Summary
Mitosis ensures equal segregation of the genome and is controlled by a variety of ubiquitylation signals on substrate proteins. However, it remains unexplored how the versatile ubiquitin code is read out during mitotic progression. Here, we identify the ubiquitin receptor protein UBASH3B as an important regulator of mitosis. UBASH3B interacts with ubiquitylated Aurora B, one of the main kinases regulating chromosome segregation, and controls its subcellular localization but not protein levels. UBASH3B is a limiting factor in this pathway, and is sufficient to drive Aurora B to microtubules prior to anaphase. Importantly, targeting Aurora B to microtubules by UBASH3B is necessary for the timing and fidelity of chromosome segregation in human cells. Our findings uncover an important mechanism defining how ubiquitin attachment to a substrate protein is decoded during mitosis.
Keywords: mitosis, ubiquitin, Aurora B, chromosome segregation, ubiquitin receptor, UBASH3B
Introduction
Mitosis ensures equal segregation of the genome to daughter cells, and defects in mitosis can lead to aneuploidy and polyploidy, frequently observed in cancers (Ganem et al., 2007). Chromosome segregation requires assembly of the mitotic spindle and its attachment to sister kinetochores of all chromosomes, and is strictly controlled by the activity of the Spindle Assembly Checkpoint (SAC) (Musacchio and Salmon, 2007). Defects in kinetochore attachment prevent SAC satisfaction and chromosome segregation. Cells with compromised/defective SAC response may segregate their chromosomes erroneously and become aneuploid or polyploid.
Aurora B kinase is one of the key mitotic factors controlling euploidy. Aurora B is a component of the Chromosomal Passenger Complex (CPC) that coordinates chromosome segregation by regulating spindle assembly, kinetochore-microtubule (KT-MT) attachments and localized activity of the SAC (van der Horst and Lens, 2014). Essential functions of Aurora B in chromosome segregation are dependent on its dynamic localization to centromeres in prometaphase and metaphase and to midzone microtubules during anaphase (Carmena et al., 2012). Centromeric recruitment of Aurora B is controlled by histone phosphorylation enhanced by positive feedback loops (van der Horst and Lens, 2014) and spindle microtubules (Banerjee et al., 2014). Relocalization of Aurora B to midzone microtubules requires mitotic kinesin MKlp2, the CPC protein INCENP, and a drop in CDK1 activity (Gruneberg et al., 2004; Hummer and Mayer, 2009). Besides phosphorylation, other postranslational modifications were shown to regulate CPC localization during mitosis (van der Horst and Lens, 2014). Previous findings identified an important role for non-proteolytic ubiquitylation of Aurora B by CUL3-based E3 ligases in its relocalization from centromeres to microtubules during mitosis (Maerki et al., 2009; Sumara and Peter, 2007, Sumara et al., 2007). However, it remains unknown how and when ubiquitylated Aurora B is targeted to the mitotic structures.
Ubiquitin attachment to substrate proteins regulates fidelity of mitosis through both proteolytic and non-proteolytic mechanisms (Bassermann et al., 2014). E3 ligases catalyze substrate ubiquitylation, ranging from addition of a single ubiquitin molecule (monoubiquitylation) to different chains of interconnected ubiquitins (Komander and Rape, 2012). Ubiquitin Binding Domain (UBD) proteins can serve as receptors, or decoders, for the specific ubiquitin signals, transferring ubiquitylated substrates to downstream signaling components and determining their cellular functions (Husnjak and Dikic, 2012). Surprisingly, despite a high number of known UBD proteins in mammalian cells, their mitotic roles remain unexplored; therefore, we carried out high-content siRNA screening and proteomic approaches to identify UBD-proteins that have a role in mitosis.
Here, we show that the UBD protein UBASH3B is important for Aurora B localization and chromosome segregation. UBASH3B directly binds to ubiquitylated Aurora B and to CUL3, and its interaction with Aurora B is dependent on CUL3 and ubiquitin. Similar to CUL3, UBASH3B does not regulate protein levels of Aurora B. Instead, UBASH3B localizes to mitotic spindles and is required for timely relocalization of Aurora B from centromeres to microtubules. Strikingly, UBASH3B forms a functional complex with MKlp2 and is sufficient to target both Aurora B and MKlp2 to microtubules, even in the presence of high CDK1 activity. Thus, UBASH3B is a limiting factor for Aurora B targeting to microtubules prior to anaphase onset. In line with this, super-resolution microscopy reveals a microtubule-associated pool of Aurora B in metaphase cells upon chromosome alignment. Taken together, our findings show that UBASH3B mediates Aurora B localization in mitosis, which relies on decoding of a non-proteolytic ubiquitin signal.
Results
UBASH3B controls chromosome segregation
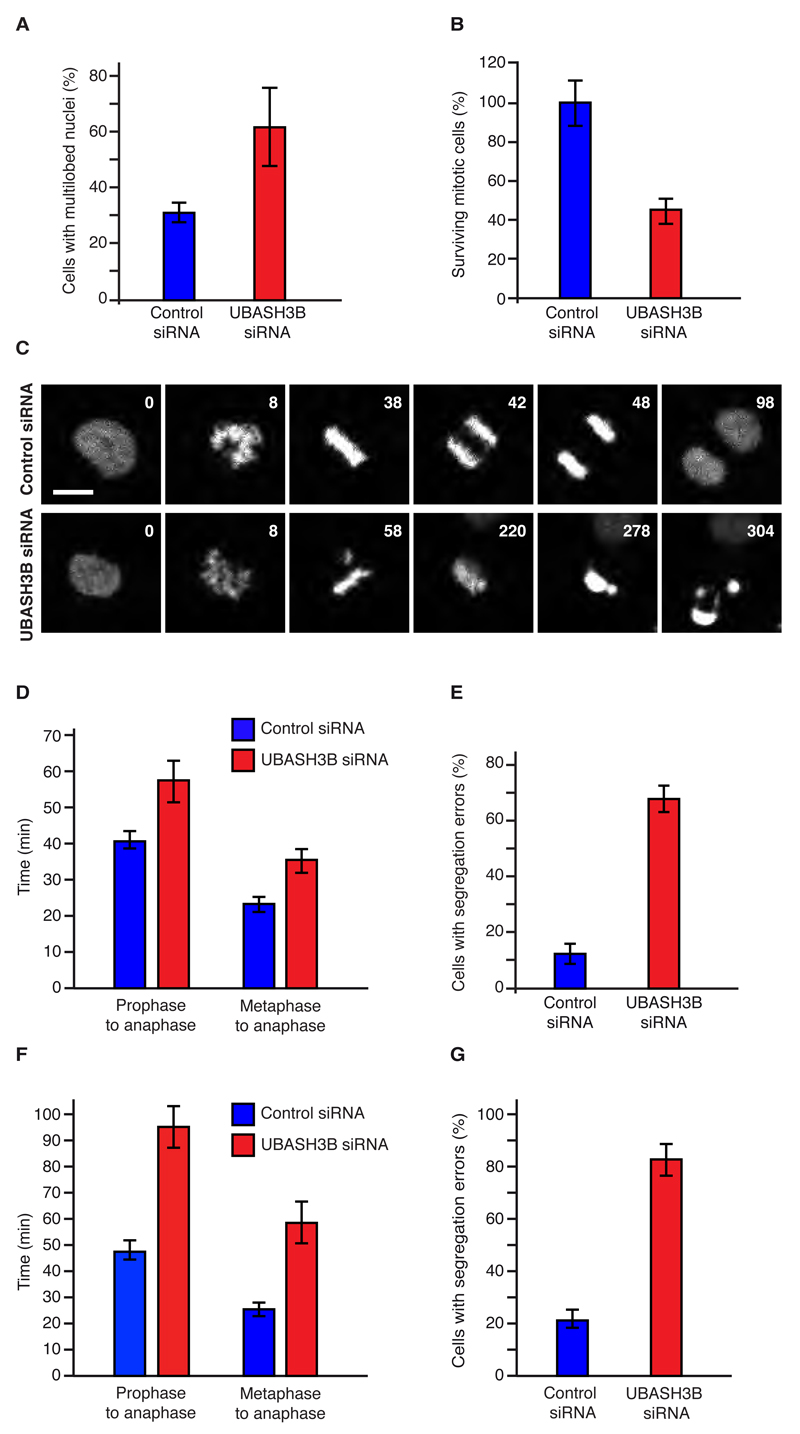
To identify ubiquitin receptors that control euploidy of human cells, we performed a high content visual siRNA screen in HeLa cells for known and predicted human UBD proteins (Table S1), and scored for defects in chromosome segregation, cytokinesis and terminal phenotypes of multilobed nuclei and multinucleation, such as observed upon downregulation of Aurora B (Figure S1A). Interestingly, one of the hits (Table S2), Ubiquitin-associated (UBA) and SH3 domain-containing protein B, UBASH3B (also known as Suppressor of T-cell receptor signaling 1, STS1 or T-cell ubiquitin ligand 2, TULA2) was also found to interact with CUL3, which was immunoprecipitated from mitotic cells and analyzed by mass spectrometry (Table S3). UBASH3B is a ubiquitously expressed protein, previously shown to bind monoubiquitylated proteins to regulate internalization of receptor protein kinases (Hoeller et al., 2006; Kowanetz et al., 2004), but it has not been linked to the regulation of mitosis. To confirm its potent role in mitotic progression, we downregulated UBASH3B by a pool of specific siRNAs (Figures S1B and C), distinct from the two pools used in the primary and secondary siRNA screens (Table S1). Both mRNA (Figure S1B) and protein levels (Figure S1C) of UBASH3B were markedly decreased upon treatment with UBASH3B-specific siRNA pool but not by the control siRNAs. In agreement with results obtained by unbiased screening (Figure S1A), downregulation of UBASH3B markedly increased the number of cells with multilobed nuclei of heterogeneous forms (Figures 1A and S1D).
Figure 1. UBASH3B is required for chromosome segregation.
(A) HeLa cells were treated with the indicated siRNAs and cells with multilobed nuclei were quantified using immunofluorescence microscopy, n= 664. Corresponding images are shown in Figure S1D. (B to E) HeLa cells expressing H2B-mCherry and EGFP-IBB were treated as in (A) and analyzed by live-cell microscopy. The percentage of surviving mitotic cells was quantified (B, n=157). The representative frames are shown in Figure 1C and S2A. Time (min) starting from prophase is indicated (1C). Hereafter, scale bar is 5 μm. The average time (min) from prophase to anaphase, from metaphase to anaphase (D,) and percentage of anaphase cells with segregation errors (E) were quantified, n=157. (F to G) HeLa cells expressing H2B-mCherry and EGFP-Tubulin were treated as in (A) and analyzed by live-cell microscopy as shown in (D, E), n=175. The representative images of anaphase cells with segregation errors are shown in Figure S2A, B. The data are shown as the mean of three independent experiments ± standard error of the mean (S.E.M.).
To understand how UBASH3B regulates mitosis, we used immunofluorescence microscopy (IF) and analyzed the distribution of different mitotic stages in synchronized cells. We observed a significant increase in a number of prometaphase cells upon downregulation of UBASH3B, suggesting defects in chromosome congression and/or timely onset of anaphase (Figure S1E). To corroborate these results, we employed live video microscopy of HeLa cells stably expressing histone marker H2B-mCherry and a probe for postmitotic nuclear reassembly, the importin-β-binding domain of importin-α, IBB–EGFP, which co-localizes with chromatin regions after reformation of a functional nuclear envelope (Schmitz et al., 2010). This analysis showed that downregulation of UBASH3B reduced survival of mitotic HeLa cells and led to death in prometaphase (was visualized by fragmentation and hypercondensation of chromatin) (Figures 1B, C and S2A). Moreover, in cells treated with siRNA against UBASH3B, the average time from prophase to anaphase was increased as compared to control siRNA-treated cells (Figures 1C, D and S2A) with the strongest delay observed from metaphase to anaphase (Figure 1D). These results suggest that UBASH3B controls the timing and fidelity of chromosome segregation. Indeed, UBASH3B-downregulated cells displayed segregation defects with frequent lagging chromosomes and chromosomal bridges, leading to unequal distribution of chromatin to daughter cells (Figures 1E and S2A, B). These observations were confirmed using a reporter cell line expressing EGFP-Tubulin and H2B-Cherry markers (Schmitz et al., 2010), where downregulation of UBASH3B also led to a delay in prophase to anaphase, as well as a strong delay in metaphase to anaphase (Figure 1F) and to chromosome segregation defects (Figures 1F, G and S2B). We occasionally observed in live video experiments that unfocused spindle poles were formed upon specific downregulation of UBASH3B, however, γ-tubulin immunofluorescence did not reveal significant defects in the bipolar spindle formation (Figure S2C, D and E). These results suggest that UBASH3B is required for chromosome segregation.
To further test the role of UBASH3B in mitosis, we analyzed its subcellular localization (Figure S3A). In interphase and prophase cells UBASH3B exhibited a largely diffuse cytosolic distribution with an enrichment at the vesicle-like structures in the vicinity of the nucleus (Figure S3A), consistent with a previous report (Raguz et al., 2007). Upon chromosome condensation, UBASH3B started accumulating on spindles, with the strongest enrichment observed in metaphase cells that had fully aligned their chromosomes (Figures S2C and S3A, B). In anaphase and telophase, a residual weak staining on microtubules was found in addition to the diffuse cytoplasmic signal (Figure S3A and C). The direct interaction of UBASH3B with microtubules was confirmed using microtubule-pelleting assays of mitotic extracts and full-length recombinant UBASH3B protein in vitro (Figure S3D and E). Taken together, our data identify UBASH3B as a spindle-associated mitotic factor important for chromosome segregation.
UBASH3B interacts with Aurora B-CUL3 complex in ubiquitin-dependent manner
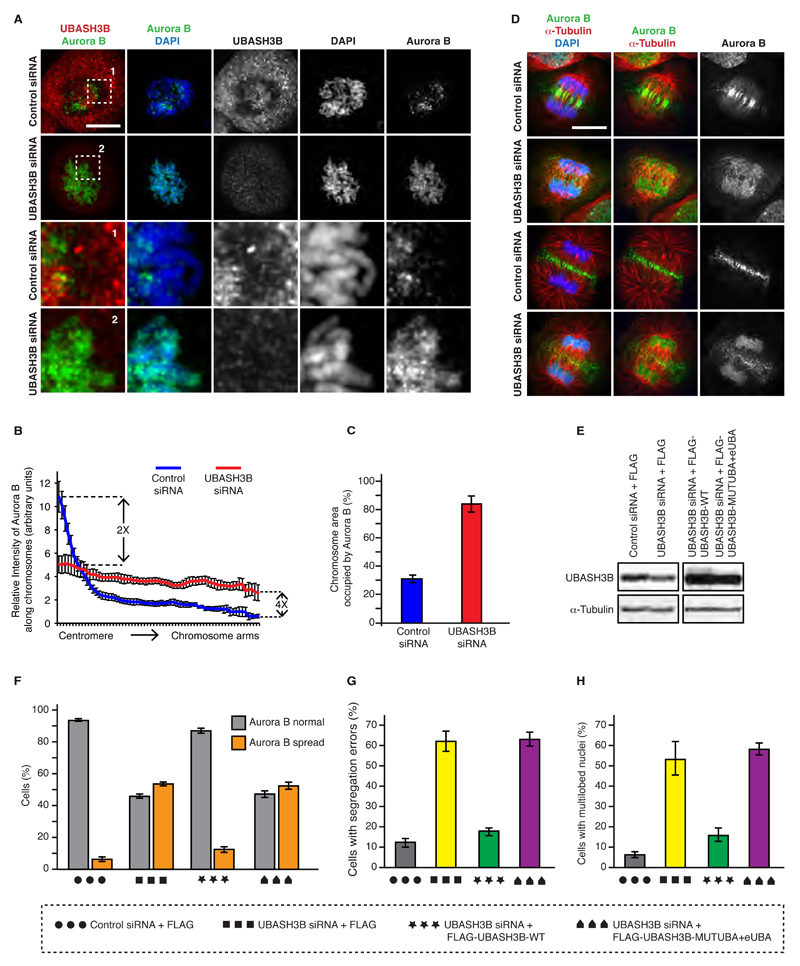
Since UBASH3B interacted with CUL3 in mitotic human cells (Table S3), we hypothesized that UBASH3B acts in the Aurora B-CUL3 pathway, possibly as a factor tethering Aurora B to microtubules (Maerki et al., 2010). If this were the case, UBASH3B should interact with Aurora B and CUL3 and regulate localization of Aurora B in mitosis. First, we tested the interactions of UBASH3B with the Aurora B-CUL3 pathway. Similar to CUL3, UBASH3B did not regulate protein levels of Aurora B or any other components of the CPC (Figure S3F) and strongly interacted with immunoprecipitated GFP-tagged Aurora B (Figure 2A). Importantly, interaction of UBASH3B with Aurora B was dependent on the presence of CUL3 protein (Figure 2B), suggesting that UBASH3B may directly regulate CUL3-modified Aurora B. Indeed, endogenous UBASH3B interacted with a slower migrating form of Aurora B detected at around 45 kDa and to a lesser extent with the 38 kDa unmodified form of Aurora B (Figure 2C). To corroborate these findings, we performed a pull-down assay using a short recombinant fragment of UBASH3B corresponding to the ubiquitin-binding domain (UBA) (Figures S3G, H and 2D), which was shown to bind monoubiquitylated proteins in vitro (Hoeller et al., 2006). Isolation of UBA-interacting proteins from the cells arrested in mitosis revealed ubiquitylated proteins, suggesting that UBASH3B can act as an intracellular ubiquitin receptor in mitosis. Interestingly, only modified, presumably mono- and to a lesser extent di-ubiquitylated form of the endogenous (Figure 2D) and the GFP-tagged form of Aurora B were found interacting with the UBA domain (Figures 2E and S3I). The UBA domain also interacted with the NEDD8-modified, active form of CUL3 but not with the other components of the CPC complex or a non-proteolytic CUL3 substrate, PLK1 kinase (Beck et al., 2013) (Figure 2E), suggesting a specific regulation of the Aurora B-CUL3 pathway by UBASH3B.
Figure 2. UBASH3B interacts directly with Aurora B-CUL3 complex in ubiquitin-dependent manner.
(A) HeLa cells expressing GFP alone (3XGFP-NLS) or GFP-Aurora B were arrested in mitosis using Taxol, immunoprecipitated using GFP-Trap beads (GFP-IP) and analyzed by Western blotting. (B) HeLa cells expressing GFP-Aurora B were treated with control (-) or CUL3 siRNAs (+), synchronized and analyzed as in (A). (C) MDA-MB-231 cells were arrested in mitosis by STLC, immunoprecipitated using UBASH3B antibody or IgG and analyzed by Western blotting. The modified and unmodified Aurora B forms are shown (SE: short exposure, LE: long exposure). The asterisk (*) indicates a non-specific signal and IgG HC points to the heavy chain of IgG. (D) Recombinant GST or GST-UBA domain of UBASH3B were incubated with extracts of mitotically synchronized HeLa cells, immunoprecipitated using glutathione-sepharose beads (GST-IP) and analyzed by Western blotting. Arrows indicate the unmodified and ubiquitin-modified Aurora B. (E) Recombinant wild-type (GST-UBA-WT), mutated form with M47A (GST-UBA-Mut), and with W72A/H76A/D79A within extended UBA (GST-eUBA-Mut) were incubated with extracts of mitotically synchronized HeLa cells expressing GFP-Aurora B and analysed as in (D) and by Coomassie blue staining and Western blotting. Arrows indicate unmodified and ubiquitin-modified GFP-Aurora B.
To confirm ubiquitin dependent function of UBASH3B, we isolated UBA-interacting proteins and performed in vitro deubiquitylation using a recombinant deubiquitinating enzyme (DUB) USP2 (Kim et al., 2011). Deubiquitylation strongly reduced the interaction of the UBA domain with ubiquitylated proteins, NEDD8-modified CUL3 and modified Aurora B (Figure S3J). Moreover, analysis of the UBASH3B sequence identified two putative ubiquitin-binding motifs. The first one corresponds to the MGF sequence (aa 47-49) within the UBA domain, previously implicated in ubiquitin binding in vitro (Hoeller et al., 2006) and containing highly conserved methionine residue (aa 47) (Figure S3K). The canonical UBA domain of UBASH3B is followed by a short C-terminal helical extension, similar to several UBA domain-containing proteins including USP5/13, UBAC2, UBXN1 (Figure S3K), and will be hereafter referred to as extended UBA (eUBA). Mutagenesis of M47 to A or all three conserved residues within the eUBA (WHD aa 72, 76, 79) to alanines dramatically reduced interaction with ubiquitylated proteins in mitotic cells (Figure 2E), suggesting that UBASH3B contacts ubiquitin molecule at two distinct sites. Importantly, both mutants lost their capacity to interact with GFP-Aurora B and CUL3 (Figure 2E). We thus conclude that UBASH3B directly interacts with Aurora B-CUL3 complex in ubiquitin-dependent manner.
UBASH3B is required for microtubule localization of Aurora B
Since localization of Aurora B is essential for its function, and Aurora B levels were not regulated by UBASH3B (Figure S3F), we next analyzed localization of Aurora B in the presence and absence of UBASH3B. Downregulation of UBASH3B by siRNA led to the redistribution of the centromeric Aurora B to the chromosome arms in prometaphase cells (Figure 3A, B) and the total chromosome area occupancy by Aurora B was increased (Figure 3C). The same localization defects of Aurora B were also observed in UBASH3B-depleted cells arrested in prometaphase by the Eg5 inhibitor STLC (Figures 3F and S4A, E). In addition, downregulation of UBASH3B in prometaphase-arrested cells also led to spreading of other components of the CPC including Survivin and INCENP to chromosomal arms (Figure S4B, C and D), consistent with their mutual co-regulation with Aurora B (Klein et al., 2006). These data suggest that UBASH3B regulates Aurora B localization in early mitotic stages. How can spindle associated UBASH3B control the chromosomal distribution of Aurora B? Interestingly, spindle microtubules had been shown to regulate centromeric focusing of Aurora B (Banerjee et al., 2014), which also requires the Aurora B-dependent positive feedback loops (van der Horst and Lens, 2014). Thus, UBASH3B may provide a molecular link between the centromeric and the microtubule-associated fractions of Aurora B. To test this assumption, we analyzed Aurora B localization in the later mitotic stages. Strikingly, downregulation of UBASH3B reduced localization of Aurora B to the midzone microtubules; the bulk of Aurora B remained associated with the arms of segregating chromosomes throughout anaphase (Figure 3D). Next, we tested if the Aurora B localization defects were specific to downregulation of UBASH3B. Importantly, re-expression of FLAG-tagged UBASH3B at nearly endogenous levels reversed the observed localization defects in prometaphase (Figures 3E, F and S4E). The ubiquitin binding of UBASH3B plays a crucial role in this process as the form of UBASH3B mutated in UBA and eUBA domains did not rescue the localization defects of Aurora B (Figures 3F and S4E). The delay from prophase to anaphase in UBASH3B downregulated cells was also reversed by re-expression of the wild-type form in UBASH3B-depleted cells (Figure S4F). The lagging anaphase chromosomes observed in UBASH3B siRNA cells were markedly decreased in cells expressing wild type but not the mutant UBASH3B (Figure 3G). Likewise, the multilobed nuclei phenotype observed upon downregulation of UBASH3B was dependent on ubiquitin binding by UBASH3B (Figure 3H). Collectively, UBASH3B specifically regulates localization of Aurora B and mitotic progression in ubiquitin-depentent manner. Interestingly, the Aurora B localization defects are very similar to those observed upon inactivation of CUL3 E3 ubiquitin ligases (Maerki et al., 2009; Sumara et al., 2007).
Figure 3. UBASH3B is required for Aurora B localization in mitosis.
(A) HeLa cells were treated with control and UBASH3B siRNAs, synchronized by double thymidine block and release in prometaphase and analyzed by immunofluorescence microscopy (IF). The magnified framed regions are shown in the corresponding panels below. Hereafter, scale bar is 5 μm. (B) Relative intensity ratios of Aurora B: DAPI along the individual chromosomes from centromeres to chromosome arms in cells shown in (A), n=220. (C) Chromosome area occupied by Aurora B in prometaphase cells shown in (A). (D) HeLa cells treated as in (A) were synchronized by double thymidine block and release in anaphase and analyzed by IF. Early (upper two rows) and late (lower two rows) anaphase cells are depicted. (E) HeLa cells treated with control and UBASH3B siRNAs, were synchronized by STLC treatment in prometaphase and analyzed by Western blotting. The siRNA-treated cells were simultaneously transfected with cDNAs encoding for FLAG, FLAG-WT-UBASH3B and the mutated form of UBASH3B with the UBA and eUBA domain (MUTUBA+eUBA). (F) Percentage of mitotic cells, shown in (E) (n= 1971 total) with Aurora B spread along the entire chromosomes (Aurora B spread, orange bars) or Aurora B concentrated in the centromeric regions (Aurora B normal, grey bars). (G) HeLa cells expressing H2B-mCherry and EGFP-Tubulin treated as in (E and F) were synchronized by double thymidine block and release and analyzed by live-cell microscopy. Percentage of anaphase cells with segregation errors was quantified, n=243. (H) HeLa cells treated as in (E and F) were analyzed by IF and the percentage of cells with multilobed nuclei was quantified (n=2000). All the data are shown as the mean of three independent experiments ± standard error of the mean (S.E.M.).
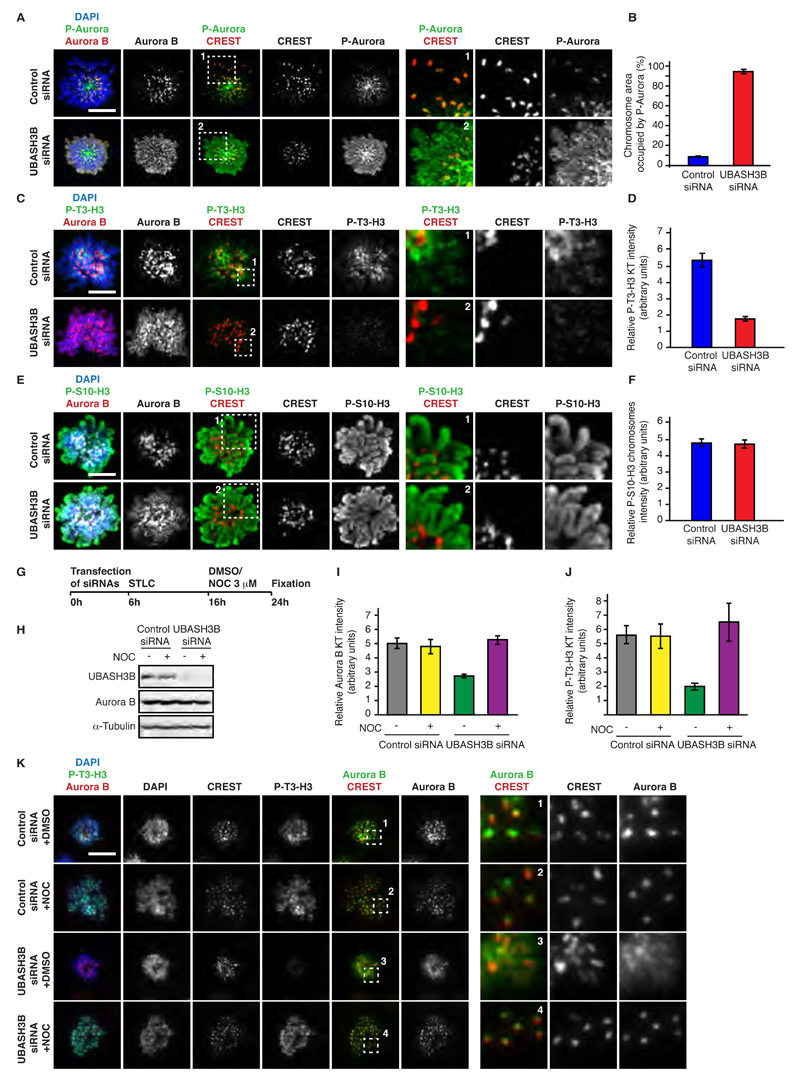
Next, we dissected the roles of the chromatin and microtubule pathways involved in the centromeric focusing of Aurora B. UBASH3B siRNA treatment led to the chromosomal spreading of active centromeric Aurora B kinase visualized by an antibody to the phosphorylated catalytic domain of Aurora kinases (P-Aurora) (Figure 4A and B), suggesting that Aurora B-mediated positive feedback loop signaling may also be dependent on UBASH3B. Specifically, Aurora B is known to phosphorylate Haspin kinase to promote generation of Threonine 3-phosphorylated Histone H3 (P-T3-H3) thereby enhancing its own recruitment to the centromeres (van der Horst and Lens, 2014). Indeed, depletion of UBASH3B markedly reduced the levels of P-T3-H3 around the centromeric regions (Figure 4C and D) but did not affect the Serine 10-phosphorylated Histone H3 found across all chromosomes (Figure 4E and F). These results suggest that deficiency of UBASH3B has a dramatic effect on localization of Aurora B by mistargeting the active kinase at the chromosome.
Figure 4. UBASH3B regulates Aurora B-depedent centromeric pathways on mitotic chromosomes in microtubule-dependent manner.
(A to F) HeLa cells were treated with control and UBASH3B siRNAs, synchronized in prometaphase using STLC and analyzed by immunofluorescence microscopy (IF). Hereafter, scale bar is 5 μm. (A, C and E) The framed regions are magnified and shown in the corresponding right panels. (B, D and F) Quantification of the relative intensity ratios of P-Aurora B: DAPI on the entire chromosomal area (B, n=270), Phospho-Threonine 3 histone H3 (P-T3-H3) : CREST (D, n=210) and Phospho-Serine 10 histone H3 (P-S10-H3) : DAPI on the entire chromosomal area (F, n=200). Blue bars represent control siRNAs, and red bars - UBASH3B siRNAs, respectively. (G to K) HeLa cells were treated with control and UBASH3B siRNAs, synchronized in prometaphase using STLC, treated with nocodazole or DMSO and analyzed by IF (I to K) and Western blotting (H). The scheme of the experiment is shown in (G). The quantifications of relative Aurora B and P-T3-H3 intensities on kinetochores are shown in (I) and (J), respectively. The framed regions in (K) are magnified and shown in the corresponding right panels. All the data are shown as the mean of three independent experiments ± standard error of the mean (S.E.M.).
We next tested if the spindle-associated UBASH3B is required for the chromosomal localization of Aurora B. Strikingly, depolymerization of microtubules with micromolar doses of nocodazole abolished the chromosomal localization defects of Aurora B observed upon downregulation of UBASH3B and the centromere levels of Aurora B and P-T3-H3 were reduced only in the presence of intact microtubules (Figure 4G-K). Thus, the microtubule-associated UBASH3B contributes to the histone marks pathways and provides a molecular link between the centromeric and the microtubule-associated fractions of Aurora B.
UBASH3B is a limiting factor sufficient for Aurora B localization to microtubules
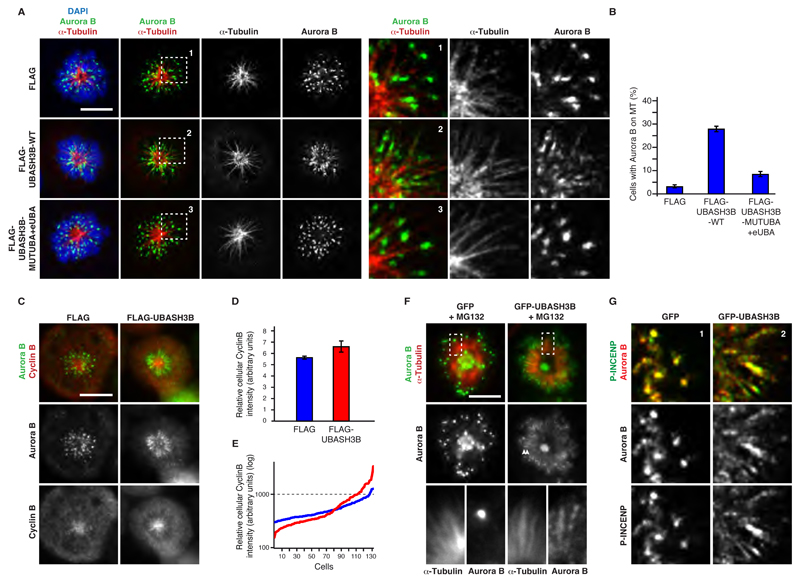
Monoubiquitylation can serve as a signal mediating reversible recruitment of the proteins to specific compartments (Komander and Rape, 2012). In agreement with previous findings (Murata-Hori et al., 2002), regulation of Aurora B localization by ubiquitin is likely to be dynamic and to contribute to Aurora B function in faithful chromosome segregation. If this were the case, UBASH3B may regulate the balance of centromeric and spindle-associated Aurora B by actively recruiting Aurora B to microtubules. To test this assumption, we overexpressed UBASH3B in cells arrested in prometaphase by STLC (Figure 5A). In control cells, the majority of Aurora B localized to the centromeric regions of circularly arranged chromosomes. Strikingly, overexpression of the FLAG-WT-UBASH3B was sufficient to drive association of endogenous Aurora B with microtubules, and both FLAG-UBASH3B and Aurora B were localized to the monopolar spindles in these cells (Figures 5A, B and S5A). In contrast, FLAG-UBASH3B mutated in both UBA and eUBA domains failed to trigger Aurora B relocalization to microtubules despite its efficient localization to microtubules (Figures 5A, B and S5A). The GFP-tagged form of Aurora B was also actively recruited to the spindle by UBASH3B and it could be visualized within the microtubule bundles (Figure S5B) in prometaphase-arrested cells. Furthermore, overexpression of UBASH3B prometaphase arrested cells increased association of Aurora B with stable microtubules (Figure S5C). Since a drop in CDK1 activity at anaphase onset was reported to contribute to Aurora B microtubule association (Hummer and Mayer, 2009), we analyzed the levels of the activatory subunit of CDK1 kinase, Cyclin B. Importantly, the average levels of Cyclin B were not dramatically influenced by overexpression of FLAG-UBASH3B (Figures 5C and D), and both low and high Cyclin B levels were observed in prometaphase cells with microtubule-associated Aurora B (Figure 5E). This result suggests that overexpression of UBASH3B may target Aurora B to microtubules also in the presence of high CDK1 activity. To corroborate these findings, we analyzed localization of Aurora B in prometaphase-arrested cells, in which degradation of Cyclin B was inhibited by the proteasome inhibitor MG132. Indeed, overexpression of GFP-UBASH3B caused relocalization of Aurora B to microtubules (Figures 5F and S5D). Next, we tested if other CPC components can be recruited to microtubules by UBASH3B. Indeed, Aurora B-phosphorylated INCENP (Serine 893 and 894) (Salimian et al., 2011) colocalized with the microtubule-associated Aurora B upon UBASH3B overexpression in prometaphase in contrast to control cells in which Aurora B and INCENP were found at the centromere (Figures 5G and S5E). Overall, our findings suggest that overexpression of UBASH3B is sufficient to localize Aurora B to microtubules in ubiquitin dependent manner prior to the anaphase onset.
Figure 5. Overexpression of UBASH3B is sufficient to localize Aurora B to microtubules.
(A and B) HeLa cells were transfected with FLAG, FLAG-WT-UBASH3B or FLAG-MUTUBA+eUBA cDNAs, synchronized in prometaphase using STLC and analyzed by immunofluorescence microscopy (IF). FLAG and DAPI signals are shown in Figure S5A. The magnified framed regions are depicted in the corresponding right panels. Hereafter, scale bar is 5 μm. (B) The percentage of cells from (A) with Aurora B localized to the monopolar spindles microtubules, n=2431. (C) HeLa cells were transfected with FLAG or FLAG-WT-UBASH3B and analyzed by IF. (D) Relative intensity of Cyclin B in cells depicted in (C), transfected with FLAG (blue bars) and FLAG-UBASH3B (red bars) n=131. The data are shown as the mean of three independent experiments ± standard error of the mean (S.E.M.). (E) Distribution of relative intensities of Cyclin B in individual cells quantified in (D). FLAG (blue lines) and FLAG-UBASH3B-expressing cells (red lines). (F) HeLa cells transfected with GFP or GFP-UBASH3B cDNAs were synchronized in prometaphase using STLC in the presence of MG132 and analyzed by IF. The magnified framed regions are shown in the corresponding right panels. GFP-UBASH3B and DAPI signals are shown in Figure S5D. (G) HeLa cells transfected with GFP or GFP-UBASH3B cDNAs were synchronized in prometaphase using STLC and analyzed by IF. The magnified framed regions of the Figure S5E are shown.
The localization of Aurora B to microtubules is established upon chromosome alignment
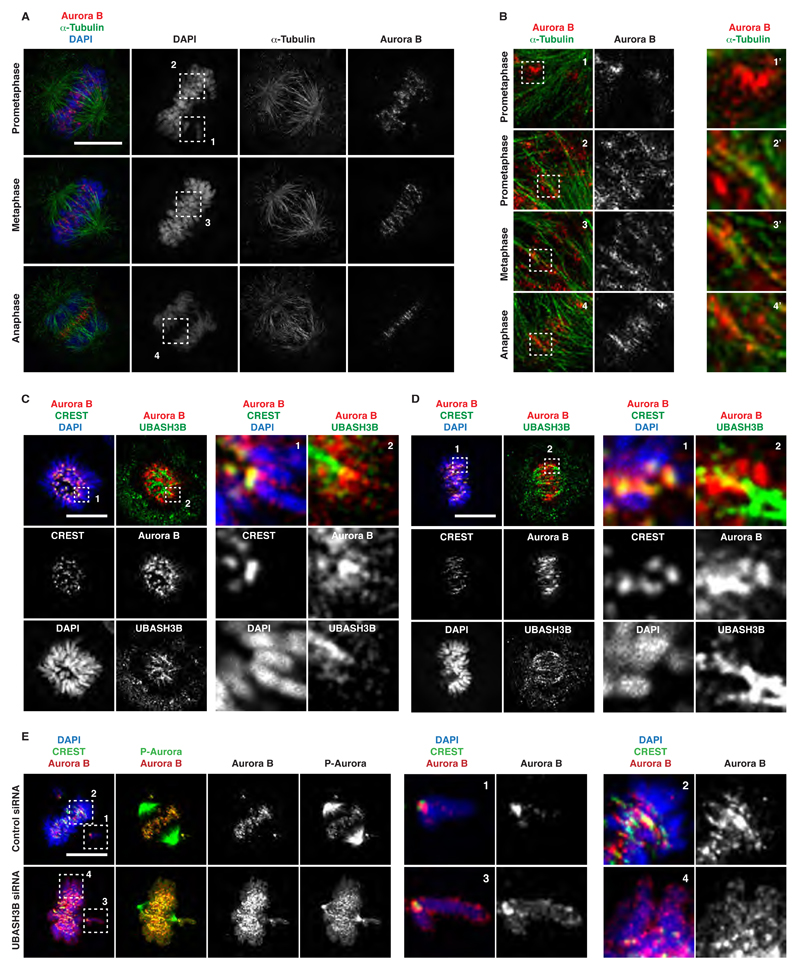
Our findings that UBASH3B overexpression targets Aurora B to microtubules prior to anaphase prompted us to investigate mitotic localization patterns of Aurora B in a greater detail. Importantly, previous studies reported localization of Aurora B on pre-anaphase spindles (Rosasco-Nitcher et al., 2008; Tseng et al., 2010). Using 3D SIM super-resolution microscopy of cells in late prometaphase, we found that a significant fraction of Aurora B co-localized with microtubules in a stripe-like pattern in the vicinity of aligned chromosomes, distinct from the typical dot-like centromeric signals of Aurora B on unaligned chromosomes (Figure 6A and B). Interestingly, Aurora B co-localized with the kinetochore microtubule marker HURP in the vicinity of the aligned chromosomes in late prometaphase stages (Figure S5F), while upon full chromosome alignment in metaphase Aurora B co-localized with the non-kinetochore microtubule protein PRC1 between stretched kinetochores (Figure S5G). This indicates that Aurora B localizes to microtubules upon establishment of stable KT-MT interactions.
Figure 6. UBASH3B mediates the microtubule localization of Aurora B upon chromosome alignment.
(A and B) HeLa cells were synchronized in different mitotic stages (prometaphase, metaphase, anaphase) by a double thymidine block and release and analysed by super-resolution microscopy. The framed and numbered regions in (A) are magnified and depicted in the corresponding panels in (B). These regions are further magnified in the corresponding right panels (indicated by the corresponding numbers with ’). Hereafter, scale bar is 5 μm. (C and D) MDA-MB-231 cells were synchronized by a double thymidine block and release in different mitotic stages (early in C and late prometaphase in D) and analysed by immunofluorescence microscopy (IF). The magnified framed and numbered regions are shown in the corresponding right panels. (E) MDA-MB-231 cells were treated with control and UBASH3B siRNAs, synchronized as in (C, D) and analysed by IF. The magnified framed and numbered regions are shown in the corresponding right panels.
To understand precisely how, and when, UBASH3B mediates the microtubule relocalization of Aurora B, we analyzed its distribution during unperturbed mitosis. UBASH3B was found on mitotic spindle and partially co-localized with Aurora B near the centromeric regions during prometaphase (Figure 6C). The co-localization of UBASH3B and Aurora B increased as cells achieved bi-orientation (Figure 6D). Consistent with the super-resolution microscopy analysis, Aurora B and the active P-Aurora displayed two distinct patterns during late prometaphase: dot-like and centromeric on unaligned chromosomes, and a stripe-like on microtubules in the vicinity of bi-oriented chromosomes (Figure 6E). Downregulation of UBASH3B abolished both localization patterns, and the spreading of Aurora B to chromosomal arms was observed under these conditions (Figure 6E). Based on these observations, we hypothesize that UBASH3B provides a spatial signal that progressively drives Aurora B localization to microtubules as chromosomes achieve bi-orientation and thus may regulate the mitotic function of Aurora B.
UBASH3B regulates the mitotic function of Aurora B
Our results show that downregulation of UBASH3B inhibits centromeric focusing of Aurora B in prometaphase and its relocalization to microtubules in metaphase leading to persistent low centromeric levels of this kinase during mitotic progression (Figure S6A). Therefore, UBASH3B may also control the function of Aurora B in correcting erroneous KT-MT attachments possibly by affecting the critical Aurora B substrates at the mitotic centromere (Nezi and Musacchio, 2009). To test this, we analyzed kinetochore levels of the microtubule kinetochore protein Astrin (Dunsch et al., 2011). Compared to control cells we observed a moderate increase of Astrin levels at kinetochores during prometaphase and a significant decrease during metaphase in UBASH3B depleted cells (Figure S6B and C). These results suggest that downregulation of UBASH3B leads to a premature stabilization of some erroneous KT-MT attachments in prometaphase and conversely to destabilization of KT-MT attachments in metaphase, which could explain the delay in the metaphase to anaphase onset (Figures 1D, F and S2A, B). Next, we tested if UBASH3B regulates the error correction function of Aurora B. Sixty minutes after release from monastrol-induced monopolar prometaphase, we observed a higher number of cells in prometaphase, a lower number of cells in metaphase and telophase and no difference in anaphase upon downregulation of UBASH3B as compared to the control siRNA-treated cells (Figure S6D). Specifically, downregulation of UBASH3B led to a lower number of monopolar prometaphases, a higher number of bipolar prometaphases and a lower number of metaphases (Figure S6D), confirming that UBASH3B does not regulate bipolar spindle formation (Figure S2C-D) but may control the correction of KT-MT attachments during mitotic progression. Moreover, cold treatment of metaphase cells showed a decrease in the formation of stable kinetochore bundles upon UBASH3B depletion (Figure S6E) and a profound reduction of the kinetochore levels of Astrin (Figure S6F and G). Collectively, these results suggest that UBASH3B also regulates the function of Aurora B at the centromere. The defects in kinetochore attachment may prevent SAC satisfaction which explains the delay in anaphase onset (Figure 1D and F) and death in prometaphase (Figures 1B, C and S2A) observed upon downregulation of UBASH3B.
UBASH3B forms a functional complex with MKlp2 to target Aurora B to microtubules prior to anaphase
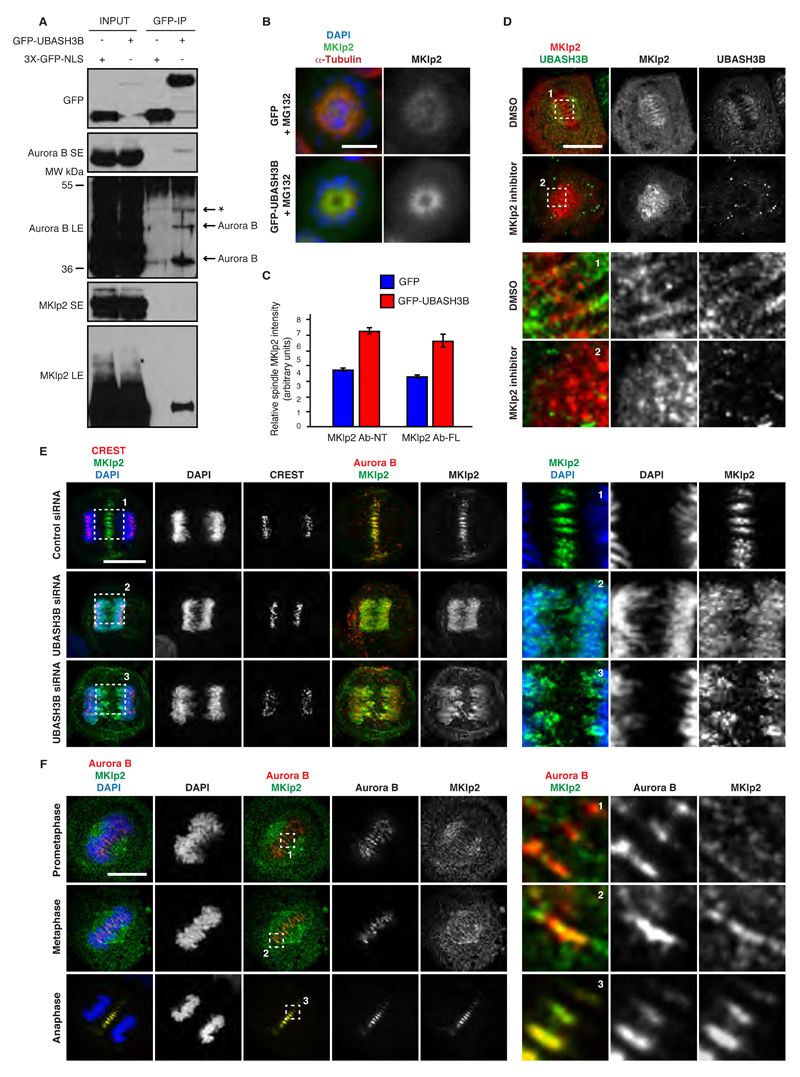
Next, we addressed the precise molecular mechanisms underlying UBASH3B-mediated microtubule targeting of Aurora B. Since the mitotic kinesin-like protein 2 (MKlp2) kinesin was demonstrated to mediate the midzone localization of Aurora B (Gruneberg et al., 2004), we hypothesized that it may act in a complex with UBASH3B prior to anaphase to shift the balance of Aurora B towards microtubules. Moreover, MKlp2 was previously found on the spindle microtubules in pre-anaphase cells (Kitagawa et al., 2014) and UBASH3B localized preferentially to the prometa- and metaphase spindles (Figure S3A and B). To test whether UBASH3B interacts with MKlp2, we immunoprecipitated GFP-tagged UBASH3B or GFP alone from the extracts of cells arrested in mitosis by Taxol. Endogenous MKlp2 visibly interacted with GFP-UBASH3B but not with GFP control (Figure 7A), suggesting that a functional complex of Aurora B, MKlp2 and UBASH3B may exist in pre-anaphase cells. Consistent with the previous findings (Figure 2C, D), we found a slower migrating, modified form of Aurora B to interact with UBASH3B. In contrast, UBASH3B bound to a presumably unmodified form of MKlp2 protein, suggesting that the UBASH3B-MKlp2 interaction is not dependent on ubiquitin recognition. To further corroborate this finding, we overexpressed GFP-tagged UBASH3B in cells arrested in prometaphase by STLC in the presence of MG132 and analyzed localization of MKlp2. Overexpression of UBASH3B was sufficient to target MKlp2 to monopolar spindles (Figures 7B and S7A). The spindles contained almost twice of the amount of MKlp2 protein as compared to the GFP-control expressing cells (Figure 7C). Moreover, inhibition of MKlp2 motor activity using paprotrain (Liu et al., 2013) led to dissociation of MKlp2 from the mitotic spindle and accumulation of UBASH3B around the centrosomes, in contrast to the control cells, which displayed uniform distribution of UBASH3B along the entire mitotic spindles (Figure S7B). In many cells, paprotrain treatment abolished microtubule localization of UBASH3B (Figure 7D), suggesting that MKlp2 and UBASH3B are mutually co-dependent for their spindle function. Accordingly, inactivation of UBASH3B by siRNA prevented microtubule and midbody association of MKlp2 in metaphase and telophase cells (Figure S7C, D). The same defects were observed in UBASH3B-depleted cells in anaphase, where MKlp2 was found on, or in the vicinity of, chromosomes instead of microtubules, similar to the Aurora B signals (Figure 7E). Our findings suggest that UBASH3B targets Aurora B to microtubules by forming a complex with MKlp2 prior to anaphase. Indeed, MKlp2 co-localization with Aurora B on the spindle microtubules was already observed in late prometaphase, and increased progressively as cells aligned their chromosomes in metaphase (Figure 7F). In accordance with these findings, downregulation of MKlp2 by siRNA inhibited centromeric focusing of Aurora B also in early mitosis (Figure S7E and F). Thus, UBASH3B cooperates with MKlp2 to regulate mitotic localization of Aurora B.
Figure 7. UBASH3B forms a functional complex with MKlp2 to target Aurora B to microtubules prior to anaphase.
(A) HeLa cells expressing GFP alone (3XGFP-NLS) or GFP-UBASH3B were arrested in prometaphase using STLC, immunoprecipitated with the GFP-Trap beads (GFP-IP) and analyzed by Western blotting. Short (SE) and long (LE) exposures of the blots are shown. The modified and unmodified Aurora B forms are indicated. The asterisk (*) indicates a non-specific signal. (B) HeLa cells transfected with GFP or GFP-UBASH3B cDNAs were synchronized in prometaphase using STLC in the presence of MG132 and analyzed by immunofluorescence microscopy (IF). UBASH3B and DAPI channels are shown in Figure S7A. Hereafter, scale bar is 5 μm. (C) Relative intensity ratios of MKlp2: α-Tubulin on mitotic spindles in cells from the experiment depicted in (B) transfected with GFP (blue bars) and GFP-UBASH3B (red bars), n=277. The antibodies against the N-terminal part (MKlp2 Ab-NT) and the full-length (MKlp2 Ab-FL) MKlp2 protein were used for IF analysis. The data are shown as the mean of three independent experiments ± standard error of the mean (S.E.M.). (D) HeLa cells were synchronized in mitosis by a double thymidine block and release, treated with the MKlp2 inhibitor paprotrain or solvent control (DMSO) and analysed by IF. Corresponding DAPI signals and other representative cells are shown in Figure S7B. The magnified framed and numbered regions are shown in the corresponding lower panels. (E) HeLa cells were treated with indicated siRNAs, synchronized in anaphase by a double thymidine block and release and analysed by IF. The magnified framed regions are shown in the corresponding right panels. (F) MDA-MB-231 cells were synchronized in different mitotic stages (prometaphase, metaphase, anaphase) by a double thymidine block and release and analysed by IF. The magnified framed regions are shown in the corresponding right panels.
Targeting of Aurora B to microtubules by UBASH3B triggers anaphase
Aurora B mediates a correction mechanism that destabilizes erroneous kinetochore attachments (Nezi and Musacchio, 2009) and thereby prevents SAC satisfaction. Since relocalization of Aurora B in anaphase was shown to prevent engagement of the SAC (Vázquez-Novelle and Petronczki, 2010) and Aurora B kinase is directly involved in maintaining checkpoint arrest independently of its upstream functions in error correction (Maldonado and Kapoor, 2011), we aimed at understanding the role of UBASH3B-mediated targeting of Aurora B to microtubules in the regulation of anaphase. For this purpose, we overexpressed UBASH3B in prometaphase-arrested cells and analyzed the protein levels and localization of the critical SAC component BubR1 to kinetochores. In contrast to the control-transfected cells, the levels of the kinetochore associated BubR1 were reduced in UBASH3B overexpressing cells (Figure S7G and H), suggesting a role of UBASH3B in Aurora B-dependent SAC silencing. Importantly, UBASH3B downregulation did not change the abundance of BubR1 or another SAC protein Mad2 (Figure S7I). To corroborate these findings, we analyzed the protein levels of Securin (Figure S7J), the target of the Anaphase Promoting Complex/Cyclosome APC/C, which is controlled by SAC (Musacchio and Salmon, 2007). Indeed, levels of Securin, but not of Aurora B, were strongly reduced in UBASH3B-overexpressing prometaphase cells. These observations are consistent with the reduced levels of Cyclin B, another target of APC/C, found in approximately 50% of UBASH3B overexpressing cells (Figure 5E). Accordingly, overexpression of GFP-tagged UBASH3B induced premature and aberrant chromosome partitioning in prometaphase-arrested cells leading to a decrease of mitotic cells and a marked increase of cells with multilobed nuclei (Figure S7K). Overexpression of UBASH3B in cells, which were not synchronized in mitosis by drugs, also induced multilobed nuclei (Figures S7L and M). These results strongly suggest that UBASH3B controls ploidy of cells by regulating microtubule localization of Aurora B and thereby its essential functions in SAC and chromosome segregation.
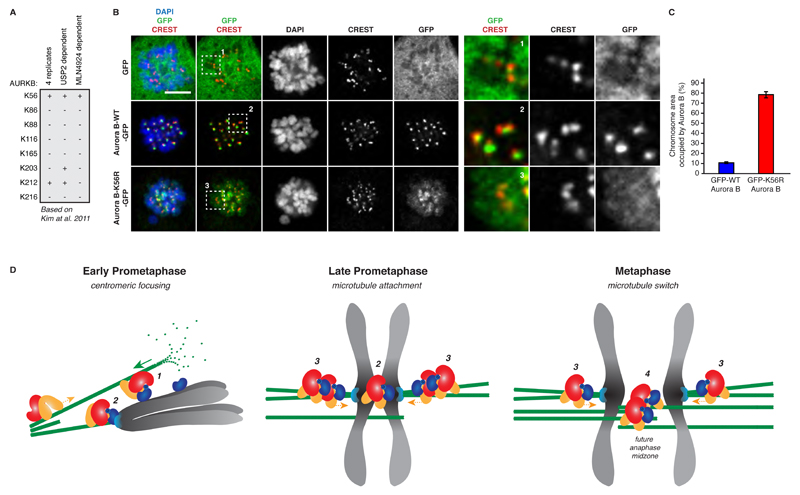
Taken together, UBASH3B is a limiting factor mediating Aurora B localization to microtubules and timely onset of chromosome segregation. Our data suggest that Aurora B microtubule targeting is mediated specifically by UBASH3B in ubiquitin-binding dependent manner (Figures 2, 3 and 5). To understand if Aurora B is a critical target of UBASH3B in mitosis, we sought to identify the ubiquitin acceptor site on Aurora B protein. Out of eight different ubiquitin-modified lysine residues found in Aurora B in human cells (Kim et al., 2011), three were shown to be sensitive to USP2 DUB treatment, and modification of a single lysine at position 56 (K56) was abolished upon treatment with the NEDD8 E1 inhibitor, which blocks activation of Cullin-based E3-ligases, including CUL3 (Kim et al., 2011) (Figure 8A). Strikingly, mutagenesis of K56 to the arginine (K56R) of the GFP form of Aurora B led to strong localization defects in prometaphase cells. Unlike WT-GFP-Aurora B, which was found at the centromeric regions, the K56R mutant spread along the entire length of the chromosomal arms being strikingly similar to Aurora B pattern observed upon downregulation of UBASH3B (Figures 8B and C). Therefore, lysine 56 is the ubiquitin acceptor site on Aurora B, which mediates the correct localization and function of this kinase.
Figure 8. The lysine 56 of Aurora B is important for its correct localization during mitosis.
(A) The list of ubiquitin-modified lysine residues (K) on Aurora B (AURKB) protein identified in human cells by (Kim et al., 2011). The sites found in four different replicate experiments and sites sensitive to the USP2 DUB or MLN4924 (NEDD8 inhibitor) treatment are indicated. (B) HeLa cells were transfected with the GFP, GFP-WT-Aurora B or the mutated form GFP-K56R-Aurora B cDNAs, synchronized in prometaphase using STLC and analysed by immunofluorescence microscopy (IF). The magnified framed regions are shown in the corresponding right panels. Scale bar, 5 μm. (C) Surface occupancy ratios of Aurora B: DAPI signals on the entire chromosomal area in prometaphase cells expressing GFP-WT-Aurora B (blue bars) and GFP-K56R-Aurora B (red bars) from experiments shown in (B), n=200. (D) A model of regulation of localization and function of Aurora B by the ubiquitin receptor UBASH3B during mitosis. In prometaphase, UBASH3B (red) interacts with the plus-end motor MKlp2 (orange) and both proteins control their microtubule targeting. The spindle microtubules (green) establish the contacts with chromosomes in a search and capture mechanism and interact with the chromosome-bound ubiquitylated Aurora B (dark blue) (1). The microtubules, which did not establish end-on contacts with the kinetochores (light blue) shrink and may contribute to the centromeric focusing of Aurora B (2). Upon formation of stable chromosome attachments in metaphase, more microtubules are bundled at the kinetochores increasing the local concentration of UBASH3B protein (3) and promoting microtubule localization of Aurora B. The plus-end motor activity of MKlp2 may participate in the formation of the anti-parallel microtubule bundles, which constitute the future anaphase midzone region (4). This mechanism may locally silence the SAC and contribute to the timing and fidelity of chromosome segregation.
Discussion
Collectively, our data suggest a model in which UBASH3B critically regulates localization and function of Aurora B and thereby fidelity of chromosome segregation (Figure 8D). By localizing to the mitotic spindles, UBASH3B mediates centromeric focusing and microtubule localization of Aurora B during mitotic progression. This role of UBASH3B depends on its ability to interact with ubiquitin, suggesting that UBASH3B acts as a non-proteolytic ubiquitin receptor in mitosis. Moreover, UBASH3B forms a complex with the plus end-directed motor protein MKlp2, and both factors co-regulate their microtubule targeting and function. Importantly, UBASH3B acts as a limiting factor for Aurora B localization, and its overexpression is sufficient to target Aurora B to microtubules as soon as chromosomes achieve bi-orientation and prior to the onset of anaphase. We propose that UBASH3B drives recruitment of ubiquitylated Aurora B to microtubules during mitosis (Figure 8D).
How do ubiquitin receptors regulate mitosis?
Directionality of mitotic progression is determined by ubiquitin-dependent degradation of numerous substrates (Min and Lindon, 2012). Moreover, the emerging non-proteolytic ubiquitin pathways were shown to control fidelity of mitosis (Beck et al., 2013; Maerki et al., 2009; Sumara et al., 2007). Ubiquitin receptors acting in the proteolytic pathways have been shown to transfer substrates for proteasomal or lysosomal degradation but UBDs can also decode non-proteolytic ubiquitin signals (Husnjak and Dikic, 2012). However, it remained unexplored how the fate of ubiquitylated mitotic substrates is determined, and very little was known about specific UBDs regulating mitosis in mammalian cells (Fournane et al., 2012). Our work presented here sheds some light on how the ubiquitin code (Komander and Rape, 2012) can be read out during mitosis, and provides an example of an intracellular ubiquitin receptor that controls a non-proteolytic ubiquitylation pathway. Our findings strongly suggest that UBASH3B controls mitotic localization of Aurora B kinase in a non-proteolytic manner (Figures 3-6, S3F). In particular, we found that UBASH3B is a limiting factor determining the outcome of CUL3-mediated ubiquitylation of Aurora B, as its overexpression is sufficient to target Aurora B to microtubules (Figure 5). Thus, our findings uncover an important mechanism how ubiquitin signals can be decoded within the cells and emphasize the critical role of the UBDs in determining the fate of ubiquitylation substrates. In future, it will be also important to investigate the precise roles of other mitotic UBD proteins identified in our study.
How does UBASH3B regulate chromosome segregation and genome integrity?
Our data suggest that UBASH3B is critically involved in the regulation of chromosome segregation. We propose that UBASH3B regulates mitosis by controlling mitotic localization of Aurora B. A reduction of the centromere-associated pool of Aurora B may compromise its function at the kinetochore and kinetochore microtubules (Funabiki and Wynne, 2013; Lampson and Cheeseman, 2011; Sarangapani and Asbury, 2014). Indeed, downregulation of UBASH3B leads to a premature stabilization of erroneous KT-MT attachments in prometaphase (Figure S6B and C). In UBASH3B-depleted cells in metaphase we observe destabilization of the KT-MT attachments (Figure S6B, C, E-G), which in turn prevents SAC satisfaction, thus inhibiting chromosome segregation (Figures 1C, D, F and S2A, B). Interestingly, we found that UBASH3B is localized to spindle microtubules during mitosis, starting in prometaphase and with the strongest signals observed upon chromosomes reaching the metaphase plates and possibly upon stabilization of the KT-MT attachments (Figure S3A and B). Since microtubule tips of the mitotic spindle were shown to control centromeric Aurora B (Banerjee et al., 2014; Rosasco-Nitcher et al., 2008), we speculate that UBASH3B regulates centromeric localization of Aurora B by its association with the spindle microtubules and may act at the transition from the lateral to the end-on KT-MT attachments (Figure 8D). The centromeric focusing of Aurora B is also controled by histone phosphorylation events enhanced by Aurora B-driven positive feedback loops (van der Horst and Lens, 2014). Indeed, we observe reduced levels of phosphorylation of threonine 3 on histone 3 in UBASH3B depleted cells (Figures 4C, D, I-K), suggesting that UBASH3B controls the Aurora B feedback loop at the centromeres. Interestingly, the centromeric and spindle pathways were recently shown to compete for the binding of the CPC (van der Horst et al., 2015). Importantly, UBASH3B-mediated localization of Aurora B depends on the presence of microtubules (Figure 4G-K). Intriguingly, a fraction of Aurora B was previously shown to localize to the mitotic spindles (Rosasco-Nitcher et al., 2008; Tseng et al., 2010), suggesting a dynamic feedback between the centromeric and the microtubule-associated fractions of Aurora B. Our data go in line with these observations and demonstrate that Aurora B localizes to microtubules already during late prometaphase and metaphase (Figure 7F and S5F, G) and that UBASH3B mediates targeting of Aurora B to microtubules attached to bi-oriented chromosomes (Figure 6C). Strikingly, overexpression of UBASH3B is sufficient to target Aurora B to microtubules even in cells arrested with monopolar spindles in prometaphase (Figure 5A, C and F). Our data can also explain how Aurora B can contribute to the assembly and stabilization of the mitotic spindle (Kelly et al., 2007; Sampath et al., 2004) and how the gradient of Aurora B activity is established before the anaphase onset (Fuller et al., 2008).
We propose a model whereby UBASH3B acts as a molecular link between the centromeric and the spindle-associated fractions of Aurora B in a rate-limiting manner and controls a mechanism promoting Aurora B association with microtubules prior to anaphase (Figure 8D). The more stable microtubule bundles are formed at the kinetochore increasing the local concentration of UBASH3B protein, the more Aurora B is extracted to microtubules. This mechanism could generate the ultrasensitive response during mitotic progression and contribute to the generation of bi-stability by the positive feedback loops (Ferrell and Ha, 2014). Indeed, downregulation of UBASH3B reduces association of Aurora B with the midzone microtubules during anaphase (Figure 3D), similar to depletion of MKlp2 (Gruneberg et al., 2004; Hummer and Mayer, 2009). Our data argue that UBASH3B forms a complex with MKlp2 targeting Aurora B to microtubules of bi-oriented chromosomes (Figure 7 and Figure S7C). Since MKlp2 protein was also previously found to be localized to metaphase spindles (Kitagawa et al., 2014), UBASH3B-mechasnim may mediate the early steps of the microtubule targeting of Aurora B while the subsequent drop in the CDK1 activity can ensure the irreversibility of this relocalization event. Our findings also suggest that relocalization of Aurora B from centromeres to microtubules allows for SAC satisfaction at the kinetochores (Figure S7G-M). This is consistent with the reported role of Aurora B in targeting the SAC proteins to kinetochores (Hauf et al., 2003) as well as with the direct role of Aurora B in SAC silencing (Maldonado and Kapoor, 2011). Therefore, both loss and gain of function of UBASH3B have effects on mitotic progression. Low levels of UBASH3B prevent SAC satisfaction, which leads to the mitotic death, while elevated levels of this protein trigger uncontrolled chromosome segregation. It is particularly intriguing that high levels of UBASH3B were observed in aggressive forms of breast and pancreatic cancers in humans and in mouse models, in which UBASH3B promoted metastasis (Lee et al., 2013). Future studies are needed to understand how the oncogenic potential of this non-proteolytic intracellular ubiquitin receptor is correlated with its role in chromosome segregation.
Experimental procedures
Microscopy, image analysis and statistics
A high content visual siRNA screen in HeLa cells was performed to identify novel UBD proteins (Table S1 and S2) that control euploidy of dividing human cells and coordinate chromosome segregation with cytokinesis. The results of the primary and secondary screens were analyzed by a multiparameter software, which was based on the Principle Component Analysis (PCA). UBASH3B function was assessed in cultured mitotic HeLa cells, plated on glass coverslips or cytospined onto glass slides. Direct and indirect immunofluorescence microscopy was used to identify localization of UBASH3B and mitotic markers with the help of Zeiss epifluorescence microscope or confocal microscope Leica/Andor/Yokogawa Spinning Disk. For live-cell microscopy HeLa cells, stably expressing indicated proteins tagged with GFP, mRFP or mCherry, were grown on LabTek II Chambered Slides (Thermo Scientific) or µ-Slide VI 0.4 (IBIDI). Live-cell microscopy was carried out using a 40X objective of confocal microscope Leica/Andor/Yokogawa Spinning Disk. Image analysis was performed using ImageJ or Metamorph software. Where indicated, analysis of the difference between two groups was performed with unpaired t test (with α≤ 0.05) using GraphPad Prism software (GraphPad Software Inc.). At least three independent experiments were performed for each comparison.
Pulldowns, immunoprecipitations and Western blotting
Preparation of HeLa cell extracts was described previously (Sumara et al., 2007). GFP-fused proteins (UBASH3B-GFP and Aurora B-GFP) were immunoprecipitated using GFP-Trap agarose beads (Chromotek). Beads were incubated with cell extracts for 2 h at 4 °C under constant rotation. Before elution, beads were washed 10 times with lysis buffer (20 mM Tris HCl pH 7.5, 100 mM NaCl, 0.5% NP40, 14 mM β-Glycerophosphate, 10% Glycerol, 1 mM NaF). GST-tagged recombinant proteins were immobilized on Glutathione Sepharose 4B, incubated with cell extracts for 2-3 h at 4°C under constant rotation in lysis buffer supplied with MG-132 (25 μM) and PR-619 (10 μM) and subsequently treated when indicated with 4 μg of Active human USP2 protein fragment (Abcam, ab125735) and USP2 Catalytic Domain (Boston Biochem, E-504) for 30 min each at 30°C in DUB buffer (150 mM NaCl, 25 mM Tris HCl pH 7.5, 10 mM DTT, 1 mM EDTA, Complete Protease Inhibitor Cocktail Tablets (Roche). Beads were washed 10 times with lysis buffer, boiled in Laemmli SDS sample buffer and subjected to SDS-PAGE.
For detection of Aurora B interaction with UBASH3B and CUL3, cells were synchronized by Taxol for 16 h. Expression of Aurora B-GFP was induced for 24 h. To detect the interaction of UBA domain of UBASH3B with Aurora B, cells were synchronized in prometaphase by treatment with monastrol or SLTC for 16-18h and subsequently treated with MG132 and PR-619 for 1 hour to enrich for ubiquitylated substrates. The ubiquitylated Aurora B was detected using rabbit polyclonal Aurora B (Abcam ab2254) or rabbit polyclonal Aurora B (Bethyl Laboratories, A300-431A, 1:500). SDS-PAGE was performed using either 8%, 9% or 10% polyacrylamide gels. Proteins were subsequently transferred from the gel to a PVDF membrane (Millipore) for immunoblotting. Membranes were blocked in 5% non-fat milk powder (BioRad) resuspended in TBS supplemented with 0.1% Tween 20 (TBS-T) for from 30 min to overnight, followed by incubation with antibodies. Membranes were developed with Luminata Forte (Millipore) or SuperSignal West Pico chemiluminescent substrate (Thermo Scientific).
Supplementary Material
The complete version of the experimental procedures can be found in the Supplemental online data, accompanying this manuscript.
Acknowledgments
We thank Jonathon Pines, Snezhana Oliferenko, Thimo Kurz, Alexander Goginashvili, Romeo Ricci, Grzegorz Sumara, Lionel Pintard, Michel Labouesse and Todd Stukenberg for helpful discussions on the manuscript. We are grateful to Daniel Gerlich, Hironori Funabiki, Michael A. Lampson and Ulrike Gruneberg for reagents. We thank Ivana Gasic, Patrick Meraldi, Alexander Goginashvili and Helena Magliarelli for help with experiments and Alexia Loynton-Ferrand and the IGBMC imaging facility for support on imaging. K.K. was supported by a PhD contract from Région Alsace and INSERM and a Labex fellowship from IGBMC and Conectus Alsace. N.P., W.R. and O.P. were supported by ANR-12-RPIB-0012-03 and ANR-10-BINF-03-02. Research in the J.R.S. lab and imaging facilities at Dundee are supported by the Wellcome Trust (095931/Z/11/Z and 097945/B/11/Z). This study was sup- ported by the grant ANR-10-LABX-0030-INRT, a French State fund managed by the Agence Nationale de la Recherche under the frame program Investisse- ments d’Avenir ANR-10-IDEX-0002-02. Research in I.S. laboratory is supported by the IGBMC, CNRS, ATIP-AVENIR program (R10076MS), Fondation ARC pour la recherche sur le cancer, La Ligue Contre le Cancer, Sanofi-Aventis and University of Strasbourg Institute of Advanced Studies (USIAS).
Footnotes
Author contribution
K.K. and I.S. designed experiments and wrote the manuscript. K.K., C.K., T.M., S.F., S.S., K.H., L.B. N.P. W.R., O.P. and I.M.P. conducted the experiments.
References
- Banerjee B, Kestner CA, Stukenberg PT. EB1 enables spindle microtubules to regulate centromeric recruitment of Aurora B. J Cell Biol. 2014;204 doi: 10.1083/jcb.201307119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassermann F, Eichner R, Pagano M. The ubiquitin proteasome system - implications for cell cycle control and the targeted treatment of cancer. Biochim Biophys Acta. 2014;1843:150–162. doi: 10.1016/j.bbamcr.2013.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck J, Maerki S, Posch M, Metzger T, Persaud A, Scheel H, Hofmann K, Rotin D, Pedrioli P, Swedlow JR, et al. Ubiquitylation-dependent localization of PLK1 in mitosis. Nat Cell Biol. 2013;15:430–439. doi: 10.1038/ncb2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M, Wheelock M, Funabiki H, Earnshaw WC. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol. 2012;13:789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunsch AK, Linnane E, Barr FA, Gruneberg U. The astrin-kinastrin/SKAP complex localizes to microtubule plus ends and facilitates chromosome alignment. J Cell Biol. 2011;192:959–968. doi: 10.1083/jcb.201008023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell JE, Ha SH. Ultrasensitivity part III: cascades, bistable switches, and oscillators. Trends Biochem Sci. 2014;39:612–618. doi: 10.1016/j.tibs.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournane S, Krupina K, Kleiss C, Sumara I. Decoding ubiquitin for mitosis. Genes Cancer. 2012;3:697–711. doi: 10.1177/1947601912473477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller BG, Lampson MA, Foley EA, Rosasco-Nitcher S, Le KV, Tobelmann P, Brautigan DL, Stukenberg PT, Kapoor TM. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 2008;453:1132–1136. doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Wynne DJ. Making an effective switch at the kinetochore by phosphorylation and dephosphorylation. Chromosoma. 2013;122:135–158. doi: 10.1007/s00412-013-0401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Gruneberg U, Neef R, Honda R, Nigg EA, Barr FA. Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J Cell Biol. 2004;166:167–172. doi: 10.1083/jcb.200403084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, Walter R, Heckel A, van Meel J, Rieder CL, Peters JM. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeller D, Crosetto N, Blagoev B, Raiborg C, Tikkanen R, Wagner S, Kowanetz K, Breitling R, Mann M, Stenmark H, et al. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat Cell Biol. 2006;8:163–169. doi: 10.1038/ncb1354. [DOI] [PubMed] [Google Scholar]
- Van der Horst A, Lens SM. Cell division: control of the chromosomal passenger complex in time and space. Chromosoma. 2014 Mar 05; doi: 10.1007/s00412-013-0437-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Horst A, Vromans MJM, Bouwman K, van der Waal MS, Hadders MA, Lens SMA. Inter-domain Cooperation in INCENP Promotes Aurora B Relocation from Centromeres to Microtubules. Cell Rep. 2015;12:380–387. doi: 10.1016/j.celrep.2015.06.038. [DOI] [PubMed] [Google Scholar]
- Hummer S, Mayer TU. Cdk1 negatively regulates midzone localization of the mitotic kinesin Mklp2 and the chromosomal passenger complex. Curr Biol. 2009;19:607–612. doi: 10.1016/j.cub.2009.02.046. [DOI] [PubMed] [Google Scholar]
- Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- Jallepalli PV, Lengauer C. Chromosome segregation and cancer: cutting through the mystery. Nat Rev Cancer. 2001;1:109–117. doi: 10.1038/35101065. [DOI] [PubMed] [Google Scholar]
- Kelly AE, Sampath SC, Maniar TA, Woo EM, Chait BT, Funabiki H. Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly. Dev Cell. 2007;12:31–43. doi: 10.1016/j.devcel.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Fung SYS, Hameed UFS, Goto H, Inagaki M, Lee SH. Cdk1 coordinates timely activation of MKlp2 kinesin with relocation of the chromosome passenger complex for cytokinesis. Cell Rep. 2014;7:166–179. doi: 10.1016/j.celrep.2014.02.034. [DOI] [PubMed] [Google Scholar]
- Klein UR, Nigg EA, Gruneberg U. Centromere targeting of the chromosomal passenger complex requires a ternary subcomplex of Borealin, Survivin, and the N-terminal domain of INCENP. Mol Biol Cell. 2006;17:2547–2558. doi: 10.1091/mbc.E05-12-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Kowanetz K, Crosetto N, Haglund K, Schmidt MH, Heldin CH, Dikic I. Suppressors of T-cell receptor signaling Sts-1 and Sts-2 bind to Cbl and inhibit endocytosis of receptor tyrosine kinases. J Biol Chem. 2004;279:32786–32795. doi: 10.1074/jbc.M403759200. [DOI] [PubMed] [Google Scholar]
- Lampson MA, Cheeseman IM. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2011;21:133–140. doi: 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ST, Feng M, Wei Y, Li Z, Qiao Y, Guan P, Jiang X, Wong CH, Huynh K, Wang J, et al. Protein tyrosine phosphatase UBASH3B is overexpressed in triple-negative breast cancer and promotes invasion and metastasis. Proc Natl Acad Sci U A. 2013;110:11121–11126. doi: 10.1073/pnas.1300873110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang Q-C, Cui X-S, Wang Z-B, Kim N-H, Sun S-C. MKlp2 inhibitor paprotrain affects polar body extrusion during mouse oocyte maturation. Reprod Biol Endocrinol RBE. 2013;11:117. doi: 10.1186/1477-7827-11-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maerki S, Olma MH, Staubli T, Steigemann P, Gerlich DW, Quadroni M, Sumara I, Peter M. The Cul3-KLHL21 E3 ubiquitin ligase targets aurora B to midzone microtubules in anaphase and is required for cytokinesis. J Cell Biol. 2009;187:791–800. doi: 10.1083/jcb.200906117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maerki S, Beck J, Sumara I, Peter M. Finding the midzone: the role of ubiquitination for CPC localization during anaphase. Cell Cycle. 2010;9:2921–2922. doi: 10.4161/cc.9.15.12740. [DOI] [PubMed] [Google Scholar]
- Maldonado M, Kapoor TM. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nat Cell Biol. 2011;13:475–482. doi: 10.1038/ncb2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min M, Lindon C. Substrate targeting by the ubiquitin-proteasome system in mitosis. Semin Cell Dev Biol. 2012;23:482–491. doi: 10.1016/j.semcdb.2012.01.015. [DOI] [PubMed] [Google Scholar]
- Murata-Hori M, Tatsuka M, Wang YL. Probing the dynamics and functions of aurora B kinase in living cells during mitosis and cytokinesis. Mol Biol Cell. 2002;13:1099–1108. doi: 10.1091/mbc.01-09-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nezi L, Musacchio A. Sister chromatid tension and the spindle assembly checkpoint. Curr Opin Cell Biol. 2009;21:785–795. doi: 10.1016/j.ceb.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Raguz J, Wagner S, Dikic I, Hoeller D. Suppressor of T-cell receptor signalling 1 and 2 differentially regulate endocytosis and signalling of receptor tyrosine kinases. FEBS Lett. 2007;581:4767–4772. doi: 10.1016/j.febslet.2007.08.077. [DOI] [PubMed] [Google Scholar]
- Rosasco-Nitcher SE, Lan W, Khorasanizadeh S, Stukenberg PT. Centromeric Aurora-B activation requires TD-60, microtubules, and substrate priming phosphorylation. Science. 2008;319:469–472. doi: 10.1126/science.1148980. [DOI] [PubMed] [Google Scholar]
- Salimian KJ, Ballister ER, Smoak EM, Wood S, Panchenko T, Lampson MA, Black BE. Feedback control in sensing chromosome biorientation by the Aurora B kinase. Curr Biol CB. 2011;21:1158–1165. doi: 10.1016/j.cub.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath SC, Ohi R, Leismann O, Salic A, Pozniakovski A, Funabiki H. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Sarangapani KK, Asbury CL. Catch and release: how do kinetochores hook the right microtubules during mitosis? Trends Genet TIG. 2014;30:150–159. doi: 10.1016/j.tig.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz MH, Held M, Janssens V, Hutchins JR, Hudecz O, Ivanova E, Goris J, Trinkle-Mulcahy L, Lamond AI, Poser I, et al. Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat Cell Biol. 2010;12:886–893. doi: 10.1038/ncb2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, Peter M. A Cul3-Based E3 Ligase Regulates Mitosis and is Required to Maintain the Spindle Assembly Checkpoint in Human CellS. Cell Cycle. 2007;6 doi: 10.4161/cc.6.24.5068. [DOI] [PubMed] [Google Scholar]
- Sumara I, Quadroni M, Frei C, Olma MH, Sumara G, Ricci R, Peter M. A Cul3-based E3 ligase removes Aurora B from mitotic chromosomes, regulating mitotic progression and completion of cytokinesis in human cells. Dev Cell. 2007;12:887–900. doi: 10.1016/j.devcel.2007.03.019. [DOI] [PubMed] [Google Scholar]
- Tseng BS, Tan L, Kapoor TM, Funabiki H. Dual detection of chromosomes and microtubules by the chromosomal passenger complex drives spindle assembly. Dev Cell. 2010;18:903–912. doi: 10.1016/j.devcel.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Novelle MD, Petronczki M. Relocation of the chromosomal passenger complex prevents mitotic checkpoint engagement at anaphase. Curr Biol CB. 2010;20:1402–1407. doi: 10.1016/j.cub.2010.06.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.