Abstract
Messenger RNA polyadenylation is one of the key post-transcriptional events in eukaryotic cells. A large number of genes in mammalian species can undergo alternative polyadenylation, which leads to mRNAs with variable 3′ ends. As the 3′ end of mRNAs often contains cis elements important for mRNA stability, mRNA localization and translation, the implications of the regulation of polyadenylation can be multifold. Alternative polyadenylation is controlled by cis elements and trans factors, and is believed to occur in a tissue- or disease-specific manner. Given the availability of many databases devoted to other aspects of mRNA metabolism, such as transcriptional initiation and splicing, systematic information on polyadenylation, including alternative polyadenylation and its regulation, is noticeably lacking. Here, we present a database named polyA_DB, through which we strive to provide several types of information regarding polyadenylation in mammalian species: (i) polyadenylation sites and their locations with respect to the genomic structure of genes; (ii) cis elements surrounding polyadenylation sites; (iii) comparison of polyadenylation configuration between orthologous genes; and (iv) tissue/organ information for alternative polyadenylation sites. Currently, polyA_DB contains 45 565 polyadenylation sites for 25 097 human and mouse genes, representing the most comprehensive polyadenylation database till date. The database is accessible via the website (http://polya.umdnj.edu/polyadb).
INTRODUCTION
Polyadenylation of eukaryotic mRNAs is a two-step process, which includes a specific cleavage at the 3′ end of a nascent mRNA and addition of a poly(A) tail (1). Polyadenylation has impacts on many aspects of mRNA metabolism in the cell, including mRNA stability, mRNA localization and translation (2). Enhanced efficiency of polyadenylation can lead to diseases such as thrombophilia (3), highlighting the importance of the regulation of polyadenylation.
Both cis elements and trans factors are involved in the regulation of polyadenylation. Cis elements can be divided into two groups based on their relative location to the cleavage site, namely upstream and downstream elements. The most well-known upstream element in metazoan cells is the polyadenylation signal (PAS) located 10–30 nt upstream of the cleavage site. Although the AAUAAA hexamer is the most common PAS, 11 single-nucleotide variants have been demonstrated or suggested to play similar roles in polyadenylation (4). PAS motifs are the binding sites for the cleavage–polyadenylation specificity factor (CPSF). U-rich and GU-rich elements are common downstream elements located 20–40 nt downstream of the cleavage site (5–7). They are the binding sites for the cleavage stimulatory factor (CstF) (8). In addition, sequence composition surrounding the cleavage site has been found to be important for defining the site in several bioinformatics studies (6,9–11). Other factors responsible for the polyadenylation process include cleavage factors I and II (CFI and CFII), and poly(A) polymerase (PAP) (12,13). Recently, several transcriptional factors and the RNA polymerase II enzyme have also been implicated in the polyadenylation process (14). Moreover, various auxiliary elements of viral or cellular origins have been shown to regulate polyadenylation (12).
It has been estimated that more than 29% of human genes have alternative polyadenylation sites [or poly(A) sites] (15). The choice of alternative poly(A) sites is believed to be related to biological conditions such as cell type and disease state (16). Alternative polyadenylation can lead to mRNAs with variable 3′ ends, or proteins with different C-termini. A growing number of genes have been found to be regulated by this mechanism. However, a public database systematically providing information on alternative polyadenylation is lacking. The availability of genomic sequences from several mammalian species as well as large numbers of expressed sequence tags (ESTs) make it feasible to comprehensively document mRNA polyadenylation configurations for genes. Although ESTs provide both sequence data and information on the biological origin of transcripts by the means of cDNA library source, they have several problems with respect to data quality, such as chimeric sequence, vector contamination and inclusion of genomic sequence. In addition, when dealing with polyadenylation, issues such as internal priming and low-quality sequences at the 5′ and 3′ ends are more palpable. Therefore, a computational approach to studying polyadenylation must take these into consideration to ensure that poly(A) sites are accurately mapped. We present here a computational pipeline that effectively utilizes genomic sequences and EST data to study polyadenylation. We applied it to human and mouse genes to build a database for polyadenylation (named polyA_DB). Currently, the database documents 45 565 poly(A) sites and various information regarding the sites, including their genome locations, evidence of cDNA/EST alignments to genomes, cis elements surrounding poly(A) sites, comparison of polyadenylation configuration between orthologs and tissue/organ information for poly(A) sites. Although only human and mouse poly(A) sites are currently documented in the database, the data process pipeline and the structure of the database are designed so as to make it easy to include other species in the future. This resource can be of great value to researchers interested in studying both the mechanism of polyadenylation and the gene regulation by alternative polyadenylation.
RESULTS
Methods and data statistics
In the polyA_DB database, genes are annotated based on LocusLink IDs (17). A computational pipeline (depicted in Figure 1) is designed to accurately map poly(A) sites on genomes:
The genomic location and the structure of a gene is determined by the alignment of its RefSeq sequence(s) and genome contigs. In the current version, human genome Build 34.2, mouse genome Build 30 and NCBI March 2004 release of RefSeq mRNAs were used. If a gene has more than one RefSeq sequence, their alignments to genomes are required to overlap, and their transcriptional orientations are required to be the same. The transcriptional orientation of a gene is determined both by its splicing and poly(A) tail information whenever possible. If a gene does not meet these two criteria, it is discarded.
To ensure that only high-quality cDNA/EST data are used, the alignment of a cDNA/EST with the genome is required to overlap with that of its corresponding RefSeq. This resulted in the discarding of 7.8% human and 10.4% mouse cDNA/ESTs that contained poly(A) or poly(T) tails, respectively. The mapping between cDNA/EST and RefSeq was obtained from the UniGene database (18).
Only those cDNA/ESTs with poly(A) tails [or poly(T) tails if in anti-sense orientation] are used to infer poly(A) cleavage sites. Poly(A) tails are required to have either eight or more consecutive As; if it has a nucleotide other than A, another eight or more consecutive As after that nucleotide are required. Possible internal priming sites are checked by examining the genomic sequence −10 to +10 nt surrounding the cleavage site. If the sequence has six continuous As or more than seven As in a 10 nt window, it is considered as an internal priming candidate. Poly(A) cleavage sites located within a 24 nt window are considered to be generated from heterogeneous cleavage of mRNA (19), and thus are clustered together.
To further ensure the mapping quality, a genuine polyadenylation site must be either supported by more than one cDNA/EST sequence, or supported by one cDNA/EST alignment together with at least one PAS within the upstream −40 to −1 nt region. An internal priming candidate can be considered as a genuine site if and only if it is supported by more than one cDNA/EST and has an upstream PAS.
Data generated from the pipeline, including genomic locations of poly(A) sites, supporting cDNA/ESTs, number of cleavage sites and PAS information are stored in a relational database using MySQL. Also in the database are the ortholog information of genes obtained from HomoloGene database (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=homologene), and the tissue/organ information of ESTs derived from cDNA library files from NCBI. Several key data statistics of the database are summarized in Table 1.
Figure 1.
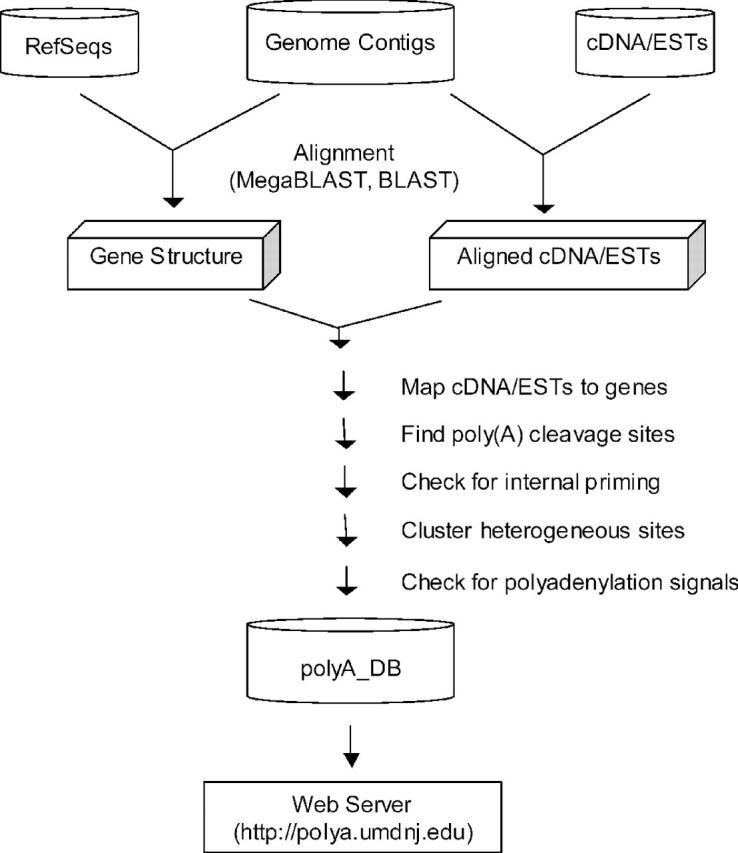
An outline of the polyA_DB building pipeline. The data flow is indicated by arrowed lines. See the main text for details.
Table 1. PolyA_DB statistics.
| Homo sapiens | Mus musculus | Total | |
|---|---|---|---|
| Aligned cDNA/ESTs | 2 103 995 | 1 181 194 | 3 285 189 |
| Poly(A) sites | 29 283 | 16 282 | 45 565 |
| Genes with one poly(A) site | 6418 | 7577 | 13 995 |
| Genes with alternative poly(A) sites | 7524 | 3578 | 11 102 |
| Total genes | 13 942 | 11 155 | 25 097 |
| Orthologous pairs | 7935 | 7935 | 7935 |
| Tissue typesa | 331 | 155 | 455 |
| Organ typesa | 107 | 47 | 133 |
aIt includes diseased tissues and organs. Some tissue and organ types occur in both human and mouse cases.
Data access and visualization
Data and documentation are available from the polyA_DB web server at http://polya.umdnj.edu/polyadb. Data can be either downloaded as flat files or queried through a web interface where graphics are dynamically generated using Bioperl modules (20). The web user interface is interactive and provides five basic views:
Gene view
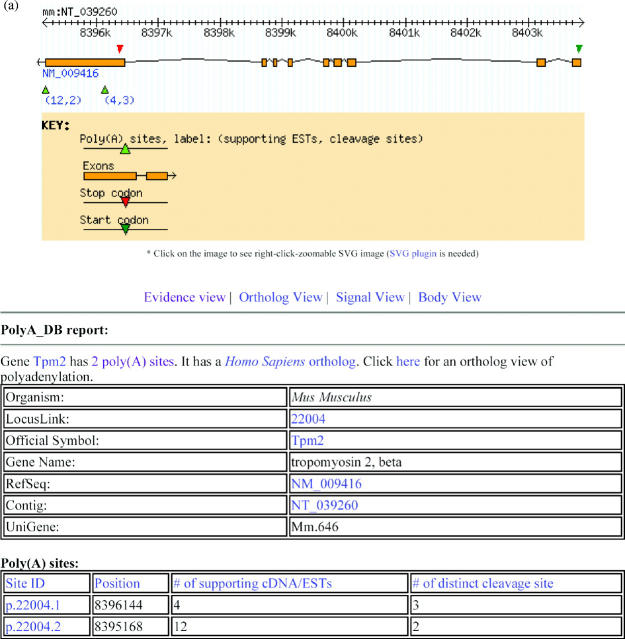
This view provides a summary of poly(A) site(s) for each queried gene, their positions relative to the RefSeq(s) and the genome contig. The gene structure inferred from the RefSeq(s) and a summary table containing cDNA/EST evidence and number of cleavage sites are also provided. Links for sequence IDs to NCBI resources are provided whenever possible. Figure 2A shows a polyA_DB gene view of mouse TPM2 gene (β-tropomyosin, LocusLink ID: 22004), with two poly(A) sites and their positions, inferred gene structure, start and stop codon positions, number of supporting ESTs and number of sites generated by heterogeneous cleavage.
Figure 2.

Views offered at the web interface of polyA_DB. (A) Gene view: Mouse gene TPM2 is used as an example. The output includes a pictorial representation of gene structure and poly(A) sites as well as two summary tables regarding the gene and the poly(A) sites. Numbers under each poly(A) site in the picture are the number of supporting cDNA/ESTs and the number of heterogeneous cleavage sites. (B) Ortholog view of human and mouse TPM2 genes. (C) Signal views of mouse and human TPM2 genes are shown in the upper panel and lower panel, respectively. The position of a signal is relative to the cleavage site, which is set to 0.
Evidence view
This view provides detailed alignment evidence from cDNA/EST sequences, which can be presented by various sorting options including the 3′ or 5′ position, exon number, cDNA/EST length and GenBank ID. A table is also provided, which lists all supporting cDNA/ESTs with links to NCBI (Supplementary Figure 1).
Ortholog view
This view provides a comparison of a pair of human and mouse orthologs. Figure 2B shows an ortholog view of mouse TPM2 gene and its human ortholog (LocusLink ID: 7169). The ortholog view readily revealed that the ortholog pair is conserved with respect to both their gene structures and polyadenylation configurations.
Signal view
This view provides information regarding cis elements in the surrounding region of a poly(A) site. Currently, we document the PAS motif (AAUAAA and its 11 single-nucleotide variants) (4) in the 1–40 nt upstream region of a poly(A) site. Figure 2B shows signal views of the mouse and human TPM2 genes, from which the conservation of signal usage of poly(A) sites can be easily discerned.
Body view
This view provides tissue/organ information for poly(A) sites (Supplementary Figure 2).
CONCLUSIONS
We present polyA_DB database—a resource for mammalian mRNA polyadenylation. This database contains comprehensive information regarding polyadenylation, including poly(A) sites in the context of gene structure, cDNA/EST evidence for poly(A) sites, PASs, conservation of polyadenylation configuration between orthologs and tissue/organ information for poly(A) site usage. We believe polyA_DB will be of great use to researchers studying both the mechanism of polyadenylation and the gene regulation by alterative polyadenylation. PolyA_DB will be continuously updated (i) when new releases of human and mouse genomes and cDNA/EST data are available, and (ii) genome and cDNA/EST data from other species are available for large-scale polyadenylation studies.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Carol S. Lutz at UMDNJ and two anonymous reviewers for their valuable suggestions, and Stephen B. Feldman at UMDNJ for technical assistance with the web server.
REFERENCES
- 1.Colgan D.F. and Manley,J.L. (1997) Mechanism and regulation of mRNA polyadenylation. Genes Dev., 11, 2755–2766. [DOI] [PubMed] [Google Scholar]
- 2.Lewis J.D., Gunderson,S.I. and Mattaj,I.W. (1995) The influence of 5′ and 3′ end structures on pre-mRNA metabolism. J. Cell Sci. Suppl., 19, 13–19. [DOI] [PubMed] [Google Scholar]
- 3.Gehring N.H., Frede,U., Neu-Yilik,G., Hundsdoerfer,P., Vetter,B., Hentze,M.W. and Kulozik,A.E. (2001) Increased efficiency of mRNA 3′ end formation: a new genetic mechanism contributing to hereditary thrombophilia. Nature Genet., 28, 389–392. [DOI] [PubMed] [Google Scholar]
- 4.Beaudoing E., Freier,S., Wyatt,J.R., Claverie,J.M. and Gautheret,D. (2000) Patterns of variant polyadenylation signal usage in human genes. Genome Res., 10, 1001–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zarudnaya M.I., Kolomiets,I.M., Potyahaylo,A.L. and Hovorun,D.M. (2003) Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res., 31, 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Legendre M. and Gautheret,D. (2003) Sequence determinants in human polyadenylation site selection. BMC Genomics, 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takagaki Y. and Manley,J.L. (1997) RNA recognition by the human polyadenylation factor CstF. Mol. Cell Biol., 17, 3907–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacDonald C.C., Wilusz,J. and Shenk,T. (1994) The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol. Cell Biol., 14, 6647–6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabaska J.E. and Zhang,M.Q. (1999) Detection of polyadenylation signals in human DNA sequences. Gene, 231, 77–86. [DOI] [PubMed] [Google Scholar]
- 10.Hajarnavis A., Korf,I. and Durbin,R. (2004) A probabilistic model of 3′ end formation in Caenorhabditis elegans. Nucleic Acids Res., 32, 3392–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graber J.H., Cantor,C.R., Mohr,S.C. and Smith,T.F. (1999) In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl Acad. Sci. USA, 96, 14055–14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao J., Hyman,L. and Moore,C. (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev., 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Proudfoot N. (1996) Ending the message is not so simple. Cell, 87, 779–781. [DOI] [PubMed] [Google Scholar]
- 14.Calvo O. and Manley,J.L. (2003) Strange bedfellows: polyadenylation factors at the promoter. Genes Dev., 17, 1321–1327. [DOI] [PubMed] [Google Scholar]
- 15.Beaudoing E. and Gautheret,D. (2001) Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res., 11, 1520–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwalds-Gilbert G., Veraldi,K.L. and Milcarek,C. (1997) Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res., 25, 2547–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wheeler D.L., Church,D.M., Federhen,S., Lash,A.E., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Sequeira,E., Tatusova,T.A. et al. (2003) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 31, 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pauws E., van Kampen,A.H., van de Graaf,S.A., de Vijlder,J.J. and Ris-Stalpers,C. (2001) Heterogeneity in polyadenylation cleavage sites in mammalian mRNA sequences: implications for SAGE analysis. Nucleic Acids Res., 29, 1690–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stajich J.E., Block,D., Boulez,K., Brenner,S.E., Chervitz,S.A., Dagdigian,C., Fuellen,G., Gilbert,J.G., Korf,I., Lapp,H. et al. (2002) The Bioperl toolkit: Perl modules for the life sciences. Genome Res., 12, 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.