Sir,
Carbapenems are declared as ‘critically important’ antibiotics by the WHO.1 Recently, carbapenemase-producing Enterobacteriaceae (CPE) arose as a major concern in human medicine, where they are increasingly isolated from patients in hospitals. However, they are only sporadically reported from non-human sources.2,3 In Europe, carbapenems are not licensed for veterinary use and no maximum residue levels are defined. Therefore, the isolation of VIM-1-producing Escherichia coli and Salmonella spp. in 2011–12 from German swine and poultry farms raised concerns that livestock might emerge as a reservoir for CPE and that pan-resistant isolates could be transmitted from this reservoir to humans.4 In fact, spread and persistence of the β-lactamase gene (bla) VIM-1 over a whole fattening period was demonstrated in the VIM-1-positive swine farm. Here we describe the presence of VIM-1-positive E. coli isolates that appear closely related to the isolates from 2011, isolated in December 2015 from a different German swine farm.4,5
In Germany, monitoring of antimicrobial resistance (AMR) in commensal E. coli is linked to the national monitoring of zoonotic agents.6 According to the Commission Implementing Decision 2013/652/EU, susceptibility is determined using the microdilution method following CLSI guidelines (CLSI M07-A9). Isolates with an ESBL/AmpC phenotype or phenotypic resistance to carbapenems are further characterized by PCR and subsequent sequencing of the PCR products.5,7
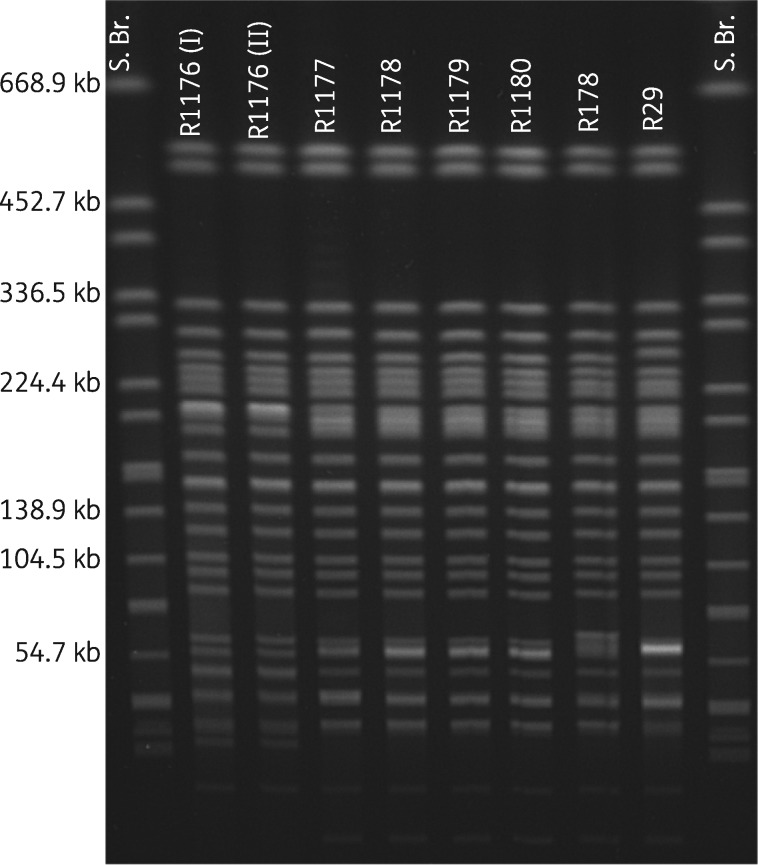
So far, no carbapenemase-producing E. coli isolates had been recorded within the scope of the national monitoring programmes. This study describes the first detection of a commensal E. coli isolate showing resistance to meropenem (MIC ≥0.5 mg/L), ertapenem (MIC ≥0.12 mg/L) and imipenem (MIC ≥2 mg/L) (isolate R1176, isolated in December 2015 from the colon content of a slaughter pig) within the monitoring programme. PCR and subsequent sequencing analysis revealed the presence of a blaVIM-1 gene. XbaI PFGE revealed a highly similar restriction pattern of R1176 to E. coli isolates described by Fischer et al.4,5 from samples collected in 2011 from a swine farm (Figure 1; R29 and R178). This indicates a clonal relationship of these VIM-1-positive E. coli, although the affected livestock farms are regionally clearly separated. In contrast to E. coli isolates R29 and R178, in R1176 neither the blaACC-1 gene nor the typical 220 kb VIM-1 plasmid of the former isolates was detected through PCR and S1 nuclease PFGE (Figure S1, available as Supplementary data at JAC Online). Supported by the failure of blaVIM-1 hybridization experiments on S1 nuclease PFGE (Figure S2) and transformation experiments, a chromosomal location of the blaVIM-1 gene in R1176 is assumed. This might be driven by an association of the blaVIM-1 gene with mobile genetic elements, as described for R178 as well.
Figure 1.
XbaI PFGE analysis. PFGE plugs of R1176 I and II were cast independently to verify reproducibility. R1176 was isolated in December 2015, R1177–R1180 were isolated in March 2016, and R178 and R29 are representatives of E. coli isolated in 2011–12. S. Br., Salmonella Braenderup (H9812) as size marker.
To verify a potential clonal persistence of VIM-1-positive E. coli within the farm that the pig originated from, in March 2016 colon content from five healthy animals in another slaughter batch from this farm was examined. This resulted in the isolation of four additional carbapenem-resistant E. coli (isolates R1177–R1180) from one of the samples. Again, XbaI PFGE patterns of these four isolates were very similar to those mentioned above (Figure 1). This hints at the presence of a specific clone on this farm and a link with isolates obtained from the farm investigated in 2011.4 S1 nuclease PFGE with subsequent hybridization experiments, described by Rodriguez et al.7 in 2009, revealed blaVIM-1 localization on 180–200 kb IncHI2 plasmids in all four isolates (Figure S1). And, indeed, the VIM-1-plasmid-harbouring isolates were also positive for the blaACC-1 gene, further resistance genes strA and strB and class-I-integron-associated resistance genes aadA1 and aacA4 and sul1, as shown for pRH-178, assuming the presence of a highly similar plasmid in these isolates compared with the ones from 2011.4,5
All five isolates described in this study belonged to ST88 and phylogenetic group A and harboured the blaVIM-1 gene on a class 1 integron with gene cassettes that were identical to those described for R29 and R178, independently of its localization on the plasmid or the chromosome.5
In the above-mentioned national AMR monitoring programmes no carbapenemase-producing E. coli had been detected until the end of 2015, indicating a very low prevalence of such bacteria in the German livestock population. However, detection of highly related VIM-1-producing E. coli isolates from an additional swine farm in Germany in this study indicates persistence of a VIM-1-producing E. coli clone in the swine population for at least 4 years. Further investigations on the persistence of this clone are currently under way. Detailed genomic analysis will be carried out to reveal a potential reason for stable maintenance of this clone and to uncover potential transmission pathways of these isolates. The understanding of transmission pathways and the persistence of CPEs among different populations to limit the spread of CPEs in livestock is of major relevance for public health. Finally, results of this study underline the importance of the carbapenemase monitoring recommended by the European Food Safety Authority (EFSA) and the European Commission.
Supplementary Material
Acknowledgements
We gratefully acknowledge the support of the regional laboratories and authorities in collecting the samples and providing the isolates in the framework of the monitoring. We also thank the team of the Institute of Animal Hygiene and Environmental Health of the Free University of Berlin for providing reference strains R29 and R178 and Dr Jens Hammerl for scientific advice.
Funding
This work was supported by the Federal Institute for Risk Assessment (BfR) (BfR-43-001) and the RESET II Project (FKZ01KI1313B; German Federal Ministry for Education and Research).
Transparency declarations
None to declare.
Supplementary data
Figures S1 and S2 are available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
References
- 1. WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance. Critically Important Antimicrobials for Human Medicine, 3rd Revision 2011. Geneva, Switzerland: WHO, 2012. http://apps.who.int/iris/bitstream/10665/77376/1/9789241504485_eng.pdf. [Google Scholar]
- 2. Tangden T, Giske CG. Global dissemination of extensively drug-resistant carbapenemase-producing Enterobacteriaceae: clinical perspectives on detection, treatment and infection control. J Intern Med 2015; 277: 501–12. [DOI] [PubMed] [Google Scholar]
- 3. Guerra B, Fischer J, Helmuth R. An emerging public health problem: acquired carbapenemase-producing microorganisms are present in food-producing animals, their environment, companion animals and wild birds. Vet Microbiol 2014; 171: 290–7. [DOI] [PubMed] [Google Scholar]
- 4. Fischer J, San Jose M, Roschanski N. et al. Spread and persistence of VIM-1 carbapenemase-producing Enterobacteriaceae in three German swine farms in 2011 and 2012. Vet Microbiol 2016; doi:10.1016/j.vetmic.2016.04.026. [DOI] [PubMed] [Google Scholar]
- 5. Fischer J, Rodríguez I, Schmoger S. et al. Escherichia coli producing VIM-1 carbapenemase isolated on a pig farm. J Antimicrob Chemother 2012; 67: 1793–5. [DOI] [PubMed] [Google Scholar]
- 6. Kaesbohrer A, Schroeter A, Tenhagen B-A. et al. Emerging antimicrobial resistance in commensal Escherichia coli with public health relevance. Zoonoses Public Health 2012; 59Suppl 2: 158–65. [DOI] [PubMed] [Google Scholar]
- 7. Rodriguez I, Barownick W, Helmuth R. et al. Extended-spectrum β-lactamases and AmpC β-lactamases in ceftiofur-resistant Salmonella enterica isolates from food and livestock obtained in Germany during 2003–07. J Antimicrob Chemother 2009; 64: 301–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.