Abstract
Objectives: Resistance-associated variants (RAVs) in Rv0678, a regulator of the MmpS5-MmpL5 efflux pump, have been shown to lead to increased MICs of bedaquiline (2- to 8- fold) and clofazimine (2- to 4-fold). The prevalence of these Rv0678 RAVs in clinical isolates and their impact on treatment outcomes are important factors to take into account in bedaquiline treatment guidelines.
Methods: Baseline isolates from two bedaquiline MDR-TB clinical trials were sequenced for Rv0678 RAVs and corresponding bedaquiline MICs were determined on 7H11 agar. Rv0678 RAVs were also investigated in non-MDR-TB sequences of a population-based cohort.
Results: Rv0678 RAVs were identified in 23/347 (6.3%) of MDR-TB baseline isolates. Surprisingly, bedaquiline MICs for these isolates were high (> 0.24 mg/L, n = 8), normal (0.03−0.24 mg/L, n = 11) or low (< 0.03 mg/L, n = 4). A variant at position −11 in the intergenic region mmpS5–Rv0678 was identified in 39 isolates (11.3%) and appeared to increase the susceptibility to bedaquiline. In non-MDR-TB isolates, the frequency of Rv0678 RAVs was lower (6/852 or 0.7%). Competition experiments suggested that rifampicin was not the drug selecting for Rv0678 RAVs.
Conclusions: RAVs in Rv0678 occur more frequently in MDR-TB patients than previously anticipated, are not associated with prior use of bedaquiline or clofazimine, and in the majority of cases do not lead to bedaquiline MICs above the provisional breakpoint (0.24 mg/L). Their origin remains unknown. Given the variety of RAVs in Rv0678 and their variable effects on the MIC, only phenotypic drug-susceptibility methods can currently be used to assess bedaquiline susceptibility.
Introduction
Resistance-associated variants (RAVs) that lead to increased MICs of bedaquiline have been described in three genes of Mycobacterium tuberculosis: atpE, Rv0678 and pepQ.1–6 RAVs in atpE, a gene coding for a transmembrane protein of the ATP synthase, target of bedaquiline, lead to 8- to 133- fold increases in bedaquiline MIC and have been isolated in vitro upon exposure to bedaquiline, but they have so far not been observed in clinical isolates from patients treated with bedaquiline.7 RAVs in pepQ, leading to 4-fold increases in bedaquiline and clofazimine MICs, have been isolated from mice treated with bedaquiline. RAVs in Rv0678, a gene regulating the expression of the MmpS5-MmpL5 efflux pump, lead to 2- to 8- fold increases in bedaquiline MIC and 2- to 4-fold increases in clofazimine MIC.1 They have been isolated in vitro upon exposure to clofazimine3 or bedaquiline,1 and have also been observed in some post-baseline isolates of patients treated with bedaquiline.7 Although RAVs in Rv0678 lead to increased MICs of clofazimine, it is not clear whether the use of clofazimine in TB patients can select for these mutants. The prevalence of these Rv0678 RAVs in clinical isolates and their impact on bedaquiline MICs and treatment outcomes are important factors to take into account in bedaquiline treatment guidelines. Upon the surprising discovery of an Rv0678 RAV in a baseline isolate from a patient without documented prior use of clofazimine or bedaquiline, we studied the prevalence of these RAVs in clinical isolates of MDR-TB and non-MDR-TB patients.
Materials and methods
MDR clinical isolates
Baseline (day 1) M. tuberculosis isolates from participants in Phase 2b studies C2088 and C2097 were included. On day 1 there were 374 isolates available, of which 363 had bedaquiline MICs available, and, out of these, DNA was available for 359 isolates (96%), which had their Rv0678 gene sequenced. Only seven of these patients had documented prior use of bedaquiline of clofazimine in their medical history.
Susceptibility testing
Bedaquiline MICs for C208 and C209 MDR-TB isolates were determined on Middlebrook 7H11 agar at 0.004, 0.008, 0.015, 0.03, 0.06, 0.12, 0.24, 0.48, 1.0, 2.0, 4.0 and 8.0 mg/L as described previously.9 Each new batch of 7H11 medium had the H37Rv reference strain (BCCM/ITM 083715) tested as quality control, with an acceptable MIC range of 0.03–0.12 mg/L. The MIC was determined as the lowest concentration of the antibiotic that resulted in 99% growth inhibition.
Susceptibility of BCA8, an H37Rv-derived Rv0678 mutant,1 to TB drugs other than pyrazinamide was determined in 7H9 broth by the resazurin microtitre assay (REMA) as previously described.10 The percentage of growth in each well was calculated using the fluorescence of the growth control minus the fluorescence of the non-growth control as 100% growth. The MIC50 was defined as the lowest concentration of antibiotic that resulted in 50% inhibition of growth. The average of the MIC50s was taken after log transforming all individual MIC50s, including the censored ones. The censor added to the average was based on the relative occurrence of the censor in the individual data. When the censor occurred in less than one-third of the data, the censor was omitted. When the censor occurred in more than two-thirds of the data, the censored was added to the average. When the censor occurred between one-third and two-thirds of the data the censored was used but an equals sign was added. To calculate the significance of the fold change, a one-sided unpaired t-test was performed with unequal variance. Pyrazinamide resistance testing was done using the MGIT-PZA kit (Becton Dickinson) as per the manufacturer’s instructions.
Rv0678, atpE and pepQ sequencing
A DNA fragment containing Rv0678 and part of the intergenic region between mmpS5 and Rv0678 of C208 and C209 MDR-TB isolates was amplified by PCR using primers CV010 and CV017 or 916R20 and 30F22 (Table S1, available as Supplementary data at JAC Online). The atpE and pepQ genes were amplified using primers atpEforward with atpEreverse, and pepQforward with pepQreverse, respectively, (Table S1) in the eight baseline isolates with MICs above the breakpoint (> 0.24 mg/L).11 The PCR products sequenced with the same primers used for amplification. For analysis of the sequences, Rv0678 sequence from M. tuberculosis H37Rv was taken as a reference12 (http://tuberculist.epfl.ch).
Data mining of non-MDR clinical isolates
A total of 941 isolates from the Hamburg non-MDR-TB cohort13 were mined to look for mutations in the same region. These samples were isolated between 2004 and 2012 and sequenced using the Illumina platforms with read lengths of 100 bp. SNP calling was undertaken using the Snippy v2.914 pipeline, which employs BWA (read-quality cleaning and mapping functions), SAMtools and FreeBayes. The genome of strain H37Rv (NC-000962.3) was used as a reference. Drug resistance and lineage assignment was determined from the genomes using the PhyResSE resistance list version 27.15
TB lineage analysis
TB lineage analysis for the clinical trial isolates was done by a real-time PCR assay targeting lineage-specific SNPs as described by Stucki et al.16 with modified probes and primers for lineage 2 (Table S2). Isolates with an unclear SNP profile were tested by spoligotyping using the MIRU-VNTRplus online application (http://www.miru-vntrplus.org/MIRU/index.faces).
Competition experiments
Two M. tuberculosis H37Rv-derived strains were used: H37Rv-rpoB, carrying a mutation in the rpoB gene (H526Y); and CV37-rpoB, carrying mutations in rpoB (H526Y) and Rv0678 (IS6110 nt 104). These strains were transformed with pND235,17 a plasmid containing green fluorescent protein (GFP) (λem 475 nm, λex 525 nm), and pND239,18 a plasmid containing DsRed2 (λem 575 nm, λex 632 nm), in order to generate fluorescence-marked strains (Table S1). To rule out any effect of the plasmids on the fitness of bacteria, competition experiments were done in two pairs: H37Rv-rpoB-pND235 (GFP marked) with CV37-rpoB-pN239 (DsRed2 marked); and H37Rv-rpoB-pND239 (DsRed2 marked) with CV37-rpoB-pN235 (GFP marked).
Competition experiments were adapted from Gullberg et al.19 Cultures were grown in 7H9/10% OADC/0.05% Tween broth to the logarithmic phase and diluted to OD = 0.1. These were further diluted 1:200 to obtain a final bacterial inoculation density of ∼5 × 104 cfu/mL per strain. Two independent flasks were inoculated with: (i) H37Rv-rpoB-pND235 and CV37-rpoB-pND239; and (ii) H37Rv-rpoB-pND239 and CV37-rpoB-pND235. Aliquots (5 mL) from each culture were put in 10 flasks; rifampicin was added to 8 of them (concentrations: 200, 100, 50, 25, 12.5, 6.25, 0.625 and 0.0625 mg/L) and 2 remained antibiotic-free. After incubation for 1 week at 37 °C, a 500 μL aliquot from all cultures was washed with 200 μL of PBS, inactivated for 90 min at 4 °C in 400 μL of 4% paraformaldehyde20 and finally resuspended in 200 μL of PBS. Serial decimal dilutions were analysed using a fluorescence-activated cell sorter in an FACS Fortessa. A minimum of 104 cells were counted per sample. The percentages of GFP-containing and DsRed2-containing bacilli were determined in each sample and the ratio of CV37-rpoB to H37Rv-rpoB was calculated for each condition. On the same day, every culture was passaged at a dilution of 1:1000 to a new flask containing the same concentration of rifampicin. These steps of sampling and passage were repeated every week for 3 weeks.
Results
MDR-TB clinical isolates at baseline contain RAVs in Rv0678, not always linked to high bedaquiline MICs
Interpretable Rv0678 sequencing results and bedaquiline MICs were available for 347 out of 359 investigated baseline isolates from MDR-TB isolates (Table 1). Their MICs ranged from 0.004 to 1.0 mg/L, with a mean MIC of 0.06 mg/L. The majority (296/347, 85.3%) had normal MICs (0.03–0.24 mg/L) (typically seen for the H37Rv reference strain), a smaller proportion (43/347, 12.4%) had low MICs (≤ 0.015 mg/L) and 8/347 (2.3%) isolates had an MIC > 0.24 mg/L, the provisional breakpoint for bedaquiline on 7H11 agar.11
Table 1.
MICs for and mutations of MDR-TB baseline clinical isolates
| Rv0678 locus | Number of isolates with bedaquiline MIC (mg/L) |
Total | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1.000 | 0.480 | 0.240 | 0.120 | 0.060 | 0.030 | 0.015 | 0.007 | 0.004 | ||
| Non-silent mutation | 1 | 5 | 2 | 3 | 1 | 1 | 13 | |||
| WT + non-silent mutation | 1 | 1 | 3 | 1 | 6 | |||||
| WT + non-silent mutation + mutation in position −11 | 2 | 2 | ||||||||
| WT + non-silent mutation + silent mutation | 1 | 1 | ||||||||
| Non-silent mutation + mutation in position −13 | 1 | 1 | ||||||||
| Intergenic mutation in position −11 or intergenic mutation in position −11 + Rv0678 silent mutation | 7 | 28 | 2 | 37 | ||||||
| Intergenic mutation, not −11 | 1 | 1 | 2 | |||||||
| Silent mutations or mixture of WT + silent | 1 | 2 | 1 | 4 | ||||||
| WT | 2 | 48 | 153 | 69 | 7 | 2 | 281 | |||
| Total analysed | 2 | 6 | 5 | 53 | 160 | 78 | 37 | 1 | 5 | 347 |
The numbers of isolates with high (> 0.24 mg/L), normal (0.03–0.24 mg/L) and low (< 0.03 mg/L) bedaquiline MICs are indicated for every type of mutation in the Rv0678 gene and the intergenic region mmpS5–Rv0678 .
Non-silent mutations refer to non-synonymous mutations and small insertions or deletions in the Rv0678 coding region.
An overview of all the Rv0678 RAVs is presented in Figure 1 and Table S3. The higher MICs could all be explained by RAVs in Rv0678 – no RAVs were found in atpE or pepQ genes of these isolates. However, 15 additional baseline isolates (4.3%) had similar Rv0678 RAVs that did not result in a high bedaquiline MIC. Taken together, 6.6% (95% CI 4.0%–9.2%) of MDR-TB baseline isolates had an RAV in the Rv0678 coding sequence. Rv0678 RAVs were not identified in any of the seven patients that had failed previous therapy with a regimen containing clofazimine prior to enrolment in the bedaquiline trial.
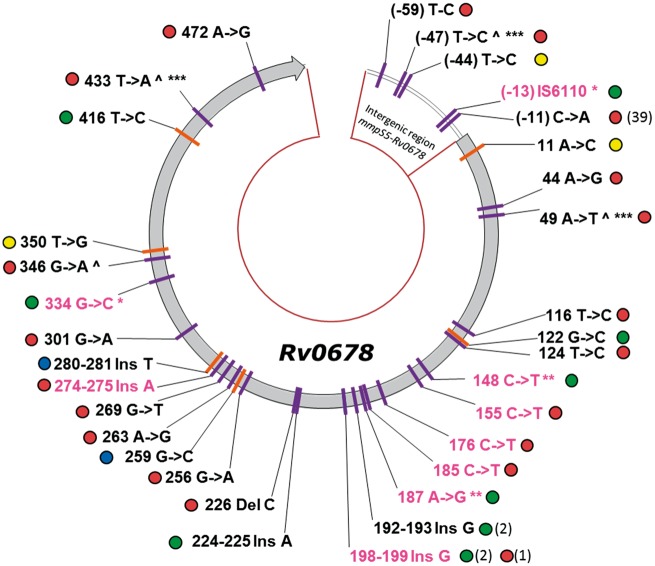
Figure 1.
Mutations in Rv0678 and the intergenic region mmpS5–Rv0678 and lineage of their respective isolates. All the different DNA mutations found in C208 and C209 baseline isolates (MDR, purple markers) and in the Hamburg cohort (non-MDR, orange markers) are shown. Silent mutations are not shown. Mutations in isolates with high MICs are coloured in pink. The circle next to every mutation indicates the lineage of the isolate: blue, lineage 1; red, lineage 2; yellow, lineage 3; and green, lineage 4. When the mutation was found in more than one isolate, the number of isolates carrying that mutation is indicated in brackets. Some isolates carry more than one mutation, and the mutations found in the same isolate are indicated with the same symbol (*, ** or ***). The mutations co-existing with the mutation in position −11 are marked with the symbol ^.
Intergenic mutations at positions −44 and −59, outside of the hypothetical binding site of Rv0678 described by Milano et al.21 were observed in < 1% of the isolates and were not associated with high bedaquiline MICs. Intergenic mutations at position −11 were observed in 39 out of the 347 isolates (11%); in 2 out of 39 this mutation appeared simultaneously with another non-silent RAV in Rv0678. The 37 isolates with only a mutation at position −11 were associated with low to moderate bedaquiline MICs (mean MIC 0.015 mg/L). An IS6110 insertion at position −13 in the intergenic region was present in combination with a missense mutation in Rv0678 in one isolate with a high MIC. The heterogeneous nature of clinical isolates explains why in some cases mixtures of WT sequences and RAVs were observed (nine isolates, 2.6%). Silent mutations in Rv0678 were observed in 1% of the isolates.
To investigate whether the presence of Rv0678 RAVs at baseline was associated with a specific lineage of M. tuberculosis, the presence of lineage-specific SNPs was analysed.16 The majority of the isolates belonged to lineage 2 (167/347, 48.1%) and lineage 4 (159/347, 45.8%), and only a few to lineage 1 (13/347, 3.7%) and lineage 3 (8/347, 2.3%). Baseline isolates carrying Rv0678 RAVs, including the intergenic region, were more abundant in lineage 2 (54/167, 32.3%). Interestingly, all the isolates containing the same mutation at position −11 in the intergenic region mmpS5–Rv0678 belonged to the Beijing family of lineage 2, which suggests that all these might be phylogenetically close. Out of the 54 isolates with Rv0678 RAVs, 19 contained a missense mutation or an intergenic mutation different from −11 C→A in Rv0678, either alone or in a mixture. Rv0678 RAVs were less frequent in lineage 3 (1/8, 12.5%) and lineage 4 (7/159, 4.4%). None of the 13 isolates belonging to lineage 1 had Rv0678 RAVs.
Out of the eight isolates with high baseline MICs (> 0.24 mg/L), five belonged to lineage 2 and three belonged to lineage 4. Low bedaquiline MICs were more frequent in lineage 2 (34/43) and normally associated with the Rv0678 RAV in position −11.
Non-MDR-TB isolates
Sequencing data were available for 941 isolates from non-MDR-TB patients of the Hamburg cohort. Read depth coverage resulting from mapping to the reference H37Rv genome averaged at 154 × (range 34 × to 740 ×) across isolates. Based on genomic SNP data, 852 isolates (90.5%) were pan-susceptible to all drugs included in the PhyResSE resistance list (fluoroquinolones, ethionamide, rifampicin, streptomycin, linezolid, amikacin, kanamycin, ciprofloxacin, isoniazid, pyrazinamide, p-aminosalicylic acid and ethambutol). Isolates with predicted drug resistance were usually resistant to isoniazid (66 isolates). Six isolates had RAVs in Rv0678 (0.7% prevalence, 95% CI 0.1%–1.2%), although no other drug resistance was detected in these isolates. These isolates were not clustered in a transmission chain (i.e. > 12 SNPs from each other22) and were from lineages 1 (2/54 isolates), 3 (2/65 isolates) and 4 (2/704 isolates) (Figure 1).
Cross-resistance of Rv0678 mutants with other TB drugs and potential selection pressures
The high prevalence of Rv0678 RAVs in baseline samples from patients without prior treatment with bedaquiline or clofazimine suggests that there is selection pressure favouring Rv0678 RAVs. The fact that many Rv0678 RAVs are present in a mixture with WT Rv0678 suggests that this selection pressure is mild, which is in accordance with a moderate fold increase in the MIC of the drug. We determined the susceptibility of BCA8, an Rv0678 mutant strain derived from H37Rv,1 to a panel of first- and second-line TB drugs to identify additional cross-resistance with bedaquiline via the same mechanism (Table 2). Bedaquiline and clofazimine had increased MICs for the Rv0678 mutant strain. Telithromycin and rifampicin also displayed slightly increased MICs for the Rv0678 mutant (5- and 3-fold, respectively), although the results were not statistically significant. We hypothesized that Rv0678 RAVs, leading to overexpression of the MmpL5 efflux pump, could benefit the survival of MDR strains in the presence of rifampicin (see the Discussion section). To test this hypothesis, we looked at the competitive advantage of the double mutant Rv0678-rpoB over the single rpoB mutant in the presence of different concentrations of rifampicin (ranging from 0.06 to 200 mg/L) through several generations. If the double mutant had a competitive advantage in the presence of rifampicin, then this population would be enriched in every generation. As can be observed in Figure S1, we found the opposite effect: the population of the double mutant Rv0678-rpoB decreased in every passage. Rifampicin is apparently not able to select for Rv0678 RAVs in an rpoB mutant background, and is therefore unlikely to be responsible for the higher prevalence of Rv0678 RAVs in MDR baseline isolates.
Table 2.
MIC50s of a panel of first- and second-line TB drugs for H37Rv and BCA8
| Compound | MIC50 (mg/L) (n) |
Fold change in MIC50 BCA8/H37Rv | P | |
|---|---|---|---|---|
| H37Rv | BCA8 | |||
| Bedaquiline base | 0.04 (11) | 0.31 (10) | 7.77* | < 0.0004 |
| Bedaquiline fumarate | 0.04 (6) | 0.28 (5) | 6.74* | < 0.0004 |
| Clofazimine | 0.11 (7) | 0.38 (5) | 3.52* | < 0.0008 |
| Thioridazine | 6.44 (6) | 10.09 (2) | 1.57* | 0.04084 |
| Telithromycin | 2.28 (7) | 11.26 (5) | 4.94 | > 0.05 |
| Rifampicin | 0.01 (13) | 0.02 (10) | 2.86 | > 0.05 |
| Amoxicillin/clavulanic acid 2/1 | 5.73 (6) | 9.61 (4) | 1.68 | > 0.05 |
| Rifapentin | ≤ 0.005 (7) | 0.01 (5) | ≥ 1.57 | > 0.05 |
| Ethambutol | 1.81 (7) | 2.38 (5) | 1.31 | > 0.05 |
| Moxifloxacin | 0.07 (7) | 0.08 (5) | 1.24 | > 0.05 |
| Erythromycin | 5.26 (7) | ≥ 6.36 (5) | ≥ 1.21 | > 0.05 |
| Clarithromycin | 0.6 (12) | 0.66 (10) | 1.09 | > 0.05 |
| Rifabutin | ≤ 0.003 (6) | 0.003 (2) | ≥ 1.01 | > 0.05 |
| Amikacin | 0.39 (6) | 0.36 (5) | 0.93 | > 0.05 |
| Ofloxacin | 0.32 (7) | 0.29 (5) | 0.92 | > 0.05 |
| Terizidone | 4.32 (6) | 3.76 (2) | 0.87 | > 0.05 |
| Levofloxacin | 0.22 (13) | 0.18 (10) | 0.79 | > 0.05 |
| Linezolid (batch 2) | 0.37 (7) | 0.29 (5) | 0.78 | > 0.05 |
| Capreomycin | 0.61 (7) | 0.45 (5) | 0.74 | > 0.05 |
| Linezolid (batch 1) | 0.47 (6) | 0.30 (5) | 0.62 | > 0.05 |
| p-Aminosalicylic acid | 0.02 (7) | 0.01 (5) | 0.5 | > 0.05 |
| Streptomycin | 0.33 (6) | 0.16 (2) | 0.48 | > 0.05 |
| Isoniazid | 0.12 (7) | 0.05 (4) | 0.46 | > 0.05 |
| Protionamide | 2.11 (5) | 0.82 (1) | 0.39 | > 0.05 |
The MIC50s for the WT (H37Rv) and an Rv0678 mutant (BCA8) are shown.
The number of times that the compound was tested is indicated in parentheses.
The MIC50 fold changes are calculated based on unrounded values.
Fold changes based on censored values become themselves censored.
The MIC50 fold changes that are statistically significant (P < 0.05) are marked with an asterisk.
Pyrazinamide was tested using the MGIT-PZA kit; both BCA8 and H37Rv were susceptible to this drug.
Discussion
Of the MDR-TB isolates, 2.3% (8/347) had bedaquiline MICs > 0.24 mg/L. All high baseline MICs could be explained by RAVs in Rv0678, and none of these isolates had an RAV in atpE or pepQ. RAVs in Rv0678 were not only identified in isolates with high baseline MICs: 4.3% of the isolates had similar RAVs (some with insertions or deletions), not resulting in bedaquiline MICs exceeding the bedaquiline breakpoint, suggesting a role of additional genes in determining the bedaquiline MIC. In total, RAVs in Rv0678 were observed in 6.6% of the MDR-TB isolates and they were associated with bedaquiline MICs exceeding the breakpoint in only 1 out of 3 cases (8 out of 23).
Intergenic mutations were observed in < 1% of the isolates and generally were not associated with high bedaquiline MICs. RAVs in the intergenic region between Rv0678 and mmpS5 at positions −9 and −10 in the hypothetical binding site of Rv0678 were previously shown to increase the expression of the MmpL5 efflux pump.21 Only one mutation was found at this binding site, and it was combined with another mutation in the gene itself, both probably contributing to the high bedaquiline MIC for the isolate. RAVs at −44 and −59, outside of the Rv0678 binding site, were identified in < 1.0% of the isolates, but did not result in high bedaquiline MICs (0.06–0.24 mg/L). RAVs at intergenic position −11 have not been described before and were observed in 10.7% of the 347 MDR-TB isolates in our sample. Interestingly, RAVs at this position were only observed in isolates from (several locations in) South Africa and they all belong to the W-Beijing lineage. They were associated with low to moderate bedaquiline MICs (0.004–0.03 mg/L, mean MIC 0.015 mg/L).
In 2.6% of the isolates we found a mixture of WT and mutant Rv0678. Mixtures could be the result of either spontaneous mutations that were further selected, or a mixed infection with two strains. Since the Sanger sequencing technique does not provide this information, we performed MIRU-VNTR typing for the mixtures and found no evidence of mixed infection, so the Rv0678 mutants were a subpopulation that was selected within the patient.
The prevalence of Rv0678 RAVs in pan-susceptible TB isolates from the population-based Hamburg cohort was 9-fold lower compared with the MDR-TB cohort, with only 6 sequences out of the 852 (0.7%) having a polymorphism in Rv0678. Bedaquiline MICs were not available for this data set.
The high prevalence of Rv0678 RAVs in baseline isolates of MDR-TB patients is surprising, given that none of these patients had been exposed to bedaquiline; nor was there any evidence for prior use of clofazimine in these patient’s medical histories (except for seven subjects). Although one cannot exclude that some of these patients were originally infected with strains already resistant to clofazimine, several factors argue against this possibility. Firstly, Rv0678 RAVs were not identified in any of the seven patients that had failed previous therapy with a regimen containing clofazimine prior to enrolment in the bedaquiline study. Secondly, the prevalence of Rv0678 RAVs was not higher in treatment-experienced MDR-TB patients or in pre-XDR- and XDR-TB patients.
Overall, Rv0678 RAVs lead to low-level resistance, and the effect on treatment outcome is not clear. Several letters and papers reported on a single patient developing an Rv0678 RAV while failing bedaquiline therapy and suggested a causal link between the RAV and the treatment failure,23–25 but in a clinical trial 12 patients whose isolates had acquired an Rv0678 RAV at the interim analysis were as likely to have converted at endpoint as those that had not developed an Rv0678 RAV.7 Although these data are limited, they do not suggest that increases in bedaquiline MICs on treatment and due to Rv0678 RAVs have a negative effect on treatment response. There was also no clear relationship between bedaquiline baseline MIC and culture conversion at the endpoint, although more efficacy data are needed for patients with baseline MICs above the provisional breakpoint. Efficacy studies in a mouse model of TB infection indicate that bedaquiline treatment is still bactericidal for such isolates, although to a lesser extent compared with the effect seen when treating an isolate without an Rv0678 RAV.1
The much lower frequency of Rv0678 RAVs in sequences from non-MDR-TB patients suggests a role of prior exposure to TB drugs. As all RAVs described in this article were isolated from patients prior to the introduction of bedaquiline, prior exposure to clofazimine was the most obvious hypothesis to explain their origin. However, none of the seven patients with prior documented use of clofazimine in this study had developed RAVs in Rv0678, and use of clofazimine in patients has not yet been reported to select for Rv0678 RAVs by other investigators. In view of these findings, prior exposure to clofazimine should not automatically lead to exclusion of bedaquiline treatment. Rv0678 RAVs did not occur more frequently in treatment-experienced MDR-TB isolates or in isolates from pre-XDR- or XDR-TB patients, providing further evidence against a role of clofazimine.
To rule out that other TB drugs than clofazimine were responsible for the selection of Rv0678 RAVs, an H37Rv isolate susceptible to first- and second-line anti-TB drugs, yet overexpressing the MmpL5 efflux pump, was used to assess any cross-resistance with a large panel of TB drugs. The only MICs that were significantly increased (> 3-fold, P < 0.05) in this strain were those of bedaquiline and clofazimine. Rifampicin turned out to be the most suspicious candidate of the list, being a first-line drug with a slightly increased MIC (2.9-fold) in the mutant overexpressing the efflux pump. In addition, increased expression of this efflux pump was described in a rifampicin-resistant (rpoB mutant) strain upon exposure to subinhibitory levels of rifampicin.26 We therefore investigated whether rifampicin could select for Rv0678 mutations in rpoB mutants by assessing the competitive advantage of the double mutant Rv0678-rpoB over the single rpoB mutant, in the presence of different concentrations of rifampicin. Since the double mutant did not grow better than the single mutant in the presence of the drug, rifampicin is unlikely to be responsible for the high prevalence of Rv0678 RAVs in MDR-TB baseline isolates.
The natural function of MmpS5/MmpL5 efflux pumps is to export siderophores.27 Expression of the efflux pump is down-regulated in high iron conditions,28 and one can expect up-regulation in low iron conditions. We checked whether the prevalence of Rv0678 RAVs was higher in isolates from female patients, but this did not appear to be the case. Iron-limiting conditions are also thought to exist in granulomatous lesions of TB. The origin of these ‘naive’ Rv0678 RAVs is therefore elusive at this point in time.
There was no evidence for Rv0678 RAVs being linked to specific TB lineages, as they were observed in lineages 2, 3 and 4. In contrast, RAVs in the intergenic region at position −11 were exclusively observed in W-Beijing isolates.
Our study has several implications for bedaquiline DST development. The absence of an algorithm to predict bedaquiline MICs based on specific Rv0678 RAVs limits the value of sequencing this specific gene—RAVs in Rv0678 were associated with bedaquiline MICs exceeding the breakpoint in only 8 out of 23 cases. Sequencing Rv0678 could be useful to rule out resistance to bedaquiline, but not to rule in resistance. Phenotypic DST tests remain the method of choice to monitor bedaquiline susceptibility
The prevalence of high baseline MICs of bedaquiline in 2.3% of patients without prior exposure to bedaquiline or clofazimine is worrisome and further surveillance studies are required to explain their origin, and to generate more data on their impact on treatment outcomes. The frequency of target-based (atpE) and non-target-based (Rv0678) RAVs for bedaquiline was similar in a set of in vitro selected isolates,4 but the latter are more likely to emerge first, as efflux-based mutations generally lead to lower levels of resistance (2- to 16-fold higher MICs) than target-based mutations (16- to 1000-fold higher MICs). This increases the likelihood that non-target-based resistance will be used as a stepping stone eventually leading to higher-level, target-based resistance in patients on a failing treatment regimen.
Supplementary Material
Acknowledgements
We thank John Mckinney and Neeraj Dhar (École Polytechnique Fédérale de Lausanne) for providing pND235 and pND239, Peggy Janssens (Janssen) for generating MIC data, Walter Van den Broeck (Janssen) for statistical analysis, Frederik Stevenaert (Janssen) for assistance with the flow cytometer, Pim de Rijk (Institute of Tropical Medicine) for molecular analyses, Stefan Niemann (National Reference Center for Mycobacteria, Borstel, Germany) and Roland Diel and Thomas Kohl (Schleswig-Holstein University Hospital, Kiel, Germany) for access to the extensive Hamburg dataset of non-MDR-TB sequences.
Funding
This work was supported by Janssen Pharmaceutica. N. C. was supported by a Baekeland PhD scholarship from the Flemish Institute for Scientific Technology (IWT 130308, Belgium). C. J. M., L. R. and B. d. J. were supported by a European Research Council Starting Grant INTERRUPTB (311725).
Transparency declarations
C. V., N. L. and K. A. are employees of Janssen Pharmaceutica. N. L. and K. A. hold shares of Johnson and Johnson. All other authors: none to declare.
Supplementary data
Tables S1 to S3 and Figure S1 are available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
References
- 1. Andries K, Villellas C, Coeck N. et al. Acquired resistance of Mycobacterium tuberculosis to bedaquiline. PLoS One 2014; 9: e102135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andries K, Verhasselt P, Guillemont J. et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 2005; 307: 223–7. [DOI] [PubMed] [Google Scholar]
- 3. Hartkoorn RC, Uplekar S, Cole ST. Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob Agents Chemother 2014; 58: 2979–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huitric E, Verhasselt P, Koul A. et al. Rates and mechanisms of resistance development in Mycobacterium tuberculosis to a novel diarylquinoline ATP synthase inhibitor. Antimicrob Agents Chemother 2010; 54: 1022–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Segala E, Sougakoff W, Nevejans-Chauffour A. et al. New mutations in the mycobacterial ATP synthase: new insights into the binding of the diarylquinoline TMC207 to the ATP synthase C-ring structure. Antimicrob Agents Chemother 2012; 56: 2326–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Almeida D, Ioerger T, Tyagi S. et al. Mutations in pepQ confer low-level resistance to bedaquiline and clofazimine in Mycobacterium tuberculosis. Antimicrob Agents Chemother 2016; 60: 4590–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pym AS, Diacon AH, Tang SJ. et al. Bedaquiline in the treatment of multidrug- and extensively drug-resistant tuberculosis. Eur Respir J 2016; 47: 564–74. [DOI] [PubMed] [Google Scholar]
- 8. Diacon AH, Pym A, Grobusch MP. et al. Multidrug-resistant tuberculosis and culture conversion with bedaquiline. N Engl J Med 2014; 371: 723–32. [DOI] [PubMed] [Google Scholar]
- 9. Torrea G, Coeck N, Desmaretz C. et al. Bedaquiline susceptibility testing of Mycobacterium tuberculosis in an automated liquid culture system. J Antimicrob Chemother 2015; 70: 2300–5. [DOI] [PubMed] [Google Scholar]
- 10. Palomino JC, Martin A, Camacho M. et al. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 2002; 46: 2720–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. EUCAST. Bedaquiline Breakpoint http://www.eucast.org/ast_of_mycobacteria/.
- 12. Cole ST, Brosch R, Parkhill J. et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998; 393: 537–44. [DOI] [PubMed] [Google Scholar]
- 13. Walker TM, Kohl TA, Omar SV. et al. Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect Dis 2015; 15: 1193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seemann T. Snippy: Fast Bacterial Variant Calling From NGS Reads 2015. https://github.com/tseemann/snippy.
- 15. Feuerriegel S, Schleusener V, Beckert P. et al. PhyResSE: a web tool delineating Mycobacterium tuberculosis antibiotic resistance and lineage from whole-genome sequencing data. J Clin Microbiol 2015; 53: 1908–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stucki D, Malla B, Hostettler S. et al. Two new rapid SNP-typing methods for classifying Mycobacterium tuberculosis complex into the main phylogenetic lineages. PLoS One 2012; 7: e41253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wakamoto Y, Dhar N, Chait R. et al. Dynamic persistence of antibiotic-stressed mycobacteria. Science 2013; 339: 91–5. [DOI] [PubMed] [Google Scholar]
- 18. Manina G, Dhar N, McKinney JD. Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe 2015; 17: 32–46. [DOI] [PubMed] [Google Scholar]
- 19. Gullberg E, Cao S, Berg OG. et al. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog 2011; 7: e1002158.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schwebach JR, Jacobs WR, Jr, Casadevall A. Sterilization of Mycobacterium tuberculosis Erdman samples by antimicrobial fixation in a biosafety level 3 laboratory. J Clin Microbiol 2001; 39: 769–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Milano A, Pasca MR, Provvedi R. et al. Azole resistance in Mycobacterium tuberculosis is mediated by the MmpS5-MmpL5 efflux system. Tuberculosis (Edinb) 2009; 89: 84–90. [DOI] [PubMed] [Google Scholar]
- 22. Walker TM, Ip CL, Harrell RH. et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis 2013; 13: 137–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Somoskovi A, Bruderer V, Homke R. et al. A mutation associated with clofazimine and bedaquiline cross-resistance in MDR-TB following bedaquiline treatment. Eur Respir J 2015; 45: 554–7. [DOI] [PubMed] [Google Scholar]
- 24. Bloemberg GV, Keller PM, Stucki D. et al. Acquired resistance to bedaquiline and delamanid in therapy for tuberculosis. N Engl J Med 2015; 373: 1986–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoffmann H, Kohl TA, Hofmann-Thiel S. et al. Delamanid and bedaquiline resistance in Mycobacterium tuberculosis ancestral Beijing genotype causing extensively drug-resistant tuberculosis in a Tibetan refugee. Am J Respir Crit Care Med 2016; 193: 337–40. [DOI] [PubMed] [Google Scholar]
- 26. de Knegt GJ, Bruning O, ten Kate MT. et al. Rifampicin-induced transcriptome response in rifampicin-resistant Mycobacterium tuberculosis. Tuberculosis (Edinb) 2013; 93: 96–101. [DOI] [PubMed] [Google Scholar]
- 27. Wells RM, Jones CM, Xi Z. et al. Discovery of a siderophore export system essential for virulence of Mycobacterium tuberculosis. PLoS Pathog 2013; 9: e1003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fang Z, Sampson SL, Warren RM. et al. Iron acquisition strategies in mycobacteria. Tuberculosis (Edinb) 2015; 95: 123–30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.