Abstract
Objectives: To investigate antimicrobial susceptibility in Staphylococcus pseudintermedius and the occurrence of methicillin-resistant S. pseudintermedius (MRSP), to explore the molecular structure of the MRSP population and to analyse risk factors for MRSP.
Methods: Susceptibility data for clinical S. pseudintermedius isolates in 2011–15 were analysed using WHONET. All MRSP isolates in 2010–14 (n = 362) were typed using PFGE. Representative isolates (n = 87) of clusters were analysed using MLST and staphylococcal cassette chromosome mec (SCCmec) typing. Risk factors were analysed using logistic regression.
Results: Of the clinical S. pseudintermedius (n = 1958; 98% from dogs), 14% were MRSP. Resistance to other antimicrobials varied between 12% and 39%. No trends were observed over time. Among clinical specimens (from infection sites) and screening specimens (from potential carriers), respectively, 2.5% (267/10 813) and 9% (211/2434) revealed MRSP. MLST revealed 42 different STs, including 19 new ones. Clonal complexes 71, 45 and 258 were the most common, but the MRSP population diversified over the years. A clinical S. pseudintermedius isolate was more likely to be MRSP if the patient was on antimicrobials at the time of sampling or was male. The presence of MRSP in screening specimens was more likely if the patient was on multiple antimicrobials at the time of sampling. Specimens from private clinics (versus the Veterinary Teaching Hospital of the University of Helsinki) had a higher likelihood of MRSP in both analyses.
Conclusions: Resistance to antimicrobials among S. pseudintermedius in Finland is high, emphasizing the importance of infection control measures and susceptibility testing prior to therapy. The diverse MRSP population indicates non-clonal spread.
Introduction
Staphylococcus pseudintermedius is a part of the normal microbiota of dogs and cats.1,2 This opportunistic bacterium can cause a wide range of infections, ranging from pyoderma and surgical wound infections to deep infections such as osteomyelitis.3–6 Over the past decade, methicillin-resistant S. pseudintermedius (MRSP) has become a worldwide problem in small animal veterinary medicine.7,8 Ever fewer effective antimicrobials are available for treatment, as MRSP isolates are commonly MDR.8–10
The epidemiology and molecular characteristics of MRSP in neighbouring countries, such as Sweden and Norway, have already been explored.11–13 There is evidence that the clonal structure of MRSP is changing in some countries.13,14 In Finland, a large veterinary hospital outbreak in 2010–11 was caused by the predominant European MRSP clone, ST71.15 The study also identified ST45 as causing a smaller cluster. These two main clones were additionally identified among Finnish guide dogs, but the presence of other unrelated STs indicated diversity in the MRSP population.16 However, no study has extensively explored the epidemiology of MRSP in Finland. The goal of this study was to investigate the epidemiology of S. pseudintermedius in Finland during 2010–15 by (i) reporting the antimicrobial susceptibility time trends of clinical isolates in 2011–15, (ii) exploring the molecular structure of the MRSP population in 2010–14, (iii) describing yearly MRSP proportions in clinical specimens and screening specimens from potential carriers and (iv) determining risk factors for MRSP among screening specimens and for a clinical S. pseudintermedius being MRSP.
Methods
Study setting
The investigation was carried out at the Clinical Microbiology Laboratory of the Faculty of Veterinary Medicine, University of Helsinki. In 2011–15, the laboratory processed 19 249 microbiological specimens.
Data collection
For analysis of antimicrobial susceptibility, results for all isolates identified as S. pseudintermedius15,17,18 between June 2011 and the end of 2015 were compiled from the laboratory information system (LIS) (Provet Net, Finnish Net Solutions, Finland). In addition, patient information data (species, sex, specimen type, presence of antimicrobial therapy at the time of sampling, and submitting clinic) were collected. Antimicrobials tested in the basic panel included clindamycin, erythromycin, fusidic acid, oxacillin, trimethoprim/sulfamethoxazole and tetracycline. Comparable data for the extended panel (amikacin, chloramphenicol, doxycycline, enrofloxacin and gentamicin) were only available for 2015. Susceptibility testing was performed by disc diffusion according to CLSI standards.19,20
Molecular methods
Suspected MRSP isolates, based on resistance to oxacillin,20 were confirmed to carry the mecA gene by PCR.16 All confirmed isolates were stored at −80°C until further studied. To detect clonal clusters, all MRSP isolates stored in 2010–14 were digested by SmaI macrorestriction (New England BioLabs, USA) and separated by PFGE21 with modifications.15 Isolates that were non-typeable by SmaI were digested using AscI (New England BioLabs, USA). PFGE clusters were illustrated with GelCompar II (v. 6.5; Applied Maths NV, Belgium) by UPGMA-based analysis using the Dice similarity coefficient with an 80% similarity cut-off; optimization and position tolerance were set at 1.2% and 1.3%, respectively. At least one isolate from each PFGE cluster was selected for further characterization by MLST and staphylococcal cassette chromosome mec (SCCmec) typing,22,23 with modifications.16 For some isolates, the MLST sequencing result for the tuf gene was poor with published primers.22 Therefore, the primers were modified as follows: tuf 19F, 5′-GTCCAATGCCACAAACTCG-3′; and tuf 19R, 5′-CCAGCTTCAGCGTAGTCTA-3′. MLST results for isolates were extrapolated to all members of the PFGE cluster. Isolates for which data had previously been published were included in the study.15,16
Data analysis
Susceptibility data for clinical S. pseudintermedius isolates (screening specimens excluded) were analysed using WHONET (v. 5.6, WHO). Non-susceptibility percentages, including resistant and intermediate isolates, with 95% CIs, were calculated and presented separately for MRSP, methicillin-susceptible S. pseudintermedius (MSSP) and all S. pseudintermedius isolates. CLSI breakpoints were used,19,20 except for fusidic acid, for which a non-susceptibility breakpoint of ≤23 mm was used.24 Yearly trends for non-susceptibility percentages were plotted and trends were investigated using a Cochran–Armitage trend test for each antimicrobial. The statistical difference in non-susceptibilities between MRSP and MSSP was investigated using Pearson’s χ2 test based on the WHONET output. P values <0.05 were considered statistically significant. Analyses were performed with the SAS® System for Windows (v. 9.3, USA). To calculate the number of MDR isolates (resistance to at least three antimicrobial classes), macrolide and lincosamide resistance was pooled due to common MLSB resistance (macrolide, lincosamide and streptogramin B).25
The proportions of MRSP in clinical and screening specimens were calculated for dogs and cats in order to derive crude prevalence estimates for MRSP in dogs and cats seeking veterinary care, from which microbiological specimens are obtained, and in high-risk populations, respectively. Patients in the latter populations have previously identified risk factors for MRSP, such as frequent antimicrobial exposure, chronic or intermittent infection, such as pyoderma, surgical site infection or previous exposure to MRSP (either in hospital or family).15 Screening is targeted at these patients.
To compare the genetic relatedness of STs, the allele sequences for each ST were added back-to-back (ack, cpn60, fdh, pta, purA, sar, tuf). The resulting sequences were aligned and a phylogenetic tree was inferred by the Bayesian Markov Chain Monte Carlo (MCMC) method implemented in BEAST (v. 1.7.2).26 Each run was continued until the effective sample size (ESS) was >200. Posterior probabilities were calculated with a 10% burn-in and values >0.7 were considered significant. Results were visualized in FigTree (v. 1.40). Additionally, goeBURST (v 1.2.1) software was used for population structure analysis of STs.27 Analysis was conducted at double- and triple-locus variant levels. Single- and double-locus variants of previously described clonal complexes (CCs) were assigned to that CC.14 The number of isolates per ST or CC per year was calculated based on the specimen collection date.
For analysis of predictors for MRSP, data were analysed separately for clinical S. pseudintermedius isolates and screening specimens by logistic regression with MRSP as the outcome variable. As data from the MRSP outbreak at the Veterinary Teaching Hospital of the University of Helsinki (VTH) in 2010–11 were likely to skew the results, data for this period were omitted.15 Due to the low number of cats in the data (n = 18 for clinical specimens and zero positive out of 145 for screening specimens), these, as well as specimens from unknown species (n = 11), were omitted from the analyses. ORs with 95% CI and P values were calculated for each variable. Variables with a P value ≤0.2 in the univariable analysis were included in the multivariable analysis. Multivariable logistic regression was performed using a backward step (Wald) method. P values <0.05 were considered statistically significant in the final model. Analyses were performed using SPSS v. 24 (IBM Inc.).
Results
Clinical S. pseudintermedius isolates
Results were available from a total of 1958 clinical S. pseudintermedius isolates. Of these, 1471 (75%) were from specimens from private clinics, while 487 were from the VTH. The isolates were mainly from dogs, comprising 1928 isolates (98%), while 18 isolates (0.9%) originated from cats. One S. pseudintermedius isolate was from a guinea pig. In 11 cases (0.6%), the species had not been recorded. The majority of specimens (n = 1507; 77%) were obtained from superficial sites, such as ears and skin, while 284 (15%) were from deep lesions (e.g. deep wounds, abscesses or synovial fluid). The bacterium was also recovered from urine (n = 98; 5%), respiratory specimens (n = 6; 0.3%) and blood cultures (n = 2; 0.1%). The rest (n = 61; 3%) were from cultured agar plates that had been sent to the laboratory for further testing.
Antimicrobial susceptibility and occurrence of MRSP
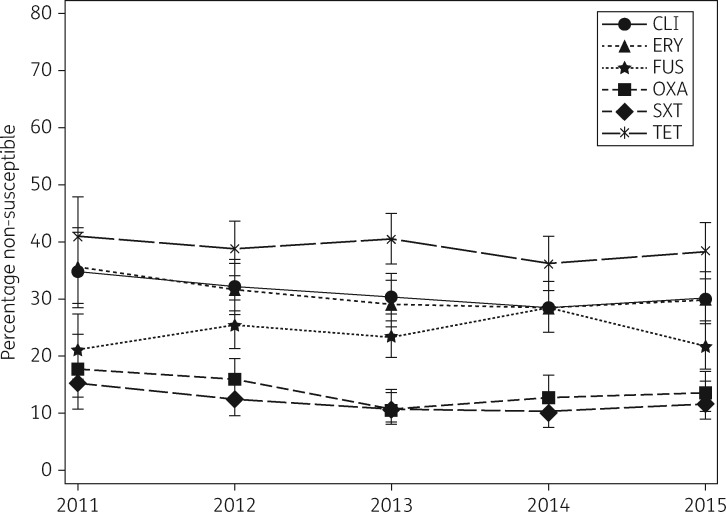
Out of the 1958 clinical S. pseudintermedius isolates, the overall proportion of oxacillin (methicillin) resistance was 14% (n = 266). Non-susceptibility data for all tested antimicrobials for MRSP, MSSP and overall are presented in Table 1. The proportion of non-susceptible isolates per year varied only slightly for each antimicrobial and no statistically significant trends were detected (Figure 1). A complete basic panel antibiogram was recorded for 1932 (99%) isolates. Of these, 17% (321 isolates) were MDR. The most common MDR profile (78/321; 24%) was lincosamides/macrolides–oxacillin –trim ethoprim/sulfamethoxazole–tetracycline. Twelve isolates were non-susceptible to all six antimicrobials investigated, while 36% (700/1932) were fully susceptible. The proportion of MRSP among clinical and screening specimens by year is presented in Table 2.
Table 1.
Antimicrobial non-susceptibility among clinical S. pseudintermedius isolates from mid-2011 to the end of 2015 in Finland
| Antimicrobial | MRSP |
MSSP |
All |
||||||
|---|---|---|---|---|---|---|---|---|---|
| % non- susceptible | 95% CI | number of isolates | % non- susceptible | 95% CI | number of isolates | % non- susceptible | 95% CI | number of isolates | |
| Basic panel | |||||||||
| clindamycina | 85.7 | 80.8 –89.6 | 265 | 22.2 | 20.2 –24.3 | 1678 | 30.8 | 28.8 –32.9 | 1947 |
| erythromycina | 85.7 | 80.8 –89.6 | 266 | 21.8 | 19.9 –23.9 | 1678 | 30.5 | 28.5 –32.6 | 1949 |
| fusidic acid | 24.5 | 19.5 –30.2 | 265 | 24.3 | 22.3 –26.4 | 1677 | 24.4 | 22.5 –26.4 | 1946 |
| oxacillina | 100 | 98.2 –100 | 266 | 0 | 0.0 –0.3 | 1682 | 13.7 | 12.2 –15.3 | 1948 |
| tetracyclinea | 74 | 68.2 –79.1 | 265 | 33.3 | 31.1 –35.6 | 1675 | 38.8 | 36.6 –41.0 | 1944 |
| trimethoprim/sulfamethoxazolea | 47.7 | 41.6 –53.9 | 266 | 6.1 | 5.0 –7.4 | 1680 | 11.8 | 10.4 –13.3 | 1951 |
|
MRSP |
MSSP |
All (2015)b |
|||||||
| % non- susceptible | 95% CI | number of isolates | % non- susceptible | 95% CI | number of isolates | % non- susceptible | 95% CI | number of isolates | |
| Extended panel | |||||||||
| amikacin | 0 | 0.0 –2.1 | 219 | 0 | 0.0 –0.9 | 547 | 0 | 0.0 –1.2 | 393 |
| chloramphenicola | 46.9 | 29.5 –65.0 | 32 | 15 | 10.6 –20.8 | 207 | 18.4 | 13.7 –24.2 | 228 |
| doxycyclinea | 28.8 | 18.6 –41.4 | 66 | 4.7 | 2.8 –7.7 | 342 | 6.1 | 4.0 –9.1 | 392 |
| enrofloxacina | 50.4 | 44.0 –56.8 | 248 | 2.6 | 1.7 –4.0 | 793 | 7.3 | 5.0 –10.4 | 395 |
| gentamicina | 44.8 | 38.5 –51.2 | 248 | 2.5 | 1.6 –3.9 | 795 | 6.6 | 4.4 –9.6 | 395 |
Statistically significant (P < 0.001) difference between MRSP and MSSP.
Consistent data only available for 2015, as the extended panel was only investigated for MRSP or otherwise MDR isolates prior to this.
Figure 1.
Non-susceptibility percentages by year with 95% CIs for clinical S. pseudintermedius isolates in Finland from mid-2011 to the end of 2015. Cochran–Armitage trend test P values were 0.14 for CLI (clindamycin), 0.14 for ERY (erythromycin), 0.86 for FUS (fusidic acid), 0.12 for OXA (oxacillin), 0.22 for SXT (trimethoprim/sulfamethoxazole) and 0.40 for TET (tetracycline).
Table 2.
Proportion of MRSP per year among clinical and screening specimens from dogs and cats in Finland in 2011–15
| Year | Clinical specimens |
Screening specimens |
||||||
|---|---|---|---|---|---|---|---|---|
| dogs |
cats |
dogs |
cats |
|||||
| MRSP % | number of specimens | MRSP % | number of specimens | MRSP % | number of specimens | MRSP % | number of specimens | |
| 2011 June→a | 3.6 | 933 | 2.3 | 130 | 9.2 | 347 | 11.5 | 52 |
| 2012 | 3.3 | 2076 | 0.3 | 358 | 8.4 | 452 | 0.0 | 59 |
| 2013 | 2.6 | 2104 | 0.0 | 368 | 6.2 | 436 | 0.0 | 28 |
| 2014 | 2.6 | 1963 | 0.3 | 382 | 10.2 | 551 | 0.0 | 24 |
| 2015 | 2.5 | 2098 | 0.2 | 401 | 11.5 | 451 | 0.0 | 34 |
| Total | 2.8 | 9174 | 0.4 | 1639 | 9.2 | 2237 | 3.0 | 197 |
The MRSP outbreak at the VTH was still ongoing in 2011.
Molecular characteristics of MRSP
Out of the 362 MRSP isolates (197 from clinical specimens, 165 from screening specimens), 279 were typeable using SmaI macrorestriction. These clustered into 19 (A–S) different clusters with four or more isolates (Figure S1, available as Supplementary data at JAC Online). Eighty-three isolates could only be typed by AscI macrorestriction and formed two clusters (T and U) and one singleton in PFGE analysis (Figure S2). In total, 87 isolates from 71 different PFGE clusters or singletons (SmaI or AscI) were investigated by MLST and SCCmec.
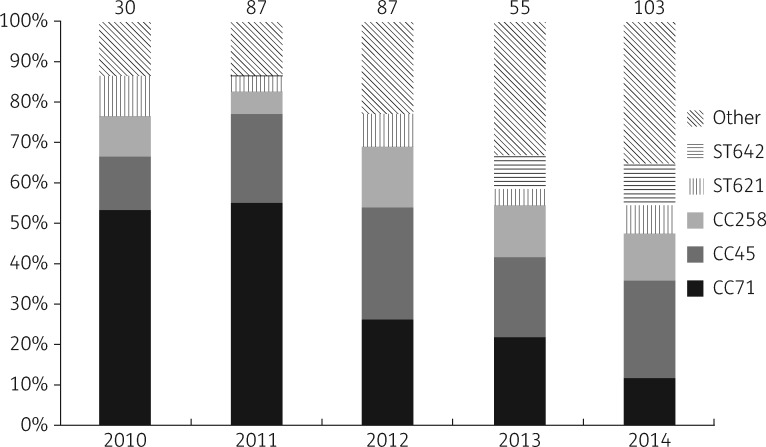
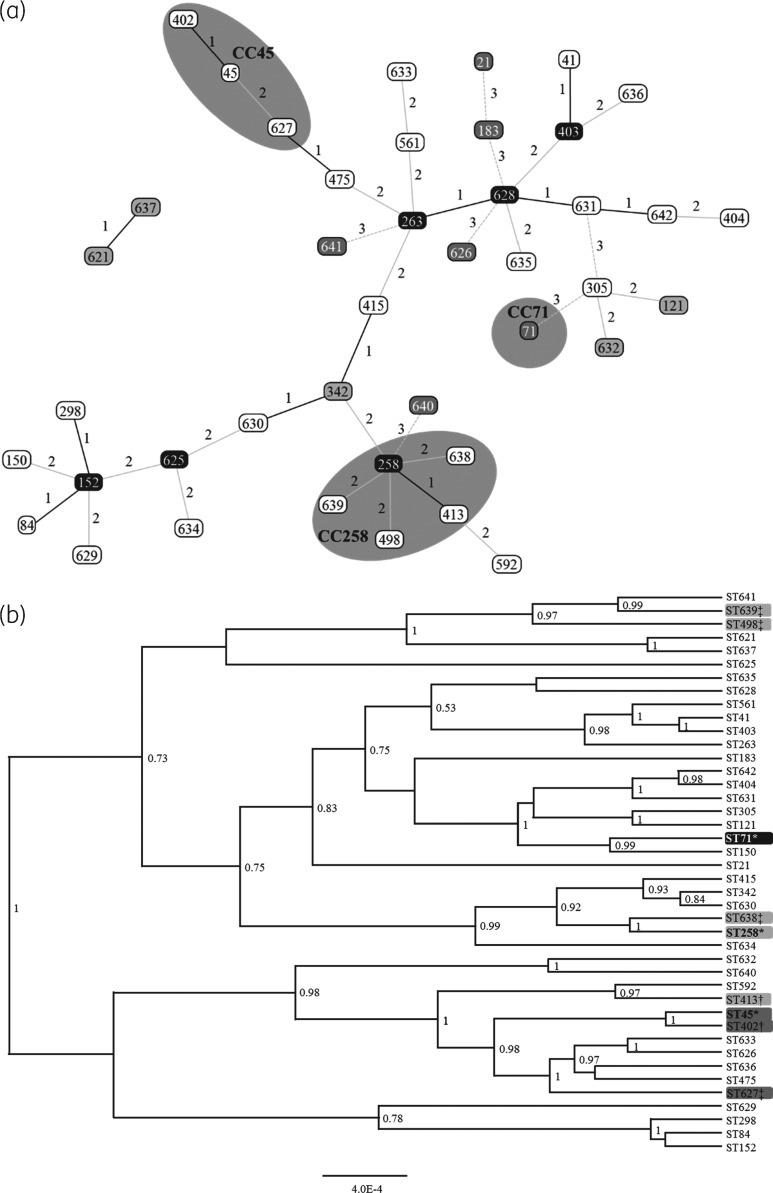
Forty-two different STs were identified, including 19 new STs (STs 621 and 625 to 642).28 All SmaI non-typeable isolates belonged to CC45. The proportion of isolates from each CC or ST changed from year to year, indicating increasing diversity in the MRSP population (Figure 2). All identified STs and their clonal and genetic relatedness are presented in Figure 3(a and b). Six STs grouped together with previously described CCs (CC45 and CC258) (Figure 3a).14 STs in the CC258 group were scattered in the sequence comparison tree, while STs in the CC45 group were more alike (Figure 3b).
Figure 2.
Proportion of CCs or STs of MRSP by year in 2010–14. The data are based on extrapolation of MLST analysis results to the corresponding PFGE cluster. Numbers at the top of the columns indicate the number of isolates. Other, miscellaneous STs.
Figure 3.
Genetic relationship of MRSP in Finland (2010–14). (a) goeBURST analysis conducted at the double-locus variant level, with triple-locus variants (connected with grey dashed lines) added to show further relatedness. Line numbers and shading indicate the number of differing loci between STs. Grey areas highlight CCs. Grey boxes with black text, group founder; black boxes with white text, sub-group founder; white boxes with black text, common node; grey boxes with white text, triple-locus variant. (b) A phylogenetic tree based on the alignment of all MLST genes of each ST. Only posterior probabilities >0.7 are shown. Grey shades indicate CC groups as assigned by goeBURST analysis. *CC founder; †single-locus variant; ‡double-locus variant.
Four different SCCmec types, covering 60 out of the 87 (69%) investigated isolates, were identified. Twenty-seven isolates (31%) were non-typeable, either due to a lack of a PCR product or because the result did not match SCCmec types I–VI (Figure S3). The number of isolates found to carry each SCCmec, as well as the STs in which these were found, are presented in Table 3.
Table 3.
Number of MRSP isolates with each identified SCCmec and the STs associated with them
| SCCmec | Number of isolates | ST(s) |
|---|---|---|
| II | 1 | 498 |
| II–III | 19 | 71 |
| IV | 37 | 258, 342, 413, 415, 475, 561, 592, 621, 625, 626, 630, 631, 632, 633, 634, 635, 638, 639, 640, 641 |
| V | 3 | 183, 404, 642 |
| Non-typeable | 27 | 21, 41, 45, 84, 121, 150, 152, 263, 298, 305, 402, 403, 561, 628, 636, 637 |
Predictors of MRSP
According to the results of multivariable logistic regression analysis, a clinical S. pseudintermedius isolate was more likely to be MRSP if the specimen was from a private clinic (14% of isolates versus 9% of isolates from the VTH), the patient was male or the patient was on any antimicrobials. Among patients who had received antimicrobials, those receiving β-lactams had higher odds of MRSP (Table 4). For screening specimens, the detection of MRSP was more likely if the patient was being treated with multiple systemic antimicrobials (most commonly a combination of ampicillin and enrofloxacin) or if the specimen was from a private clinic (Table 5).
Table 4.
Risk factors associated with the discovery of MRSP among canine clinical S. pseudintermedius isolates taken during 2012–15
| MRSP (n = 227) |
MSSP (n = 1501) |
Univariable logistic regression |
Multivariable logistic regression |
|||||
|---|---|---|---|---|---|---|---|---|
| na | %a | na | %a | unadjusted OR (95% CI) | univariate P | adjusted OR (95% CI) | Wald P | |
| Private clinic versus university teaching hospital | 192 | 84.6 | 1140 | 75.9 | 1.74 (1.19–2.54) | 0.004 | 1.88 (1.25–2.81) | 0.003 |
| Gender: male versus female | 150 | 67.6 | 761 | 52.3 | 1.90 (1.41–2.56) | <0.001 | 1.83 (1.34–2.51) | <0.001 |
| Deep lesion specimen | 31 | 13.7 | 234 | 15.6 | 0.86 (0.57–1.28) | 0.452 | ||
| Superficial lesion specimen | 173 | 76.2 | 1147 | 76.4 | 0.99 (0.71–1.37) | 0.946 | ||
| Urine specimen | 8 | 3.5 | 81 | 5.4 | 0.64 (0.31–1.34) | 0.238 | ||
| Other specimens | 15 | 6.6 | 39 | 2.6 | 2.65 (1.43–4.90) | 0.002 | 1.65 (0.77–3.51) | 0.196b |
| Antimicrobial treatment during sampling | 84 | 40.2 | 264 | 19.1 | 2.84 (2.09–3.87) | <0.001 | 2.67 (2.09–3.93) | <0.001 |
| Antimicrobial groupsc | ||||||||
| systemic β–lactams | 60 | 71.4 | 150 | 57.3 | 1.87 (1.10–3.18) | 0.022 | 1.87 (1.10–3.18) | 0.022 |
| multiple systemic antimicrobials | 1 | 1.2 | 6 | 2.3 | 0.51 (0.06–4.33) | 0.541 | ||
| other systemic antimicrobials | 15 | 17.9 | 63 | 23.9 | 0.25 (0.37–1.30) | 0.252 | ||
| topical antimicrobials | 8 | 9.5 | 40 | 15.3 | 0.58 (0.26–1.30) | 0.189 | 0.90 (0.35–2.29) | 0.825b |
P values in bold indicate variables included in multivariate analysis (P ≤ 0.2).
n refers to the number of dogs with the factor in question, while % refers to the proportion out of the total number of dogs from which data were available, for example, antimicrobial treatment data were available from 209 dogs among MRSP cases (84/0.402).
Variables eliminated from the final models.
Antimicrobial group variables were analysed separately for patients that had received antimicrobials.
Table 5.
Risk factors associated with the discovery of MRSP among canine screening specimens taken during 2012–15
| MRSP positive (n= 173) |
MRSP negative (n= 1717) |
Univariable logistic regression |
Multivariable logistic regression |
|||||
|---|---|---|---|---|---|---|---|---|
| na | %a | na | %a | unadjusted OR (95% CI) | univariate P | adjusted OR (95% CI) | Wald P | |
| Private clinic versus university teaching hospital | 67 | 38.7 | 521 | 30.3 | 1.45 (1.05–2.00) | 0.024 | 1.54 (1.09–2.18) | 0.015 |
| Gender: male versus female | 92 | 54.4 | 849 | 50.2 | 1.19 (0.86–1.63) | 0.295 | ||
| Antimicrobial treatment during samplingb | 45 | 29.4 | 446 | 28.5 | 1.04 (0.73–1.50) | 0.815 | ||
| Systemic β-lactams | 20 | 13.1 | 219 | 14.0 | 0.92 (0.57–1.51) | 0.751 | ||
| Multiple systemic antimicrobials | 10 | 6.5 | 56 | 3.6 | 1.88 (0.94–3.77) | 0.074 | 2.14 (1.06–4.32) | 0.034 |
| Other systemic antimicrobials | 7 | 4.0 | 108 | 6.3 | 0.63 (0.29–1.37) | 0.243 | ||
| Topical antimicrobials | 6 | 3.9 | 45 | 2.9 | 1.38 (0.58–3.28) | 0.470 | ||
P values in bold indicate variables included in multivariate analysis (P ≤ 0.2).
n refers to the number of dogs with the factor in question, while % refers to the proportion out of the total number of dogs from which data were available, for example, antimicrobial treatment data were available from 153 dogs among MRSP cases (45/0.294).
Antimicrobial treatment in general was not significant in the univariable analysis and antimicrobial groups were therefore included in the model.
Discussion
The proportion of MRSP among clinical S. pseudintermedius isolates was fairly high, being nearly 14%. The high percentage (17%) in 2011 was influenced by the MRSP outbreak at the VTH.15 Interestingly, the proportions of MRSP among isolates from private clinics and the VTH were significantly different. This difference may be due to dissimilar patient populations. While the VTH is a national referral veterinary hospital, it also treats first-opinion patients. Numerous clinical specimens originated from private clinics that are treating many dermatological patients, a group at risk of MRSP due to frequent antimicrobial therapy and veterinary visits.15,16,29–31 A threshold to obtain bacteriological specimens early in the infection process is probably higher among private clinic veterinarians. The VTH has had a strict policy of obtaining specimens in all cases of suspected bacterial infection since the MRSP outbreak in 2010–11.15 It is also likely that screening criteria for risk patients are wider at the VTH compared with private clinics, which, in our experience, mainly screen dermatological patients. As the specimens from private clinics may predominantly originate from chronic cases, a higher rate of resistant bacteria would be expected. A comparable difference in patient populations was the likely explanation for unequal methicillin resistance rates between two laboratories in the UK.32 At the microbiology laboratory of the Royal Veterinary College (RVC), 14% of Staphylococcus intermedius group isolates in 2006–12 were oxacillin resistant, while oxacillin resistance in another laboratory was only 1%. This difference was attributed to the high resistance among referral animals of the RVC.32
Non-susceptibility to antimicrobials was significantly higher among MRSP isolates than MSSP isolates, except for fusidic acid, for which non-susceptibility was around 24% in both. Similar findings apply to MRSA and MSSA.33 Resistance to fusidic acid among S. pseudintermedius has varied between studies. In Sweden, 20% of isolates were resistant to fusidic acid in 2015 (MIC ≥1 mg/L),13 which is similar to our results. In Norway, nearly half of S. pseudintermedius isolates investigated were fusidic acid resistant (MIC ≥1 mg/L).34 On the other hand, an Italian study did not find fusidic acid resistance in S. intermedius.35
A recent report from Sweden details resistance among S. pseudintermedius collected from skin lesions.13 Although our data also include isolates from other sources, the difference in oxacillin resistance, in particular, is exceptional, being 2% in the Swedish data and 14% in our data for 2015. Similar differences can be seen among other tested antimicrobials, which is concerning. These differences may reflect contrasts in the use of antimicrobials and infection control policies in these countries. Due to the high resistance for commonly used antimicrobials in S. pseudintermedius in Finland, clinicians are strongly encouraged to take specimens for bacterial cultures early in the disease process to ensure the efficacy of the intended treatment. This is also stated in the newly published national guidelines on the use of antimicrobials in animals.36 In addition, since S. pseudintermedius is a common finding in infections associated with dermatological diseases, it is vital that any underlying disease process is properly controlled to avoid the unnecessary use of antimicrobials. More research is required to determine the value of antimicrobial therapy in patients when underlying conditions have been controlled. Furthermore, the high resistance among S. pseudintermedius could warrant making MRSP a notifiable animal disease in Finland, as it is in Sweden.13
Regarding other risk factors, clinical and screening specimens were investigated separately, as they were thought to represent different populations (see Methods section). We are unaware of any other country where routine MRSP screening, similar to screening for MRSA, for instance, in the Netherlands,37 would be part of an MRSP control scheme. The specimens are taken from patients that are deemed to have a higher risk of MRSP.15 Currently, many of the private clinics that submit specimens to our laboratory screen particularly dermatological patients. While antimicrobial therapy has been indicated as a risk factor for MRSP in multiple studies,15,16,30,38 no study, to our knowledge, has yet reported gender as potentially being one. S. pseudintermedius from clinical specimens of males were more likely to be MRSP (16.4% versus 9.4%). The reason for this is unclear. Studies have not found sex to be a predisposing factor for atopic dermatitis or food allergy.39–41 It is possible that there is some unknown variable that could explain the result. It may also be only due to chance, as gender was not observed to be a risk factor for MRSP in screening specimens. Clinical specimens from dogs revealed more S. pseudintermedius and MRSP than those from cats. Furthermore, not a single MRSP was isolated from feline screening specimens after 2011 (n = 145). These results are unsurprising, as both S. pseudintermedius and MRSP colonization are less common in cats.42,43 Screening of cats for MRSP carriage, even if they have risk factors, is thus deemed unnecessary.
MRSP was detected in 2.5% of all clinical specimens (regardless of whether they revealed S. pseudintermedius), a proportion similar to the MRSP prevalence in the Finnish guide dog population (3%).16 While this figure of 2.5% is not a true prevalence, it may be used as a crude approximation of the prevalence in an average small-animal population from which bacterial cultures have been taken, in order to design future prevalence studies.
The identification of 23 previously identified and 19 new STs is a testament to the diverse nature of the MRSP population in Finland. A changing trend was evident, as the population diversified during the time frame investigated. Note, however, that in 2010–11 many isolates originated from the VTH outbreak.15 Before the outbreak, MRSP was a very uncommon finding in the VTH, although previously published surveillance data indicate that MRSP arrived in force in Finland in 2009, when a sharp increase in MRSP proportion was observed.44 A Swedish study (isolates from 2008 to mid-2010) showed a fairly homogeneous MRSP population, with 96% (216/226) of the isolates representing the predominant European lineage, ST71.11 This clone was also the most common ST in Finland in 2010–11, but has diminished since (Figure 2). Similarly to our data, a change in the MRSP population has also been observed in Sweden after 2010.13 There, 20 out of 58 MRSP isolates (34%) were ST258 in 2015.13 CC258 was the third most common CC in our study and was the largest clone in Norway in a recent study.12 Additionally, CC258 has recently been reported as a major CC in both the Netherlands14 and Denmark.45 These studies also reported a marked diversity in the MRSP population in recent years.14,45 It appears that ST71 has lost its sustainability and CC258 is only filling the gap as a more successful lineage. ST71 carries SCCmec II–III, which has only been found in related STs and ST354.45,46 The immobility of this element may be an underlying reason for the demise of the clone. In contrast, our data indicate that SCCmec IV, in particular, is readily transferred between different clones, or is received from other staphylococci, as it was identified in 20 different STs. The acquisition of SCCmec elements by MSSP from MRSP, CoNS or MRSA may explain the plethora of STs in MRSP. Our findings indicate that the epidemiology of MRSP is changing; clonal spread is becoming less significant and the spread of SCCmec elements is more common. This could make it more difficult to control the spread of MRSP.
Contrary to CC71, CC45 has maintained a steady proportion, with an annual share of the MRSP isolates of ∼20% in 2011–14. Representatives of this CC have been detected at least in Sweden,47 the Netherlands14 and in Israel and Thailand,48,49 but have been rather rare in reports from Europe.14,47 Representatives of ST45 are typically non-typeable by SmaI PFGE,48 as was the case in our study. Interestingly, 80% (66/83) of CC45 isolates showed the same basic antibiogram, only being susceptible to trimethoprim/sulfamethoxazole and fusidic acid (Figure S2). The distribution of STs among clinical isolates and screening isolates was similar, with ST71 being the most common and ST45 being the second most common ST in both groups (data not shown).
The sequence alignment and eBURST analyses grouped the identified STs quite differently. For example, STs 258 and 413, single-locus variants, were placed in different groups in the maximum likelihood tree (six nucleotide differences in one locus), while ST638, a double-locus variant of ST258, was deemed nearly identical to ST258 (four nucleotide differences in two loci) in the maximum likelihood tree (Figure 3a and b). Performing eBURST analysis of MLST data has become common when analysing the clonality of MRSP. However, it does seem to be a crude way of assigning genetic relatedness. A single locus, e.g. cpn60 alleles 1 and 10, may have 26 single-nucleotide differences. All other alleles being identical, two isolates would be considered single-locus variants and be assigned the same CC. On the other hand, two isolates that are triple-locus variants, but only by one nucleotide in each locus (e.g. ack 1 and 2, fdh 1 and 5, and sar 1 and 7), would not be assigned to the same CC, even though the total number of nucleotide differences is much smaller (3–26). It could be beneficial to determine genetic relatedness based on the actual sequences, as was done by Kjellman et al.,12 rather than by comparing combinations of allele numbers. It is, however, likely that WGS will replace eBURST analysis once its costs decrease.
There are some limitations in this study. The specimens mainly originated from southern Finland and may thus not reflect the resistance situation in the entire country. On the other hand, the proportion of private clinic specimens rose from 16% in 2011 to 47% in 2015, with nearly 200 submitting clinics, which increased the geographical coverage. In addition, information on risk factors, such as antimicrobial therapy, is prone to reporting bias, as this information is more likely to be reported if the animal is receiving treatment. This is unlikely to have impacted our results, as the minimum data coverage for risk factors was 88%. In addition, susceptibility data may contain more than one isolate from the same animal, which may have caused bias in the data. The bias caused by this is, however, likely to be minimal due to the large number of isolates and because such isolates would probably be distributed evenly among susceptible and resistant populations, and over the years. Furthermore, PFGE was used as a screening method to find candidates for MSLT and SCCmec typing without having to type all MRSP isolates, which was not feasible. It is therefore possible that there are other, as yet unidentified STs among the isolates. However, apart from one instance where multiple isolates had been typed from a PFGE cluster, all belonged to the same CC.
Conclusions
To conclude, the high resistance to methicillin and other antimicrobials among S. pseudintermedius is of concern. Currently, MRSP in Finland may be spreading via gene transfer rather than clonally, which makes it more difficult to predict resistance patterns in S. pseudintermedius and prevent MRSP. Additionally, routine screening of cats for MRSP is not necessary due to the low occurrence, even among risk patients. We emphasize the need to obtain a bacteriological specimen whenever antimicrobial therapy is indicated. The importance of infection control in veterinary premises should also be stressed as a means of preventing the spread of resistance. The use of non-antimicrobial therapy should be considered whenever feasible.
Supplementary Material
Acknowledgements
Jouni Junnila is acknowledged for his expertise with statistical analyses. We thank Marja Matikka and Gina Meder for their excellent technical assistance and we thank Roy Siddall for revising the language of the manuscript. We thank the Alfred Kordelin Foundation for their generous grant to T. G. and the Helvi Knuuttila Foundation for a grant for material expenses that made the molecular typing possible.
Funding
This work was supported by the Alfred Kordelin Foundation (grant number 140171 to T. G.) and the Helvi Knuuttila Foundation (material grant).
Transparency declarations
None to declare.
Supplementary data
Figures S1 to S3 are available as Supplementary data at JAC Online.
References
- 1.Hajek V. Staphylococcus intermedius, a new species isolated from animals. Int J Syst Bacteriol 1976; 26: 401–8. [Google Scholar]
- 2.Igimi S, Atobe H, Tohya Y. et al. Characterization of the most frequently encountered Staphylococcus sp. in cats. Vet Microbiol 1994; 39: 255–60. [DOI] [PubMed] [Google Scholar]
- 3.Cabassu J, Moissonnier P.. Surgical treatment of a vertebral fracture associated with a haematogenous osteomyelitis in a dog. Vet Comp Orthop Traumatol 2007; 20: 227–30. [DOI] [PubMed] [Google Scholar]
- 4.Huerta B, Maldonado A, Ginel PJ. et al. Risk factors associated with the antimicrobial resistance of staphylococci in canine pyoderma. Vet Microbiol 2011; 150: 302–8. [DOI] [PubMed] [Google Scholar]
- 5.Fitzgerald JR. The Staphylococcus intermedius group of bacterial pathogens: species re-classification, pathogenesis and the emergence of meticillin resistance. Vet Dermatol 2009; 20: 490.. [DOI] [PubMed] [Google Scholar]
- 6.Weese JS, van Duijkeren E.. Methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius in veterinary medicine. Vet Microbiol 2010; 140: 418–29. [DOI] [PubMed] [Google Scholar]
- 7.Loeffler A, Linek M, Moodley A. et al. First report of multiresistant, mecA-positive Staphylococcus intermedius in Europe: 12 cases from a veterinary dermatology referral clinic in Germany. Vet Dermatol 2007; 18: 412–21. [DOI] [PubMed] [Google Scholar]
- 8.van Duijkeren E, Catry B, Greko C. et al. Review on methicillin-resistant Staphylococcus pseudintermedius. J Antimicrob Chemother 2011; 66: 2705–14. [DOI] [PubMed] [Google Scholar]
- 9.Ruscher C, Lubke-Becker A, Semmler T. et al. Widespread rapid emergence of a distinct methicillin- and multidrug-resistant Staphylococcus pseudintermedius (MRSP) genetic lineage in Europe. Vet Microbiol 2010; 144: 340–6. [DOI] [PubMed] [Google Scholar]
- 10.Perreten V, Kadlec K, Schwarz S. et al. Clonal spread of methicillin-resistant Staphylococcus pseudintermedius in Europe and North America: an international multicentre study. J Antimicrob Chemother 2010; 65: 1145–54. [DOI] [PubMed] [Google Scholar]
- 11.Borjesson S, Landen A, Bergstrom M. et al. Methicillin-resistant Staphylococcus pseudintermedius in Sweden. Microb Drug Resist 2012; 18: 597–603. [DOI] [PubMed] [Google Scholar]
- 12.Kjellman EE, Slettemeas JS, Small H. et al. Methicillin-resistant Staphylococcus pseudintermedius (MRSP) from healthy dogs in Norway - occurrence, genotypes and comparison to clinical MRSP. Microbiol Open 2015; 4: 857–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.SWEDRES-SVARM 2015 . Use of Antimicrobials and Occurrence of Antimicrobial Resistance in Sweden. Solna/Uppsala, Sweden, 2016. http://www.sva.se/globalassets/redesign2011/pdf/om_sva/publikationer/swedres_svarm2015.pdf.
- 14.Duim B, Verstappen KM, Broens EM. et al. Changes in the population of methicillin-resistant Staphylococcus pseudintermedius and dissemination of antimicrobial-resistant phenotypes in the Netherlands. J Clin Microbiol 2016; 54: 283–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grönthal T, Moodley A, Nykäsenoja S. et al. Large outbreak caused by methicillin resistant Staphylococcus pseudintermedius ST71 in a Finnish veterinary teaching hospital—from outbreak control to outbreak prevention. PLoS One 2014; 9: e110084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grönthal T, Ollilainen M, Eklund M. et al. Epidemiology of methicillin resistant Staphylococcus pseudintermedius in guide dogs in Finland. Acta Vet Scand 2015; 57: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devriese LA, Vancanneyt M, Baele M. et al. Staphylococcus pseudintermedius sp. nov., a coagulase-positive species from animals. Int J Syst Evol Microbiol 2005; 55: 1569–73. [DOI] [PubMed] [Google Scholar]
- 18.Devriese LA, Hermans K, Baele M. et al. Staphylococcus pseudintermedius versus Staphylococcus intermedius. Vet Microbiol 2009; 133: 206–7. [DOI] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals—Fourth Edition: Approved Standard VET01-A4 CLSI, Wayne, PA, USA, 2013.
- 20.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals: Second Informational Supplement VET01-S2 CLSI, Wayne, PA, USA, 2013.
- 21.Murchan S, Kaufmann ME, Deplano A. et al. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J Clin Microbiol 2003; 41: 1574–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solyman SM, Black CC, Duim B. et al. Multilocus sequence typing for characterization of Staphylococcus pseudintermedius. J Clin Microbiol 2013; 51: 306–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kondo Y, Ito T, Ma XX. et al. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother 2007; 51: 264–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Institute of Health and Welfare. Vanha FiRe-standardi (the old FiRe-standard) https://www.thl.fi/fi/web/infektiotaudit/seuranta-ja-epidemiat/mikrobilaakeresistenssi/fire-suomalainen-mikrobilaakeresistenssin-tutkimusryhma/menetelmat/vanha-fire-standardi.
- 25.Giguère S. Lincosamides, pleuromutilins, and streptogramins In: Giguère S, Prescott JF, Dowling PM, eds. Antimicrobial Therapy in Veterinary Medicine. Oxford, UK: John Wiley & Sons, Inc, 2013; 199–231. [Google Scholar]
- 26.Drummond AJ, Rambaut A.. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 2007; 7: 214.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Francisco M, Bugalho M, Ramirez M. et al. goeBURST homepage http://www.phyloviz.net/goeburst/.
- 28.PubMLST. Staphylococcus pseudintermedius MLST Databases http://pubmlst.org/spseudintermedius/.
- 29.Soares Magalhaes RJ, Loeffler A, Lindsay J. et al. Risk factors for methicillin-resistant Staphylococcus aureus (MRSA) infection in dogs and cats: a case-control study. Vet Res 2010; 41: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nienhoff U, Kadlec K, Chaberny IF. et al. Methicillin-resistant Staphylococcus pseudintermedius among dogs admitted to a small animal hospital. Vet Microbiol 2011; 150: 191–7. [DOI] [PubMed] [Google Scholar]
- 31.Lehner G, Linek M, Bond R. et al. Case-control risk factor study of methicillin-resistant Staphylococcus pseudintermedius (MRSP) infection in dogs and cats in Germany. Vet Microbiol 2014; 168: 154–60. [DOI] [PubMed] [Google Scholar]
- 32.Beever L, Bond R, Graham PA. et al. Increasing antimicrobial resistance in clinical isolates of Staphylococcus intermedius group bacteria and emergence of MRSP in the UK. Vet Rec 2015; 176: 172. [DOI] [PubMed] [Google Scholar]
- 33.Livermore D, James D, Duckworth G. et al. Fusidic-acid use and resistance. Lancet 2002; 360: 806.. [DOI] [PubMed] [Google Scholar]
- 34.Norstrom M, Sunde M, Tharaldsen H. et al. Antimicrobial resistance in Staphylococcus pseudintermedius in the Norwegian dog population. Microb Drug Resist 2009; 15: 55–9.19216647 [Google Scholar]
- 35.Vanni M, Tognetti R, Pretti C. et al. Antimicrobial susceptibility of Staphylococcus intermedius and Staphylococcus schleiferi isolated from dogs. Res Vet Sci 2009; 87: 192–5. [DOI] [PubMed] [Google Scholar]
- 36.Finnish Food Safety Authority and the Faculty of Veterinary Medicine at the University of Helsinki. Mikrobilääkkeiden käyttösuositukset eläinten tärkeimpiin tulehdus- ja tartuntatauteihin (Guidelines for Use of Antimicrobials for the Most Important Infectious Diseases in Animals) Finnish Food Safety Authority, 2016, Helsinki, Finland. www.evira.fi.
- 37.van Trijp MJ, Melles DC, Hendriks WD. et al. Successful control of widespread methicillin-resistant Staphylococcus aureus colonization and infection in a large teaching hospital in the Netherlands. Infect Control Hosp Epidemiol 2007; 28: 970–5. [DOI] [PubMed] [Google Scholar]
- 38.Eckholm NG, Outerbridge CA, White SD. et al. Prevalence of and risk factors for isolation of meticillin-resistant Staphylococcus spp. from dogs with pyoderma in northern California, USA. Vet Dermatol 2013; 24: 154–61 e34. [DOI] [PubMed] [Google Scholar]
- 39.Bizikova P, Pucheu-Haston CM, Eisenschenk MN. et al. Review: role of genetics and the environment in the pathogenesis of canine atopic dermatitis. Vet Dermatol 2015; 26: 95–e26. [DOI] [PubMed] [Google Scholar]
- 40.Nodtvedt A, Bergvall K, Sallander M. et al. A case-control study of risk factors for canine atopic dermatitis among boxer, bullterrier and West Highland white terrier dogs in Sweden. Vet Dermatol 2007; 18: 309–15. [DOI] [PubMed] [Google Scholar]
- 41.Verlinden A, Hesta M, Millet S. et al. Food allergy in dogs and cats: a review. Crit Rev Food Sci Nutr 2006; 46: 259–73. [DOI] [PubMed] [Google Scholar]
- 42.Hanselman BA, Kruth SA, Rousseau J. et al. Coagulase positive staphylococcal colonization of humans and their household pets. Canadian Vet J 2009; 50: 954–8. [PMC free article] [PubMed] [Google Scholar]
- 43.Ruscher C, Lubke-Becker A, Wleklinski CG. et al. Prevalence of methicillin-resistant Staphylococcus pseudintermedius isolated from clinical samples of companion animals and equidaes. Vet Microbiol 2009; 136: 197–201. [DOI] [PubMed] [Google Scholar]
- 44.FINRES-Vet 2010-2012 Finnish Veterinary Antimicrobial Resistance Monitoring and Consumption of Antimicrobial Agents. Finnish Food Safety Authority Evira, 2015, Helsinki, Finland. www.evira.fi. [Google Scholar]
- 45.Damborg P, Moodley A, Aalbaek B. et al. High genotypic diversity among methicillin-resistant Staphylococcus pseudintermedius isolated from canine infections in Denmark. BMC Vet Res 2016; 12: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishihara K, Koizumi A, Saito M. et al. Detection of methicillin-resistant Staphylococcus pseudintermedius ST169 and novel ST354 SCCmec II-III isolates related to the worldwide ST71 clone. Epidemiol Infect 2016; 144: 434–42. [DOI] [PubMed] [Google Scholar]
- 47.SWEDRES-SVARM 2014 : Use of Antimicrobials and Occurrence of Antimicrobial Resistance in Sweden. Solna/Uppsala, Sweden, 2015. http://www.sva.se/globalassets/redesign2011/pdf/om_sva/publikationer/swedres_svarm2014.pdf.
- 48.Perreten V, Chanchaithong P, Prapasarakul N. et al. Novel pseudo-staphylococcal cassette chromosome mec element (ΨSCCmec57395) in methicillin-resistant Staphylococcus pseudintermedius CC45. Antimicrob Agents Chemother 2013; 57: 5509–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kadlec K, Weiss S, Wendlandt S. et al. Characterization of canine and feline methicillin-resistant Staphylococcus pseudintermedius (MRSP) from Thailand. Vet Microbiol 2016; 194: 93–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.