origins of replication and log2 ratios of single S-phase cells follow the patterns of early and late replication domains (Van der Aa et al., 2013; Dimitriadou et al., 2014). Hence, at a given time point the genetic CN profiling of an S-phase cell will demonstrate different loci with CN status of 2, 3 or 4, depending on the replication status of this specific locus. This can possibly result in false positive and false negative CN determinations if a sufficient portion of the chromosome is replicating and depending on the criteria used to define the CN status. Hence, DNA imbalances may, on the one hand, be falsely interpreted as genuine aberrations in the S-phase cell's CN profile and hence lead to diagnostic error. This is especially relevant for cleavage stage embryos, where, fast dividing cells such as human blastomeres or some tumour cells are expected to reside more often in S-phase, and thus the chances are higher to isolate a cell in S-phase from such a population. This is another potential source of error poorly acknowledged in existing studies, even thought no direct evidence has been reported so far that whole chromosome aneuploidy call can be compromised by the cell cycle phase.
The above-mentioned sources of false positive errors also provide a possible explanation for data reported regarding the generation of karyotypically normal embryonic stem cells from supposed aneuploid embryos after a FISH based PGD-A on single blastomeres (Bazrgar et al., 2013).
Blastocyst stage
Studies of mosaicism in blastocysts have reported much lower levels of compared to the cleavage stage. This observation has commonly been interpreted as a selection against embryos with mitotic derived aneuploidy between the cleavage and blastocyst stage of development. According to this model, it is indeed possible that blastomeres affected by a mitotic chromosome segregation error might harbour additional cellular defects causing a developmental arrest. However, since all types of uniform aneuploidies can survive to the blastocyst stage (Fragouli et al., 2011; Franasiak et al., 2014), including complex aneuploidies, an alternative explanation for the observed difference between cleavage and blastocyst stage mosaicism rate can be found in the improved accuracy achieved when evaluating multiple cells instead of single cells. This includes the fact that S-phase artefacts are less likely to impact predictions from a TE biopsy. Of course, the reduced rate of mosaicism may also be due to masking of aneuploidy by euploid cells or by cells with the reciprocal aneuploidy within the same biopsy, resulting in overestimation of euploidy.
The ideal approach to investigate the incidence and prevalence of mosaicism in human blastocysts would entail dissection of the entire embryo into single cells and the use of a robust CCS platform with a very low and well-defined error rate. However, this approach has been unattainable as a result of absence of an effective method to disaggregate blastocysts down to single cells. Alternative approaches to estimate mosaicism in blastocysts are single cell analysis by FISH or by CCS analysis of multiple blinded biopsies (Fig. 1).
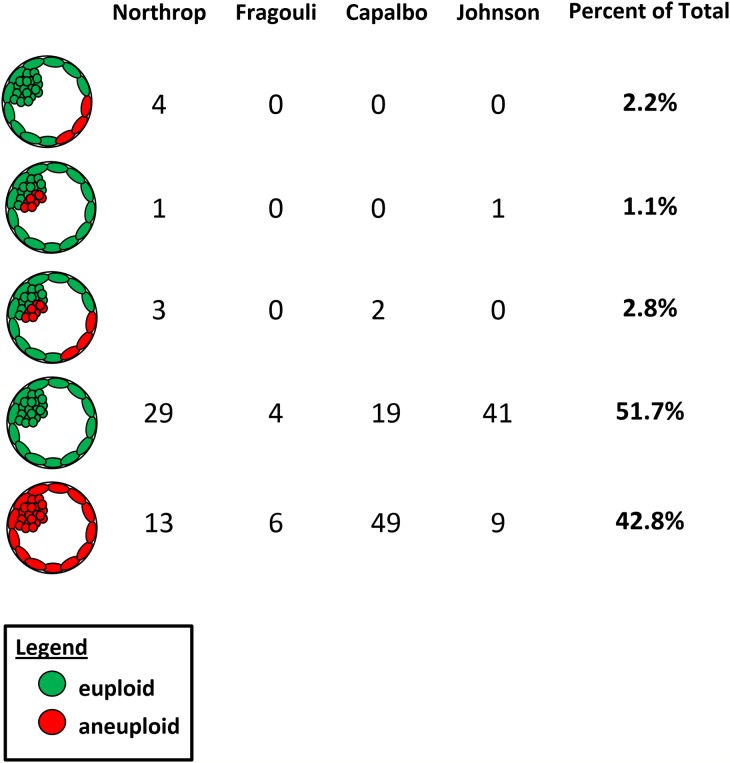
A number of recent studies have investigated the frequency and distribution of mosaicism within the blastocyst using FISH or contemporary methods of CCS. Johnson et al. (2010) observed no sign of mosaicism for the whole chromosome CN within 51 blastocysts and again no evidence of preferential segregation to ICM or TE lineage. Fragouli et al. (2011) found 17% mosaic diploid/aneuploid among 52 blastocysts and no sign of preferential allocation (Fragouli et al. 2008), and Northrop et al. (2010) found 14% euploid/aneuploid mosaic blastocysts among 50 embryos, also demonstrating a lack of preferential segregation (Fig. 2).
Figure 2.
Summary of data from relevant CCS studies on the cytogenetic constitution of ICM and TE samples from disaggregated human blastocysts. No sign of preferential allocation or confinement of chromosomally abnormal cells to the TE or ICM lineage was observed. The fact that results were almost identical for samples from the TE and ICM indicates that data obtained from a sample of a few cells biopsied from the blastocyst TE can generally be considered representative of the ICM chromosomal complement. Data taken from Northrop et al. (2010), Fragouli et al. (2008), Johnson et al. (2010) and Capalbo et al. (2014).
We have reported an extensive study on chromosome mosaicism at the blastocyst stage (Capalbo et al., 2014) using FISH to analyse three TE sections and the ICM of 70 blastocysts. In this study, ICM isolation methodology using TE specific biomarkers was also validated. Furthermore, a mosaicism classification was made only if >10% of the nuclei presented with the same abnormality and if the same aneuploid signal was present in at least two cells from the same embryonic section. This criterion is currently one of the more straightforward approaches in cytogenetics to distinguish true aneuploidy from FISH artefacts due to false positive results and mosaicism diagnosis, when multiple nuclei are tested together and control material is lacking. Indeed, Vysis’ multicentre study of FISH using AneuVysion applied to nuclei from uncultured amniocytes (Vysis, Inc., Downers Grove, IL, USA, 1997) indicated that a non-mosaic disomic result had >90% of nuclei with two signals and can be expected to have up to 10% of nuclei with deviant patter (i.e. not two signals, mostly one or three signals and skewed towards the former). Since in the analysis of few nuclei per sample, for instance eight, only one deviant pattern is required to surpass the 10% threshold, more than one deviant pattern was also included as criteria to provide evidence of mosaicism. That is the reason why a 10% threshold and the observation of at least two consistent aneuploid results in the analysis of the whole dissected blastocyst was used, trying to maximize the detection of genuine mosaicism cases, even though some low grade mosaicism cases would be missed. Using these criteria of analysis, mosaic chromosomal errors were observed in 15.7% of the blastocysts tested, but only two cases (2.9%) embryos showed a mix of normal and abnormal cells (mosaic diploid/aneuploid). Of note, we observed that the proportion of abnormal cells within the blastocyst predicted their distribution, i.e. embryos showing higher mosaicism rates had abnormal cells present across all blastocyst sections. Accordingly, high grade mosaicism cases that are expected to be of clinical relevance are expected to show abnormal cells in all blastocysts area, thus increasing the likelihood of detection in TE based PGD-A. This was confirmed in the study by comparing the original CCS diagnosis obtained from clinical TE biopsies with the actual chromosomal constitution of the dissected blastocysts (Capalbo et al., 2014). Furthermore, no evidence of preferential segregation of aneuploidy in the ICM or TE was observed. It has to be underlined that, as for all FISH-based studies, the analysis of nine chromosomes might have lowered the sensitivity of detection.
Finally, an our more recent study on multiple TE biopsies including 161 blastocysts, showed >99% (3468/3473; 95% CI: 0.99–1) consistent chromosome CN diagnosis, suggesting no major diagnostic impact of mosaicism when testing at the blastocyst stage (Capalbo et al., 2015). Furthermore, owing to the high overall consistency of chromosome diagnosis reported in this study using different CCS methods on multiple TE biopsies from the same embryos, blastocyst-stage aneuploidy screening proved to be a highly reliable and effective approach for PGD-A.
To summarize, considering that in prenatal specimens chromosomal mosaicism is detected in ~1–2%, that mosaicism associated with ART persists beyond the preimplantation embryo at a rate similar to that associated with pregnancies conceived spontaneously (Huang et al., 2009) and the possible methodological and technical flaws described above to assess its real incidence in preimplantation embryos, it is reasonable to speculate that chromosome mosaicism can be regarded as a relatively uncommon event in IVF embryos and should not have a major diagnostic impact on PGD-A cycles. The most extensive studies showed around 4–5% of blastocysts being mosaic diploid/aneuploid (Fig. 2). Also, since there is no difference in the prevalence of mosaicism at the end of the first trimester in pregnancies conceived spontaneously compared with those with infertility treatments, the suggestion that inadequacies of embryo culture play a role in the genesis of this problem, increasing the risk of chromosome malsegregation during mitosis remains highly speculative at this time.
Detection within a TE biopsy with contemporary CCS methods
One of the common misconceptions surrounding the ability of CCS technology to detect mosaicism is that an intermediate alteration in the log2 ratio is definitive for making a prediction. Many groups have suggested that altered log2 ratios when observed in a TE biopsy can be used to diagnose mosaicism (Greco et al., 2015; Munné et al., 2016). Unfortunately, this criterion is insufficient as there are many possible alternative explanations for such observations, first and foremost being artifacts introduced by whole genome analysis (WGA) and aCGH or next generation sequencing (NGS) analyses on low input samples. There is a considerable risk of making an inaccurate prediction of mosaicism, resulting in a false positive diagnosis. This is a possible alternative explanation of the clinical cases reported in the recent report by Greco et al., (2015), where the authors suggest that embryos with mosaicism can implant and result in delivery of normal babies. In fact, the authors fail to acknowledge an alternative explanation for the observations made, that the original diagnosis of mosaicism was incorrect and uniformly euploid were erroneously classified as mosaic.
First, the authors disregarded the available data from their cell line mixture models on the rate of false positive mosaicism predictions. That is, they did not evaluate how often a chromosome was predicted mosaic when it should not have been. The specificity (the proportion of negatives, non-mosaic samples, that are correctly identified as such) of the assay was therefore not established. Furthermore, the authors failed to validate the ability to predict mosaicism by analyzing additional biopsies from the same embryo to establish predictive value. A more rigorous method for finding mosaicism in an embryo would be to identify reciprocal aneuploidy of the same chromosome in multiple biopsies (Fig. 1). For example, finding some cells with monosomy and other cells with trisomy of the same chromosome. Without appropriate clinical evaluation of the accuracy of using log2 ratios to predict mosaicism, it may be prudent to modify the interpretation of such observations to indicate that the pattern is consistent with possible mosaicism, rather than stating that the embryo is indeed mosaic.
Furthermore, it should be important to evaluate reliability of each method to detect mosaicism in TE biopsies for every single chromosome, as each one can have its own and different performance in the analysis.
There is currently very limited data on mixture models to evaluate reliability, sensitivity and specificity of detecting aneuploidy within a TE biopsy. A previous study using SNP array analysis investigated performance on detecting monosomy X in mixtures of male and female cells (Northrop et al., 2010). While this study found that detection was possible at 40% male to 60% female ratios, the sample size investigated was insufficient to establish reliability or to adequately evaluate specificity. Another study evaluated mixtures of euploid and trisomy 10 cells using aCGH and found that detection was possible at approximately a 50:50 ratio (Mamas et al., 2012). More recently, Greco et al. suggested that aCGH was capable of detection at 20% aneuploidy levels. However, the reliability of detecting this level of aneuploidy was not evaluated, nor was the rate of false positive prediction considered. Another study evaluated the performance of targeted NGS based CCS on predicting aneuploidy in mixture models. This study found detection was possible at 16% aneuploidy, but with reduced reliability overall when compared to 50% aneuploidy, where detection success reached 100% (Goodrich et al. 2015). In addition, no false positives were identified demonstrating good predictive value. A recent randomized, double blinded study on cell mixture model also illustrates that some methods of CCS, such as qPCR, already inherently identify aneuploidy in embryos that are mosaic and that new criteria are not necessary (Goodrich et al., 2016). The same study highlighted that WGA and technical artifacts can result in altered log2 ratios that can be reported as mosaic when poorly validated scoring criteria are used. Additional studies evaluating NGS methodology have not been reported to date. However, genetic laboratories have communicated higher rates of putative mosaicism in TE biopsies when moving from aCGH protocols to commercially available NGS technologies for PGS. With the improved resolution and sensitivity of NGS, it is expected that while there is higher sensitivity of mosaicism detection the artificial fluctuation of log2 ratio can be even more pronounced and reported as mosaicism, thus a further cautionary note is necessary for the clinical application of these new technologies and more preclinical studies are warranted before systematic clinical application.
Another set of data that should be required before clinical application of new criteria for predicting mosaicism is a non-selection clinical trial (Scott et al., 2012). That is, embryos predicted euploid by conventional methods of CCS are selected without interpreting data for mosaicism until after embryo transfer. Once embryos, which implant are identified by DNA fingerprinting the mosaicism predictions can be made to determine whether it would have successfully predicted a negative clinical outcome. As it is unlikely that mosaicism predictions in the preimplantation embryo will always successfully predict a negative clinical outcome, these data are critical to establish the true predictive value of methods used and to provide evidence-based information to patients who may utilize such methods.
Management in the clinical practice
As discussed before, mosaicism is expected to affect a minority of blastocysts, thus the development of CCS technologies able to quantify the rate of abnormal cells in a mosaic euploid/aneuploid embryo is anticipated to provide a marginal improvement in the clinical practice of PGD-A. However, important implications may arise by the systematic reporting of fluctuating log2 ratios as mosaicism in routine clinical practice, as is already the case for many PGD laboratories.
The contemporary inability to adequately distinguish between technical and biological contributions to altered log2 ratios necessitates avoiding reporting mosaicism in clinical cases at present time. Until evidence is presented that demonstrates one or the other is the predominant source of altered log2 ratios, both sources should be acknowledged as possible contributions to the observations made. Without any confirmation from additional biopsies from the same embryo, the interpretation as mosaicism of discordant altered profiles is simply not justified. In addition, since grading embryonic mosaicism from a single biopsy is significantly flawed, the rate of normal to abnormal cells estimated from a TE sample has also a potential for reporting misleading data to patients and in our opinion should be avoided. An alternative option could include suggesting that a given pattern is ‘consistent with possible mosaicism’. If prospective non-selection studies are able to demonstrate that significantly reduced success rates result from the transfer of such embryos, perhaps the additional information could be used to prioritize embryos available for transfer.
In any case, if mosaicism is reported, extensive genetic counselling should also be provided, including discussion of the technical limitations of defining the presence and extent of mosaicism from a single embryo biopsy, as well as the potential clinical consequences with respect to the actual chromosome involved. Clearly, utilization of methods that overestimate mosaicism will lead to inappropriate discard of healthy reproductively competent embryos. In contrast, if patients elect to transfer an embryo with predicted mosaicism there is a real risk for an abnormal pregnancy. For example, some have considered a strategy involving the selective transfer of embryos mosaic for monosomy. This is a major concern since a population of trisomic cells can be present due to a mitotic non-disjunction event and could result in a negative clinical outcome, such as a miscarriage, or, in the case of a viable pregnancy, a live born with severe congenital abnormalities.
At present time, we recommend that the uncertainness and limitations of mosaicism diagnosis should be acknowledged in consent forms and represent an integral part of the genetic/reproductive counselling process if there is intention to report these observations in clinical cases.
Conclusion
Mosaicism has been likely overestimated by the imperfect nature and lack of robustness of methods of testing and it does not seem to be increased in IVF cycles compared to natural conceptions. Accordingly, chromosome mosaicism is not expected to be a major biological limitation for the systematic application of PGD-A in IVF. Methods of detecting aneuploidy within a mosaic TE biopsy are under development but may only provide marginal improvements to the clinical application of PGD-A due to the expected low incidence of this biological phenomenon in blastocysts and pregnancies and the fact that there will always exist a sampling error when estimating the presence of mosaicism from a single biopsy. The use of altered log2 ratio data to predict mosaicism must be considered with criticism in order to avoid erroneous discard of embryos with reproductive potential. When results indicate the potential for mosaicism to exist, it is inappropriate to report a genuine diagnosis of mosaicism but it should instead be classified as a pattern ‘consistent with possible mosaicism’ and be accompanied by extensive genetic counselling which recognizes the many limitations of such a prediction. Future work should also focus on developing level I evidence of the clinical predictive value of new mosaicism classification schemes.
Authors' roles
A.C. and N.R.T. developed and wrote the article. F.M.U., L.R. and R.S. provide a critical review of concepts.
Funding
No specific funding was used for this article.
Conflict of interest
None declared.
References
- Bazrgar M, Gourabi H, Valojerdi MR, Yazdi PE, Baharvand H. Self-correction of chromosomal abnormalities in human preimplantation embryos and embryonic stem cells. Stem Cells Dev 2013;22:2449. [DOI] [PubMed] [Google Scholar]
- Capalbo A, Bono S, Spizzichino L, Biricik A, Baldi M, Colamaria S et al. . Sequential comprehensive chromosome analysis on polar bodies, blastomeres and trophoblast: insights into female meiotic errors and chromosomal segregation in the preimplantation window of embryo development. Hum Reprod 2013. a;28:509. [DOI] [PubMed] [Google Scholar]
- Capalbo A, Wright G, Elliott T, Ubaldi FM, Rienzi L, Nagy ZP. FISH reanalysis of inner cell mass and trophectoderm samples of previously array-CGH screened blastocysts shows high accuracy of diagnosis and no major diagnostic impact of mosaicism at the blastocyst stage. Hum Reprod 2013. b;28:2298. [DOI] [PubMed] [Google Scholar]
- Capalbo A, Rienzi L, Cimadomo D, Maggiulli R, Elliott T, Wright G, Nagy ZP, Ubaldi FM. Correlation between standard blastocyst morphology, euploidy and implantation: an observational study in two centers involving 956 screened blastocysts. Hum Reprod 2014;29:1173. [DOI] [PubMed] [Google Scholar]
- Capalbo A, Treff NR, Cimadomo D, Tao X, Upham K, Ubaldi FM et al. . Comparison of array comparative genomic hybridization and quantitative real-time PCR-based aneuploidy screening of blastocyst biopsies. Eur J Hum Genet 2015;23:901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. Sorting out chromosome errors. Science 2002;296:2164. [DOI] [PubMed] [Google Scholar]
- Daphnis DD, Delhanty JD, Jerkovic S, Geyer J, Craft I, Harper JC. Detailed FISH analysis of day 5 human embryos reveals the mechanisms leading to mosaic aneuploidy. Hum Reprod 2005;1:129. [DOI] [PubMed] [Google Scholar]
- Delhanty JD, Harper JC, Ao A, Handyside AH, Winston RM. Multicolour FISH detects frequent chromosomal mosaicism and chaotic division in normal preimplantation embryos from fertile patients. Hum Genet 1997;99:755. [DOI] [PubMed] [Google Scholar]
- Dimitriadou E, Van der Aa N, Cheng J, Voet T, Vermeesch JR. Single cell segmental aneuploidy detection is compromised by S phase. Mol Cytogenet 2014;7:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragouli E, Alfarawati S, Daphnis DD, Goodall NN, Mania A, Griffiths T et al. . Cytogenetic analysis of human blastocysts with the use of FISH, CGH and aCGH: scientific data and technical evaluation. Hum Reprod 2011;26:480. [DOI] [PubMed] [Google Scholar]
- Fragouli E, Lenzi M, Ross R, Katz-Jaffe M, Schoolcraft WB, Wells D. Comprehensive molecular cytogenetic analysis of the human blastocyst stage. Hum Reprod 2008;23:2596. [DOI] [PubMed] [Google Scholar]
- Franasiak JM, Forman EJ, Hong KH, Werner MD, Upham KM, Treff NR et al. . The nature of aneuploidy with increasing age of the female partner: a review of 15 169 consecutive trophectoderm biopsies evaluated with comprehensive chromosomal screening. Fertil Steril 2014;101:656–63 e1. [DOI] [PubMed] [Google Scholar]
- Goodrich D, Tao X, Bohrer C, Bedard J, Landis JN, Scott RT, Treff NR. Evaluation of next generation sequencing (NGS) based comprehensive chromosome screening (CCS) sensitivity to mosaicism. Fertil and Steril 2015;104:e280–e281. [Google Scholar]
- Goodrich D, Tao X, Bohrer C, Lonczak A, Xing T, Zimmerman R, Zhan Y, Scott RT Jr, Treff NR. A randomized and blinded comparison of qPCR and NGS-based detection of aneuploidy in a cell line mixture model of blastocyst biopsy mosaicism. J Assist Reprod Genet 2016. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco E, Minasi MG, Fiorentino F. Healthy babies after intrauterine transfer of mosaic aneuploid blastocysts. N Engl J Med 2015;373:2089. [DOI] [PubMed] [Google Scholar]
- Gueye NA, Devkota B, Taylor D, Pfundt R, Scott RT Jr, Treff NR. Uniparental disomy in the human blastocyst is exceedingly rare. Fertil Steril 2014;101:232. [DOI] [PubMed] [Google Scholar]
- Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2001;2:280. [DOI] [PubMed] [Google Scholar]
- Huang A, Adusumalli J, Patel S, Liem J, Williams J 3rd, Pisarska MD. Prevalence of chromosomal mosaicism in pregnancies from couples with infertility. Fertil Steril 2009;91:2355. [DOI] [PubMed] [Google Scholar]
- Jacobs K, Mertzanidou A, Geens M, Nguyen HT, Staessen C, Spits C. Low-grade chromosomal mosaicism in human somatic and embryonic stem cell populations. Nat Commun 2014;5:4227. [DOI] [PubMed] [Google Scholar]
- Johnson A, Wapner RJ, Davis GH, Jackson LG. Mosaicism in chorionic villus sampling: an association with poor perinatal outcome. Obstet Gynecol 1990;75:573. [PubMed] [Google Scholar]
- Johnson DS, Cinnioglu C, Ross R, Filby A, Gemelos G, Hill M et al. . Comprehensive analysis of karyotypic mosaicism between trophectoderm and inner cell mass. Mol Hum Reprod 2010;16:944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamas T, Gordon A, Brown A, Harper J, Sengupta S. Detection of aneuploidy by array comparative genomic hybridization using cell lines to mimic a mosaic trophectoderm biopsy. Fertil Steril 2012;4:943. [DOI] [PubMed] [Google Scholar]
- Malvestiti F, Agrati C, Grimi B, Pompilii E, Izzi C, Martinoni L et al. . Interpreting mosaicism in chorionic villi: results of a monocentric series of 1001 mosaics in chorionic villi with follow-up amniocentesis. Prenat Diagn 2015;35:1117. [DOI] [PubMed] [Google Scholar]
- Mertzanidou A, Spits C, Nguyen HT, Van de Velde H, Sermon K. Evolution of aneuploidy up to Day 4 of human preimplantation development. Hum Reprod 2013. a;28:1716. [DOI] [PubMed] [Google Scholar]
- Mertzanidou A, Wilton L, Cheng J, Spits C, Vanneste E, Moreau Y et al. . Microarray analysis reveals abnormal chromosomal complements in over 70% of 14 normally developing human embryos. Hum Reprod 2013. b;28:256. [DOI] [PubMed] [Google Scholar]
- Mastenbroek S, Repping S. Preimplantation genetic screening: back to the future. Hum Reprod 2014;29:1846. [DOI] [PubMed] [Google Scholar]
- Mantikou E, Wong KM, Repping S, Mastenbroek S. Molecular origin of mitotic aneuploidies in preimplantation embryos. Biochim Biophys Acta 2012;12:1921. [DOI] [PubMed] [Google Scholar]
- Munné S, Grifo J, Wells D. Mosaicism: ‘survival of the fittest’ versus ‘no embryo left behind’. Fertil Steril 2016. [DOI] [PubMed] [Google Scholar]
- Munné S, Velilla E, Colls P, Garcia Bermudez M, Vemuri MC, Steuerwald N et al. . Self-correction of chromosomally abnormal embryos in culture and implications for stem cell production. Fertil Steril 2005;84:1328. [DOI] [PubMed] [Google Scholar]
- Northrop LE, Treff NR, Levy B, Scott RT Jr. SNP microarray-based 24 chromosome aneuploidy screening demonstrates that cleavage-stage FISH poorly predicts aneuploidy in embryos that develop to morphologically normal blastocysts. Mol Hum Reprod 2010;16:590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RT Jr, Ferry K, Su J, Tao X, Scott K, Treff NR. Comprehensive chromosome screening is highly predictive of the reproductive potential of human embryos: a prospective, blinded, nonselection study. Fertil Steril 2012;97:870. [DOI] [PubMed] [Google Scholar]
- Scriven PN, Bossuyt PM. Diagnostic accuracy: theoretical models for preimplantation genetic testing of a single nucleus using the fluorescence in situ hybridization technique. Hum Reprod 2010;25:2622. [DOI] [PubMed] [Google Scholar]
- Taylor TH, Gitlin SA, Patrick JL, Crain JL, Wilson JM, Griffin DK. The origin, mechanisms, incidence and clinical consequences of chromosomal mosaicism in humans. Hum Reprod Update 2014;20:571–581. [DOI] [PubMed] [Google Scholar]
- Treff NR, Levy B, Su J, Northrop LE, Tao X, Scott RT Jr. SNP microarray-based 24 chromosome aneuploidy screening is significantly more consistent than FISH. Mol Hum Reprod 2010;16:583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treff NR, Su J, Tao X, Levy B, Scott RT Jr. Accurate single cell 24 chromosome aneuploidy screening using whole genome amplification and single nucleotide polymorphism microarrays. Fertil Steril 2010;94:2017. [DOI] [PubMed] [Google Scholar]
- Van der Aa N, Cheng J, Mateiu L, Zamani Esteki M, Kumar P, Dimitriadou E et al. . Genome-wide copy number profiling of single cells in S-phase reveals DNA-replication domains. Nucleic Acids Res 2013;41:e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C et al. . Chromosome instability is common in human cleavage-stage embryos. Nat Med 2009;15:577. [DOI] [PubMed] [Google Scholar]
- Wapner RJ, Simpson JL, Golbus MS, Zachary JM, Ledbetter DH, Desnick RJ et al. . Chorionic mosaicism: association with fetal loss but not with adverse perinatal outcome. Prenat Diagn 1992;12:347. [DOI] [PubMed] [Google Scholar]
- Wells D, Delhanty JD. Comprehensive chromosomal analysis of human preimplantation embryos using whole genome amplification and single cell comparative genomic hybridization. Mol Hum Reprod 2000;6:1055. [DOI] [PubMed] [Google Scholar]
- Wilkins-Haug L, Roberts DJ, Morton CC. Confined placental mosaicism and intrauterine growth retardation: a case-control analysis of placentas at delivery. Am J Obstet Gynecol 1995;172:44–50. [DOI] [PubMed] [Google Scholar]
- Youssoufian H, Pyeritz RE. Mechanisms and consequences of somatic mosaicism in humans. Nat Rev Genet 2002;3:748. [DOI] [PubMed] [Google Scholar]