Abstract
S-palmitoylation (S-acylation) is an emerging dynamic post-translational modification of cysteine residues within proteins. Current assays for protein S-palmitoylation involve either in vivo labelling or chemical cleavage of S-palmitoyl groups to reveal a free cysteine sulfhydryl that can be subsequently labelled with an affinity handle (acyl-exchange). Assays for protein S-palmitoylation using acyl-exchange chemistry therefore require blocking of non-S-palmitoylated cysteines, typically using N-ethylmaleimide, to prevent non-specific detection. This in turn necessitates multiple precipitation based clean-up steps to remove reagents between stages, often leading to variable sample loss, reduced signal or protein aggregation. These combine to reduce the sensitivity, reliability and accuracy of these assays and also requires a substantial amount of time to perform. By substituting these precipitation steps with chemical scavenging of N-ethylmaleimide by 2,3-dimethyl-1,3-butadiene in an aqueous Diels-Alder 4+2 cyclo-addition reaction it is possible to greatly improve sensitivity and accuracy while reducing hands-on and overall time required for assays.
Keywords: S-acylation, palmitoylation, S-palmitoylation, acyl biotin exchange, acyl-RAC, biotin switch, maleimide, N-ethylmaleimide, cysteine, thiol, ABE, Diels-Alder
Introduction
Protein S-palmitoylation (S-acylation) involves the addition of long chain fatty acids to cysteine residues within proteins though a thioester bond. Recent work has demonstrated that, far from being a passive way of anchoring proteins to membranes, S-palmitoylation is a dynamic regulator of protein function (1,2). In particular S-palmitoylation has been shown to affect many integral membrane proteins, with nearly 40% of the membrane proteome estimated to be S-palmitoylated (3–5). Given the importance of membrane proteins as drug targets, acting as regulators of trans -membrane transport and transducing signals across membranes understanding membrane protein function, and in particular their regulation, is a high priority. As a result rapid, reproducible and accurate methods for examining S-palmitoylation are much in demand.
Two complementary approaches are currently used for assaying protein S-palmitoylation; acyl-exchange (6) and alkyne fatty acid labelling (7). Both routes to studying S-palmitoylation have allowed researchers to achieve in days what previously took weeks or months using traditional 3H palmitate labelling methods (6,7). The alkyne fatty acid labelling method uses in vivo feeding of cells with alkyne derivatized fatty acids (generally stearate and palmitate mimics) followed by extraction and azide-alkyne click chemistry to tag and subsequently detect proteins of interest (7). This method is however restricted to tissue culture with cells able to take up the applied label, and is therefore of limited use in the context of studying whole organisms or tissue specific in vivo processes. Acyl-exchange assays on the other hand can be used to assess levels of protein S-palmitoylation from samples of any origin. Subsequent refinement of acyl-exchange assays for use in a variety of species, at proteomic scales (3,4) or using more convenient reagents (acyl-Resin Assisted Capture; acyl-RAC) (8) have improved matters further. None-the-less there are still improvements to be made. The existing protocols still require a lot of time and are less than ideal for quantitative analysis, particularly of large or aggregation prone proteins. The many handling and precipitation steps required to remove reagents from samples before progressing to the next stage also introduce sample to sample variation and therefore impacts upon accuracy and reproducibility. Here we present an optimised protocol that reduces sample handling and eliminates all precipitation steps for acyl-RAC style assays and reduces the number of precipitations to one for acyl-biotin exchange (ABE) assays.
Materials and Methods
All standard reagents and chemicals were purchased from Sigma-Aldrich (Gillingham, UK) or Fisher Scientific (Loughborough, UK) unless noted otherwise. Catalogue numbers for specified reagents are given in parentheses.
Organism growth conditions
Arabidopsis Col-0 accession was grown on 0.5x MS medium, 0.8% phytagar under 16:8 hour light:dark cycles at 20°C in MLR-350 growth chambers (Panasonic, Loughborough, UK). 10 days post germination seedlings were harvested and flash frozen in liquid nitrogen before processing. Rat hearts were briefly perfused with Krebs–Henseleit solution in the Langendorff mode to remove contaminating blood then flash frozen in liquid nitrogen and ground to a fine powder.
Western blotting
SDS-PAGE of ABE samples was performed using 7.5% gels run under standard Laemmli conditions and proteins transferred to PVDF using Towbin buffer (65V, 2.5 hours, 4°C). Membranes were blocked with 5% skimmed milk powder in TBS 0.05% Tween20 (TBS-T) for 1 hour at room temperature. Rabbit polyclonal anti-FLS2 antisera was diluted 1:5000 in blocking solution and membranes incubated for 1 hour at room temperature. Membranes were washed 3 times for 5 minutes each with TBS-T before incubation for 45 minutes with anti-rabbit HRP conjugate (Insight Biotechnology, Wembley, UK, sc-2004) diluted 1:5000 in blocking buffer. Membranes were washed 3 time with TBS-T and twice with TBS before being exposed to a G:box storm XT4 imaging system (Syngene, Cambridge, UK) using a mixture of supersignal west femto and pico (ThermoFisher, Paisley, UK) in a 1:3 ratio. SDS-PAGE of acyl-RAC samples was performed on 6-18% gradient gels and transferred to PVDF using a semi-dry transfer system. Membranes were blocked for 1 hour in 5% skimmed milk powder in PBS 0.1% Tween20 (PBS-T) and probed overnight at 4°C with anti-NCX1 (P11-13, Swant, Marly, CH) at 1:1000or anti-flotillin 2 (610383, BD Biosciences, Oxford, UK) at 1:5000. After 6 washes with PBS-T membranes were incubated with HRP-linked anti-mouse (315-035-045, Jackson ImmunoResearch, PA, USA) at 1:2000 for 1 hour in blocking buffer, washed 6 more times, and images acquired using a ChemiDoc XRS imaging system (BioRad, Hemel Hempstead, UK) using Immobilon HRP substrate (EMD Millipore, Watford, UK).
Acyl-biotin exchange (ABE) procedure
ABE assays were performed essentially as described previously (4,9,10). Briefly samples were ground to a fine powder in liquid nitrogen and resuspended in 2ml of lysis buffer (50mM Tris.HCl pH 7.2, 5mM EDTA, 150mM NaCl, 2.5% SDS, 25mM N-ethylmaleimide (AC156100100, Fisher Scientific), 5μl.ml-1 protease inhibitor (P9599, Sigma-Aldrich), 100mg PVPP). Samples were solubilised at room temperature for 10 minutes with gentle mixing, centrifuged at 3000 xg for 1 minute, filtered through 2 layers of miracloth (EMD Millipore) and re-centrifuged at 16 000 xg for 10 minutes. The clarified supernatant was retained and protein concentration determined using BCA assay ( 23225, ThermoFisher). 2mg of protein was combined with lysis buffer (no PVPP) to a final volume of 1 ml and incubated with gentle mixing for 1 hour. Samples were chloroform/methanol precipitated, briefly air dried and resuspended in 1ml 50mM Tris.HCl pH7.2, 2% SDS, 8M urea, 5mM EDTA by gentle mixing. Samples were then split in two; one aliquot was combined with 0.5ml 1M hydroxylamine (159417, Sigma-Aldrich) while the other was combined with 0.5ml 1M NaCl (negative control). 50μl was removed from each as a loading control and incubated at RT for 1 hour before being chloroform/methanol precipitated. 100μl of 2mM biotin HPDP (21341, ThermoFisher) in DMSO was added to the remaining sample and gently mixed for 1 hour before being chloroform/methanol precipitated, briefly air dried and resuspended in 150µl 50mM Tris.HCl pH7.2, 2% SDS, 8M urea, 5mM EDTA by gentle mixing. Samples were diluted 1:10 with PBS and mixed with 20µl of a PBS washed 50% slurry of high capacity neutravidin beads (29202, ThermoFisher) for 1 hour. Beads were washed 3x with PBS 1% SDS and dried by aspiration. Bead bound samples and loading control precipitates were resuspended in 20μl of 2x reducing SDS-PAGE sample buffer containing 6M urea at 37°C for 30 minutes with frequent mixing.
Acyl-RAC procedure
Acyl-RAC was performed essentially as described previously (10–12). Powdered left ventricle was lysed directly in 2.5% SDS, 1mM EDTA, 100mM HEPES, 25mM NEM pH 7.5 and incubated at 40°C for 4 hours to block free thiols. NEM was removed by acetone precipitation followed by multiple washes with 70% acetone and resolubilised in 1% SDS, 1mM EDTA, 100mM HEPES pH 7.5 at 40°C. Samples were split in two and S-palmitoylated proteins were captured on thiopropyl Sepharose 6b (T8387, Sigma-Aldrich) in the presence of 250mM neutral hydroxylamine or water (negative control) by agitating for 2.5 hours at room temperature. Loading controls were collected before addition of thiopropyl Sepharose 6b beads. Following 5 washes with 1% SDS, 1mM EDTA, 100mM HEPES pH 7.5 proteins were eluted from thiopropyl Sepharose 6b beads in 50µl SDS-PAGE sample buffer supplemented with 100mM DTT.
Removal of N-ethylmaleimide from solutions during Acyl-RAC and ABE procedures
This step replaces the first precipitation step to remove NEM in acyl-RAC and ABE protocols: Following the NEM blocking stages of Acyl-RAC or ABE 2,3-dimethyl-1,3-butadiene (Sigma-Aldrich 145491) was added to 100mM and incubated with vigorous mixing for 1 hour at 25°C. Subsequently 1/10th volume of chloroform was added, vortexed for 1 minute and centrifuged at maximum speed. The supernatant was retained and used in subsequent hydroxylamine treatment steps (continuing after resolubilisation steps of standard protocols).
Quantification of N-ethylmaleimide
NEM concentration in solution was measured by spectrophotometry at 300 nm using an Ultrospec 2100 pro (GE Healthcare, Little Chalfont, UK). The limit of detection by spectrophotometry in our buffer system was determined to be ˜40 μM. To calculate NEM concentrations in the range of 0.1-100 μM a fluorometric maleimide quantification kit (ab112141, Abcam, Cambridge, UK) was used in conjunction with a Varioskan LUX plate reader (ThermoFisher).
Results and discussion
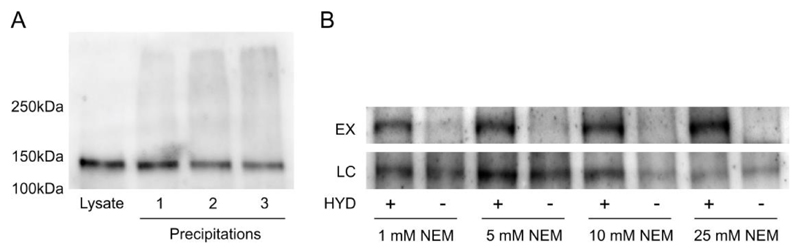
Established acyl switch methods for assaying protein palmitoylation state rely on using hydroxylamine to cleave away S-acyl groups to reveal a cysteine that can then be labelled with biotin (6,9) or bound directly by sulfhydryl reactive resin (8). This therefore requires that all other cysteines in the protein are blocked to prevent false positives. Traditionally N-ethylmaleimide (NEM), a sulfhydryl reactive reagent, has been the favoured reagent to irreversibly block all free cysteines in a protein (6). NEM subsequently has to be removed by one or more precipitation steps to avoid downstream competition with reagents required to label the previously S-acylated cysteine. While working with a number of difficult, low abundance or aggregation prone plant proteins we noticed that protein precipitation led to smearing on western blots or protein remaining trapped in gel wells. This is likely due to precipitated proteins forming insoluble aggregates or not fully unfolding during resolubilisation. As a result trying to perform quantitative analyses to examine changes in S-palmitoylation state is almost impossible under these conditions. The Arabidopsis receptor-like kinase FLS2 typifies many of these problems and is particularly prone to aggregation and smearing after precipitation (Fig1A) and during sample processing for acyl-biotin exchange (Fig1B). Despite using different detergents, chaotropes and resolubilisation regimes we were never able to fully eliminate this problem. We therefore sought ways to eliminate the precipitation and resuspension steps in our protocol without interfering with downstream processes.
Figure 1.
(A) FLS2 aggregates and smears on 7.5% SDS-PAGE gels after multiple precipitations. 50µg of total protein was loaded per lane. FLS2 appears heavier than the predicted 130 kDa due to extensive glycosylation of the leucine-rich repeats. (B) Minimum concentrations of N-ethylmaleimide (NEM) required for elimination of false positive identification of FLS2 S-palmitoylation levels in acyl-biotin exchange assays. Acyl-biotin exchange assays were performed using the indicated concentrations of NEM during the blocking stage. All other steps in the protocol were as described in the methods. HYD indicates presence (+) or absence (-) of hydroxylamine required for acyl group cleavage during the biotin labelling step. Samples were bound to neutravidin beads following biotin labelling and enriched proteins were eluted and represent S-palmitoylated proteins (EX). Prior to neutravidin binding a sample was removed as an input loading control (LC). All samples were run on 7.5% SDS-PAGE gels and blotted for FLS2.
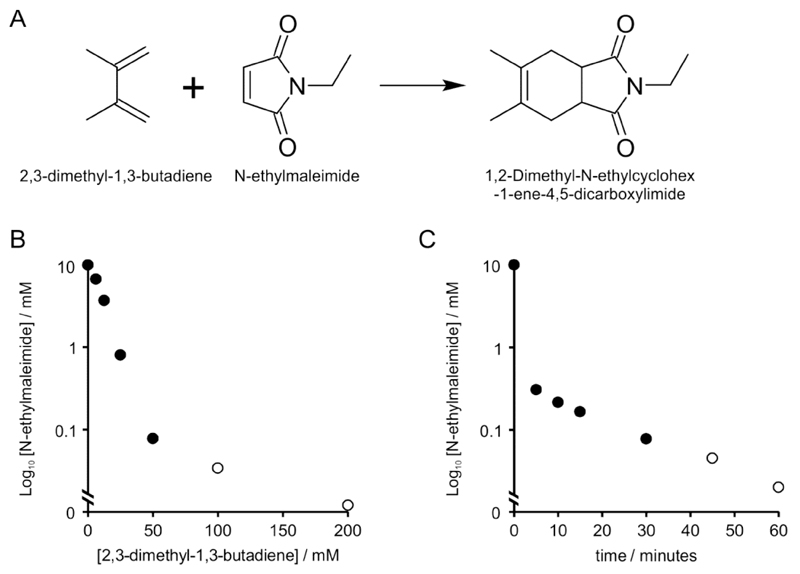
Maleimides, such as NEM used as the blocking reagent in acyl-biotin exchange (ABE) and acyl resin assisted capture (acyl-RAC) S-palmitoylation assays, are electrophiles. Maleimides also form one of the classic components of Diels-Alder 4+2 cyclo-addition reactions where a substituted alkene known as a dienophile, in this case NEM, reacts with a conjugated diene (Figure 2A). Interestingly, although classically thought of as organic chemistry, many Diels-Alder reactions not only work (13) but appear to proceed faster in aqueous reaction buffers (14). We therefore speculated that a suitable diene would be able to scavenge NEM that had not reacted with sulfhydryls and remove the need for precipitation clean up. We have been unable to find reports of conjugated dienes reacting with protein functional groups, making it unlikely that proteins would be modified by this treatment. To evaluate this method we first optimised minimum NEM concentrations required for full blocking of sulfhydryls in protein extracts. FLS2 contains 16 cysteines, 4 of which are likely involved in disulfide bonds (15) and 2 are S-acylated (4), making it an excellent test case for examining thiol blocking efficiency in S-palmitoylation assays. Using Arabidopsis seedlings a range of NEM concentrations were tested for efficient blocking of FLS2 cysteines in ABE assays. We determined that 10mM was sufficient to eliminate false positives while still maintaining rapid and reproducible blocking of cysteines (Figure 1B). We then tested the ability of 2,3-dimethyl-1,3-butadiene, as a classic Diels-Alder diene, to react with NEM in our typical assay buffer. We found that a 10-fold molar excess of 2,3-dimethyl-1,3-butadiene was able to reduce NEM concentration from 10 mM to undetectable levels (< 50 µM NEM in our assay buffer using spectrophotometry at 300 nm) after 1 hour of vigorous mixing at 25°C (Figure 2B). Using a fluorescent assay to detect NEM we were able to determine that NEM concentrations were reduced to ˜ 35 µM using a 10-fold excess of 2,3-dimethyl-1,3-butadiene. A 20-fold molar excess reduced NEM concentrations to ˜15 µM. 2,3-dimethyl-1,3-butadiene is volatile, flammable and has an unpleasant odour; removal of excess 2,3-dimethyl-1,3-butadiene after reaction is therefore a sensible precaution. As 2,3-dimethyl-1,3-butadiene is immiscible with water it can be separated from aqueous components by centrifugation after reaction. However, 2,3-dimethyl-1,3-butadiene is less dense than water and therefore floats on top of the aqueous phase. Careful pipetting or allowing 2,3-dimethyl-1,3-butadiene to evaporate in a fume hood is required to collect sample without disturbing the upper organic phase. Neither of these options are desirable for a number of practical reasons and we therefore investigated alternatives. As chloroform is denser than water and a good water immiscible organic solvent we speculated that it would be able to remove unreacted 2,3-dimethyl-1,3-butadiene from aqueous solutions by phase partitioning. Adding 1/10th of the aqueous reaction volume of chloroform efficiently extracted all visible 2,3-dimethyl-1,3-butadiene into the chloroform layer that, after centrifugation, formed at the bottom of the tube. A diffuse white layer formed at the interphase but further analysis indicated that this layer did not appear to contain detectable protein. As this interphase layer was also formed when using buffer only it is likely formed from detergent in the sample buffer. Chloroform is also an effective solvent for NEM and 1/10th of the sample volume of chloroform alone was capable of reducing NEM concentration in the aqueous phase by nearly 70% (69.8%, SD = 0.021, n = 3) and is therefore a worthwhile additional procedure in its own right. The time required for removal of NEM from samples by 2,3-dimethyl-1,3-butadiene and chloroform was optimised next. We found that 10mM NEM was depleted by ˜90% after 15 minutes treatment with 100mM 2,3-dimethyl-1,3-butadiene at 25°C with vigorous shaking. NEM was at the limits of detection using absorbance at 300 nm after 30 minutes and was not able to be determined in samples after 45 and 60 minutes treatment (Figure 2C). Using the more sensitive fluorescent assay we determined that NEM concentrations were ˜50 µM (45 minutes) and ˜15 µM (60 minutes). These levels of NEM are well below the concentrations of sulfhydryl capture reagents typically used in acyl -RAC and ABE assays and are therefore unlikely to interfere in subsequent steps. While use of 200mM 2,3-dimethyl-1,3-butadiene would arguably be most effective, it is difficult to fully remove using 1/10th volume chloroform, meaning that it is carried over into subsequent steps and increased chloroform volume led to protein precipitation. Further refinements of the method, potentially using different dienes or water immiscible solvents, may be able to circumvent this limitation. To ensure maximal removal of NEM from solutions and prevent interference with downstream reactions we therefore decided to use 1 hour treatment with 100mM 2,3-dimethyl-1,3-butadiene followed by chloroform phase separation for further experiments.
Figure 2.
(A) Reaction scheme of N-ethylmaleimide combining with 2,3-dimethyl-1,3-butadiene. (B) Determination of 2,3-dimethyl-1,3-butadiene concentration required for elimination of 10 mM N-ethylmaleimide (NEM) from lysis buffer in 1 hour. A standard curve of NEM was prepared in lysis buffer and absorbance at 300 nm measured. A 10mM solution of NEM was treated with a range of 2,3-dimethyl-1,3-butadiene concentrations for 1 hour at 25°C and remaining NEM concentration were determined from standard curves by spectrophotometric (filled circles) or fluorometric (open circles) methods. Data points are averages of 3 technical replicates. (C) Determination of time required for elimination of 10mM NEM by 100 mM 2,3-dimethyl-1,3-butadiene and chloroform extraction in lysis buffer. A standard curve of NEM was prepared in lysis buffer and absorbance at 300 nm measured. A 10 mM solution of NEM was treated with 100 mM 2,3-dimethyl-1,3-butadiene for the times indicated at 25°C and phase partitioned using chloroform. Remaining NEM concentration were determined from standard curves by spectrophotometric (filled circles) or fluorometric (open circles) methods. Data points are averages of 3 technical replicates.
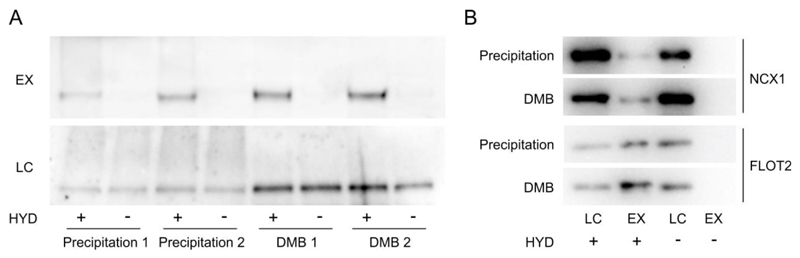
Using these new procedures we examined the ability to reproduce and improve upon our previous data on FLS2 palmitoylation using ABE and 2,3-dimethyl-1,3-butadiene treatment instead of protein precipitation for NEM removal. As shown (Figure 3) our new procedure appears to be completely compatible with the pyridyl disulfide chemistry found in acyl-RAC and ABE procedures. The new method produced stronger, sharper and more reproducible intensity bands of FLS2 on western blots with reduced smearing, thereby making future quantification and interpretation of data easier and more accurate. To ensure that the new protocol was applicable to other proteins, organisms and variations in procedure we assessed the palmitoylation state of the NCX1 sodium/calcium exchanger (16) and FLOT2 (17) in rat hearts using the original and new acyl-RAC procedure. No difference was observed in assessment of palmitoylation state of either protein between protocols. However, with the new protocol, hands-on and total time required were reduced. These data combined indicate that this method will be useful for assessment of palmitoylation of all proteins, not just aggregation prone or large proteins. This will allow faster or higher-throughput analyses to be performed, including automation, and benefit large scale proteomic analyses of S-palmitoylation where sample handling errors can impact upon quantification. With the increasing interest in dynamic S-palmitoylation, sensitive, accurate and reproducible assays capable of dealing with multiple samples are required. The refinements presented here to NEM based acyl-exchange assays fulfil these requirements. Other sulfhydryl reactive reagents commonly used for analysis of cysteine modifications include pyridyl disulfides, haloacetamides, haloacetic acids, methyl methanethiosulfonate (MMTS), vinyl sulfones and 4-vinyl pyridine. With the exception of some vinyl group containing compounds, none of these compounds are reported to undergo reactions with (conjugated) dienes such as 2,3-dimethyl-1,3-butadiene. 2,3-dimethyl-1,3-butadiene is therefore unlikely to be able to remove these other functionalities from reaction mixtures. The suitability of the reported procedure for removal of vinyl sulfones and 4-vinyl pyridine needs to be empirically determined. By using maleimides alongside these alternative cysteine labelling reagents, and 2,3-dimethyl-1,3-butadiene as a scavenger for maleimides, it should be feasible to perform “1 pot” differential labelling reactions for identification of modified cysteines in a similar manner to what is described here for ABE and acyl -RAC assays. For example, PEG-shift/APE assays for protein palmitoylation state, currently requiring more than 3 precipitation clean-up steps (18), could be performed using maleimides to block unmodified cysteines, 2,3-dimethyl-1,3-butadiene to scavenge maleimides followed by hydroxylamine and iodoacetamide derivatized PEG treatment to label previously S-palmitoylated cysteines, all without any precipitations being required. Although developed for the analysis of S-palmitoylation this procedure should also be compatible with methods that require the removal of maleimides such as NEM from aqueous solution under mild conditions. This includes analysis of other cysteine post-translational modifications such as S-nitrosylation, S-glutathionylation, sulfenylation, sulfinylation, sulfonylation and disulphide mapping.
Figure 3.
(A) Use of 2,3-dimethyl-1,3-butadiene in place of precipitation to remove NEM from solution greatly improves detection of FLS2 S-palmitoylation by Acyl-biotin exchange (ABE). The ABE protocol presented in the methods was followed using either chloroform/methanol precipitation or 2,3-dimethyl-1,3-butadiene (DMB) treatment for NEM removal. The same initial extract was split into four and processed in parallel. HYD indicates presence (+) or absence (-) of hydroxylamine required for acyl group cleavage during the biotin labelling step. Samples were bound to neutravidin beads following biotin labelling and enriched proteins were eluted and represent S-palmitoylated proteins (EX). Prior to neutravidin binding a sample was removed as an input loading control (LC). All samples were run on 7.5% SDS-PAGE gels and blotted for FLS2. (B) Use of 2,3-dimethyl-1,3-butadiene in place of precipitation to remove NEM from solution is compatible with acyl -RAC procedures. The acyl-RAC protocol presented in the methods was followed either using acetone precipitation or 2,3-dimethyl-1,3-butadiene treatment for NEM removal. The same initial extract was split into two and processed in parallel. HYD indicates presence (+) or absence (-) of hydroxylamine required for acyl group cleavage during the thiopropyl Sepharose 6b capture step. Thiopropyl Sepharose 6b enriched proteins were eluted and represent S-palmitoylated proteins (EX). Prior to Thiopropyl Sepharose 6b capture a sample was removed as an input loading control (LC).
Methods Summary.
A precipitation free method to remove maleimides from aqueous solutions allows for rapid analysis of S-palmitoylation. Assays using this new method show greater signal intensity, accuracy and reproducibility compared to methods requiring precipitation.
Acknowledgements
This work was funded by Biotechnology and Biological Sciences Research Council (BBSRC) grant BB/M024911/1 to PAH and British Heart Foundation (BHF) PhD studentship FS/14/68/30988 to WF.
Footnotes
Author contributions: PAH conceived and developed the work. CHH, DT, FP, WF and PAH performed the experiments and analysed the results. PAH and WF wrote the manuscript with input from DT, CHH and FP.
Competing Interests Statement: The authors declare no competing interests.
References
- 1.Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. Global profiling of dynamic protein palmitoylation. Nat Methods. 2011;9:84–89. doi: 10.1038/nmeth.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sorek N, Poraty L, Sternberg H, Bar E, Lewinsohn E, Yalovsky S. Activation status-coupled transient S acylation determines membrane partitioning of a plant Rho-related GTPase. Mol Cell Biol. 2007;27:2144–2154. doi: 10.1128/MCB.02347-06. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, Phinney BS, Yates JR, 3rd, Davis NG. Global analysis of protein palmitoylation in yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hemsley PA, Weimar T, Lilley KS, Dupree P, Grierson CS. A proteomic approach identifies many novel palmitoylated proteins in Arabidopsis. New Phytologist. 2013;197:805–814. doi: 10.1111/nph.12077. [DOI] [PubMed] [Google Scholar]
- 5.Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, Bailey AO, Thompson JX, Roth AF, et al. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drisdel RC, Green WN. Labeling and quantifying sites of protein palmitoylation. Biotechniques. 2004;36:276–285. doi: 10.2144/04362RR02. [DOI] [PubMed] [Google Scholar]
- 7.Martin BR, Cravatt BF. Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forrester MT, Hess DT, Thompson JW, Hultman R, Moseley MA, Stamler JS, Casey PJ. Site-specific analysis of protein S-acylation by resin-assisted capture. J Lipid Res. 2010;52:393–398. doi: 10.1194/jlr.D011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hemsley PA, Taylor L, Grierson CS. Assaying protein palmitoylation in plants. Plant methods. 2008;4:2. doi: 10.1186/1746-4811-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forrester MT, Hess DT, Thompson JW, Hultman R, Moseley MA, Stamler JS, Casey PJ. Site-specific analysis of protein S-acylation by resin-assisted capture. J Lipid Res. 2011;52:393–398. doi: 10.1194/jlr.D011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wypijewski KJ, Howie J, Reilly L, Tulloch LB, Aughton KL, McLatchie LM, Shattock MJ, Calaghan SC, Fuller W. A separate pool of cardiac phospholemman that does not regulate or associate with the sodium pump: multimers of phospholemman in ventricular muscle. J Biol Chem. 2013;288:13808–13820. doi: 10.1074/jbc.M113.460956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howie J, Reilly L, Fraser NJ, Vlachaki Walker JM, Wypijewski KJ, Ashford ML, Calaghan SC, McClafferty H, et al. Substrate recognition by the cell surface palmitoyl transferase DHHC5. Proc Natl Acad Sci U S A. 2014;111:17534–17539. doi: 10.1073/pnas.1413627111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diels O, Alder K. Synthesen in der hydroaromatischen Reihe. XII. Mitteilung. („Dien-Synthesen” sauerstoffhaltiger Heteroringe. 2. Dien-Synthesen des Furans.) Justus Liebigs Annalen der Chemie. 1931;490:243–257. [Google Scholar]
- 14.Rideout DC, Breslow R. Hydrophobic acceleration of Diels-Alder reactions. Journal of the American Chemical Society. 1980;102:7816–7817. [Google Scholar]
- 15.Sun W, Cao Y, Jansen Labby K, Bittel P, Boller T, Bent AF. Probing the Arabidopsis flagellin receptor: FLS2-FLS2 association and the contributions of specific domains to signaling function. Plant Cell. 2012;24:1096–1113. doi: 10.1105/tpc.112.095919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reilly L, Howie J, Wypijewski K, Ashford ML, Hilgemann DW, Fuller W. Palmitoylation of the Na/Ca exchanger cytoplasmic loop controls its inactivation and internalization during stress signaling. FASEB J. 2015;29:4532–4543. doi: 10.1096/fj.15-276493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neumann-Giesen C, Falkenbach B, Beicht P, Claasen S, Luers G, Stuermer CA, Herzog V, Tikkanen R. Membrane and raft association of reggie-1/flotillin-2: role of myristoylation, palmitoylation and oligomerization and induction of filopodia by overexpression. Biochem J. 2004;378:509–518. doi: 10.1042/BJ20031100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yokoi N, Fukata Y, Sekiya A, Murakami T, Kobayashi K, Fukata M. Identification of PSD-95 Depalmitoylating Enzymes. J Neurosci. 2016;36:6431–6444. doi: 10.1523/JNEUROSCI.0419-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]