Abstract
Objectives: To investigate the context of the ribosomal RNA methyltransferase gene armA in carbapenem-resistant global clone 2 (GC2) Acinetobacter baumannii isolates from Singapore.
Methods: Antibiotic resistance was determined using disc diffusion; PCR was used to identify resistance genes. Whole genome sequences were determined and contigs were assembled and ordered using PCR. Resistance regions in unsequenced isolates were mapped.
Results: Fifteen GC2 A. baumannii isolated at Singapore General Hospital over the period 2004–11 and found to carry the armA gene were resistant to carbapenems, third-generation cephalosporins, fluoroquinolones and most aminoglycosides. In these isolates, the armA gene was located in a third chromosomal resistance island, previously designated AbGRI3. In four isolates, armA was in a 19 kb IS26-bounded transposon, designated Tn6180. In three of them, a 2.7 kb transposon carrying the aphA1b gene, designated Tn6179, was found adjacent to and sharing an IS26 with Tn6180. However, in these four isolates a 3.1 kb segment of the adjacent chromosomal DNA has been inverted by an IS26-mediated event. The remaining 11 isolates all contained a derivative of Tn6180 that had lost part of the central segment and only one retained Tn6179. The chromosomal inversion was present in four of these and in seven the deletion extended beyond the inversion into adjacent chromosomal DNA. AbGRI3 forms were found in available GC2 sequences carrying armA.
Conclusions: In GC2 A. baumannii, the armA gene is located in various forms of a third genomic resistance island named AbGRI3. An aphA1b transposon is variably present in AbGRI3.
Introduction
Aminoglycoside antibiotics, particularly amikacin, gentamicin and tobramycin, are important treatment options when Acinetobacter baumannii infections are resistant to the first-line carbapenem antibiotics. Resistance to aminoglycosides can occur in a number of ways but most concerning is the emergence of acquired 16S rRNA methyltransferases that confer resistance to most aminoglycoside antibiotics. These enzymes provide ribosomal protection through methylation of 16S rRNA, which hampers the binding of aminoglycosides to the 30S subunit.1 ArmA is a 16S rRNA methyltransferase that was first characterized from a Klebsiella pneumoniae isolated in 2000 in France and shown to confer resistance to the 4,6-disubstituted deoxystreptamines, gentamicin, kanamycin, amikacin, tobramycin, isepamicin, netilmicin and sisomicin.2
The earliest A. baumannii isolates shown to carry the armA gene were recovered in 2003 in Korea.3 Since then, this gene has been reported in strains from North America,4–6 Japan,7 China,8–10 Malaysia,11 Nepal,12 India13 and Italy.14 It was also found in isolates from Norway recovered from patients that had previously been hospitalized in Asia.15 Where MLST data using the Institut Pasteur scheme are available, most are ST2 and are consequently members of global clone 2 (GC2), one of the globally disseminated clones of A. baumannii. In the earliest available completed genomes of GC2 isolates carrying armA, the gene is located in a 19 kb IS26-bounded transposon interrupting a chromosomal gene encoding a putative GNAT family acetyltransferase5,6 (corresponding to ABK1_1290 in 1656-2, GenBank accession number CP001921), designated atr hereafter.
Most resistant GC2 A. baumannii contain a form of both the AbGRI1 and AbGRI2 resistance islands. AbGRI1 characteristically contains the sul2 (confers sulphonamide resistance), strA-strB (spectinomycin resistance) and tetA(B) (resistance to tetracycline) resistance genes.16,17 AbGRI2 variants on the other hand generally contain some or all of the following genes, blaTEM (resistance to ampicillin), aphA1b in Tn6020 (kanamycin and neomycin resistance)18 and sul1, aacC1 and aadA1 in a class 1 integron (conferring resistance to sulphonamides, gentamicin and streptomycin, respectively).19–21 Hence, the armA transposon represents a third chromosomally located resistance island, and for simplicity and consistency we recently called it AbGRI3.20 The three resistance islands are widely separated on the chromosome (see Figure 1 in Blackwell et al.20).
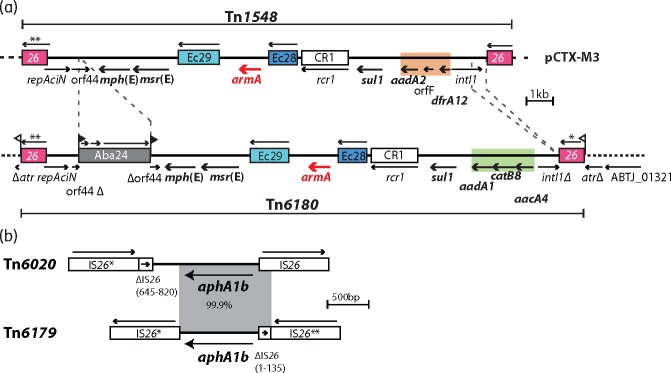
To date, the content and organization of the armA transposon in GC2 isolates has been described only as similar to that of Tn1548.5,6 However, there are several differences that distinguish it and we have named it Tn6180 (Figure 1a). Compared with Tn1548, the Tn6180 integron carries a different set of cassettes aadA1-catB8-aacA4, conferring resistance to streptomycin and spectinomycin, chloramphenicol and gentamicin and tobramycin, respectively, and a deletion at the right end of the transposon has removed 216 bp of the intI1 gene. Another difference is the insertion of ISAba24, a novel member of the IS66 family, in orf44 in Tn6180. In three completed GC2 genomes,9,10 Tn6180 together with an aphA1b-containing segment has been reported as a plasmid. However, as the only putative rep gene present, repAciN, is incomplete and is known to be non-functional,22 this configuration is questionable.
Figure 1.
Transposon structures. (a) Comparison between Tn1548 and Tn6180. (b) Comparison between Tn6020 and Tn6179. Arrows below show the extent and orientation of the genes. Antibiotic resistance gene names are bold. IS and CR elements are depicted as coloured boxes and the numbers inside indicate the identity of the element. Arrows above the IS show the orientation of the tnp transposase gene. ISAba24 is 2421 bp in length with inverted repeats of 22 bp (21/22 bp) and creates an 8 bp TSD. An asterisk indicates the IS26 is different to the standard sequence of IS26 at three positions [G(459)A, G(613)A and G(614)A], and two asterisks indicates two differences [G(613)A and G(614)A]. Dashed lines indicate surrounding plasmid or chromosomal sequence. Thick black solid lines indicate sequence within the transposons. In (a), orange and green boxes highlight the different cassettes in each transposon. Flags indicate TSDs. Tn1548 is drawn from pCTX-M3 (GenBank accession number AF550415). Tn6180 is drawn to scale as in MDR-TJ (GenBank accession number CP003500) and the locus tag for the gene adjacent to atr is provided. In (b), grey blocks of shading represent sequence of 99.9% nucleotide identity. Tn6020 is drawn to scale from GenBank accession number FJ172370 and Tn6179 is drawn from GenBank accession number KX011025. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
In this study, 20 carbapenem-resistant GC2 isolates collected at Singapore General Hospital (SGH) between 1996 and 2011 were examined for the presence of armA and its context was analysed. Whether circular forms are present at detectable levels was also examined.
Materials and methods
Bacterial isolates
Twenty extensively antibiotic-resistant (XAR) A. baumannii isolates recovered from SGH and assigned to GC2 using tri-locus typing23 were examined. Fifteen of these, isolated over the period 2004–11, were found to contain the armA gene and were further characterized (Table 1). All isolates were screened for resistance to 30 antibiotics using a disc diffusion method as described previously.21 Colistin susceptibility was determined by Etest (bioMérieux, Durham, NC, USA).
Table 1.
Aminoglycoside resistance of the GC2 isolates carrying armA from SGH used in this study
| Isolate | Sourcea | Year | Amioglycoside resistanceb | aacA4 | aphA1b | AbGRI3 version | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SGH0702 | blood | 2007 | Ak | Gm | Km | Net | – | Sm | Sp | Tm | + | – | 1i |
| SGH0402 | ETTA | 2004 | Ak | Gm | Km | Net | Nm | Sm | Sp | Tm | + | + | 2i |
| SGH0403 | ETTA | 2004 | Ak | Gm | Km | Net | Nm | Sm | Sp | Tm | + | + | 2i |
| SGH0701 | blood | 2007 | Ak | Gm | Km | Net | Nm | Sm | Sp | Tm | + | + | 2i |
| SGH1111 | sputum | 2011 | Ak | Gm | Km | Net | – | Sm | Sp | Tm | + | – | 3i |
| SGH1112 | sputum | 2011 | Ak | Gm | Km | Net | – | Sm | Sp | Tm | + | – | 3i |
| SGH0905 | blood | 2009 | Ak | Gm | Km | Net | Nm | Sm | Sp | Tm | – | + | 4 |
| SGH0907 | blood | 2009 | Ak | Gm | Km | Net | Nm | Sm | Sp | Tm | – | + | 4 |
| SGH0908 | blood | 2009 | Ak | Gm | Km | Net | Nm | Sm | – | Tm | – | + | 4 |
| SGH1010 | sputum | 2010 | Ak | Gm | Km | Net | – | Sm | Sp | Tm | – | – | 4 |
| SGH1011 | blood | 2010 | Ak | Gm | Km | Net | – | Sm | Sp | Tm | – | – | 4 |
| SGH1012 | ETTA | 2010 | Ak | Gm | Km | Net | – | Sm | Sp | Tm | – | – | 4 |
| SGH1113 | blood | 2011 | Ak | Gm | Km | Net | Nm | Sm | Sp | Tm | – | + | 4 |
| SGH0825 | trach | 2008 | Ak | Gm | Km | Net | Nm | Sm | Sp | Tm | + | + | 5i |
| SGH0602 | ETTA | 2006 | Ak | Gm | Km | Net | – | Sm | Sp | Tm | + | – | +c |
Tracheal aspiration (trach) and endotracheal aspiration (ETTA).
Codes are as follows: amikacin, Ak; gentamicin, Gm; kanamycin, Km; netilmicin, Net; neomycin, Nm; streptomycin, Sm; spectinomycin, Sp; tobramycin, Tm.
The junction on the left is the same as SGH0701 but then not positive for any other PCR.
DNA isolation and PCR amplification
Genomic DNA for WGS was extracted using a Qiagen DNA extraction kit. Genomic DNA for PCR was prepared as described previously.24 For screening of the inverted arrangements, genomic DNA from single colonies was isolated by heating at 100ºC for 5 min as described previously.25 PCR was used to screen for resistance genes and to assemble the AbGRI3 region and the amplicons were sequenced. The primers used to detect the aphA1b and aphA6,24 ISAba1 upstream of ampC26 and oxa2327 genes have been reported previously. RH2012 (5'-TCCATTCCCTTCTCCTTTCC-3') and RH2013 (5'-GGGGGTCTTACTATTCTGCCTA-3') were designed to detect armA. The primers used to assemble AbGRI3 are listed in Table 2. All primers can be used with an annealing temperature of 60°C. The PCR conditions used to detect short amplicons (<2.5 kb) were as described previously using Taq DNA polymerase and a minimum extension time of 1 min per 1 kb of predicted amplicon length.24
Table 2.
Primers used to assemble AbGRI3 variants
| Primera | Primer locationb | Sequence (5'-3') |
|---|---|---|
| RH2001 | ABK1_1291 | GGAGTTGGTTTTGGTACAGCA |
| RH2002 | repAciN | TATAAGCCACCTCGCTCACC |
| RH2003 | intI1 | GCCTTGATGTTACCCGAGAG |
| RH2005 | ABK1_1288 | CACTGATCTGCTGGCTTTCA |
| RH2006 | ABK1_1287 | TGACGAGCTTTGTTTAGGTGTG |
| RH2010 | ISAba24 | TTTCGTGACACTCTCGCTTG |
| RH2011 | aacA4 | CATAGAGCATCGCAAGGTCA |
| RH2015 | ABK1_1289 | AATGTGGTTGGCGGTTTTTA |
| RH2016 | ABK1_1289 | GCAGCCTCAAAGTGGAAAAC |
| RH880 | aphA1b | CAACGGGAAACGTCTTGCTC |
| RH831 | aphA1b | TATACCCATATAAATCAGCATCC |
DNA sequencing and sequence analysis
Genomic DNA of four isolates was sequenced using Illumina HiSeq at the Wellcome Trust Sanger Institute (associated data shown in Table S1, available as Supplementary data at JAC Online). Paired-end reads of 100 bp were assembled as described previously.28 Contigs of the draft genomes containing antibiotic resistance genes were identified using ResFinder (https://cge.cbs.dtu.dk//services/ResFinder/) and contigs containing fragments of IS26 were recovered using standalone BLAST.29 Sequencher 5.2.3 (Gene Codes Corporation, Ann Arbor, MI, USA) was used to assemble the resistance regions and gene construction kit version 4 (Textco, West Lebanon, NH, USA) was used to draw figures to scale. In addition, MLST using the Pasteur and Oxford schemes (http://pubmlst.org/abaumannii/) was performed for the isolates with genome sequence available.
GenBank accession numbers
The sequences of AbGRI3 variants and surrounding chromosomal sequence from SGH0701, SGH0908 and SGH1111 have been deposited into GenBank under accession numbers KX011025, KX011026 and KX011027, respectively. The reads and draft genomes have been deposited in the European Nucleotide Archive under Project Number ERP001080 (see Table S1 for individual accession numbers).
Results
Resistance phenotype and genotype
Twenty GC2 isolates from SGH were screened for the presence of a number of resistance determinants. All the isolates contained oxa23, conferring resistance to carbapenems and ISAba1 was found upstream of ampC conferring resistance to third-generation cephalosporins. The armA gene was found in 15 isolates, indicating that AbGRI3 may be present. The presence of the aphA1b (confers resistance to kanamycin and neomycin) and aacA4 (amikacin, kanamycin and neomycin resistance) genes was variable (Table 1).
The 15 A. baumannii isolates carrying armA (Table 1) were all found to be XAR, using published criteria.30 They were resistant to carbapenems (imipenem, meropenem and doripenem), extended-spectrum cephalosporins (cefotaxime, ceftriaxone, ceftazidime and cefepime), penicillins and β-lactamase inhibitors (ticarcillin/clavulanic acid, ampicillin/sulbactam and piperacillin/tazobactam), quinolones (nalidixic acid), fluoroquinolones (ciprofloxacin and levofloxacin), folate pathway inhibitors (trimethoprim and sulfamethoxazole) and tetracyclines (tetracycline and doxycycline). All isolates were resistant to streptomycin, gentamicin, kanamycin, netilmicin, tobramycin and amikacin but varied in their susceptibility to neomycin and one isolate was susceptible to spectinomycin (Table 1). Resistance to neomycin correlated with the presence of aphA1b. SGH1010 had reduced susceptibility to colistin (MIC 3 mg/L) but the other isolates were all susceptible.
Features of the sequenced isolates
The genomes of four isolates carrying armA were sequenced (bold in Table 1), and for these isolates, MLST was performed using both the Institut Pasteur and Oxford schemes (Table 3). Three isolates were ST2 (cpn60-2, fusA-2, gltA-2, pyrG-2, recA-2, rplB-2, rpoB-2) using the Institut Pasteur scheme and SGH0908 was a single locus variant (SLV) of ST2 (cpn60-1). Hence, all isolates belong to GC2, confirming the original screening. Using the Oxford scheme, two were ST208 (cpn60-2, gdhB-3, gltA-1, gpi-97, gyrB-3, recA-2, rpoD-3). SGH0701 was ST218, an SLV of ST208 (gpi-102). SGH0908 had a novel ST that was assigned the number ST1166. ST1166 is a triple locus variant (cpn60-4, gpi-106 and rpoD-2). The gpi gene lies within the K locus31 and the differences in the gpi allele reflect the fact that the isolates carry three different sets of capsule biosynthesis genes (Table 3).
Table 3.
Properties of sequenced GC2 isolates from SGH
| Isolate | MLST (Pasteur) | MLST (Oxford) | gpi allele | K locusa | Resistance regions |
||
|---|---|---|---|---|---|---|---|
| locationb | resistance genes | no. of contigsc | |||||
| SGH0701 | 2 | 218d | 102 | 7 | AbGRI1 | sul2, strA-strB, tetA(B) | 2 |
| AbGRI2 | none | ||||||
| AbGRI3 | armA, msr(E), mph(E), catB8, aadA1, aacA4, sul1, aphA1b | 2 | |||||
| other | oxa23 | 1 | |||||
| SGH0908e | 98f | 1166g | 106 | 49 | AbGRI1 | sul2, strA-strB, tetA(B) | 2 |
| AbGRI2 | aphA1b, blaTEM | 2 | |||||
| AbGRI3 | armA, msr(E), mph(E) | 1 | |||||
| other | oxa23 | 1 | |||||
| SGH1111 | 2 | 208 | 97 | 2 | AbGRI1 | strA-strB, tetA(B) | 1 |
| AbGRI2 | aacC1 | 1 | |||||
| AbGRI3 | armA, msr(E), mph(E), catB8, sul1, aacA4, aadA1 | 1 | |||||
| other | oxa23 | 1 | |||||
| SGH1112 | 2 | 208 | 97 | 2 | AbGRI1 | strA-strB, tetA(B) | 1 |
| AbGRI2 | aacC1 | 1 | |||||
| AbGRI3 | armA, msr(E), mph(E), catB8, sul1, aacA4, aadA1 | 1 | |||||
| other | oxa23 | 1 | |||||
The structures of KL2, KL7 and KL49 are in GenBank accession numbers JN968483, KX011025 and KT359616, respectively.
Other: genes that have been detected in plasmids and/or at various locations in the chromosome. SGH0701 and SGH0908 contain two and three copies of the oxa23 gene, respectively, with one copy in AbGRI1.
Contigs containing resistance genes only.
SLV of ST208.
Spectinomycin susceptible as no aadA gene is present.
SLV of ST2, has cpn60-1.
Trilocus variant of ST208, has cpn60-4, rpoD-2.
For each sequenced isolate, the resistance genes identified by ResFinder, and listed in Table 3, have been separated into four different lines based on where they are found, in AbGRI1, AbGRI2, AbGRI3 or elsewhere. Each of these isolates contains a version of AbGRI1 and AbGRI2 but SGH0701 has no resistance genes in AbGRI2. These resistance islands will be described in more detail elsewhere. The aphA1b gene in Tn6020 is frequently found in AbGRI2.18–20 However, in the sequenced aphA1b-positive strains, the context of this gene is different (see Figure 1b). We named this 2706 bp transposon Tn6179. We found Tn6179 in the genome of UH9907 (GenBank accession number AY0H00000000),5 adjacent to Tn6180 and sharing one IS26 with the right IS of Tn6180. This AbGRI3 structure, version 2, is shown in the second line of Figure 2(a). Hence, Tn6179 was assigned to AbGRI3 in the SGH isolates sequenced here.
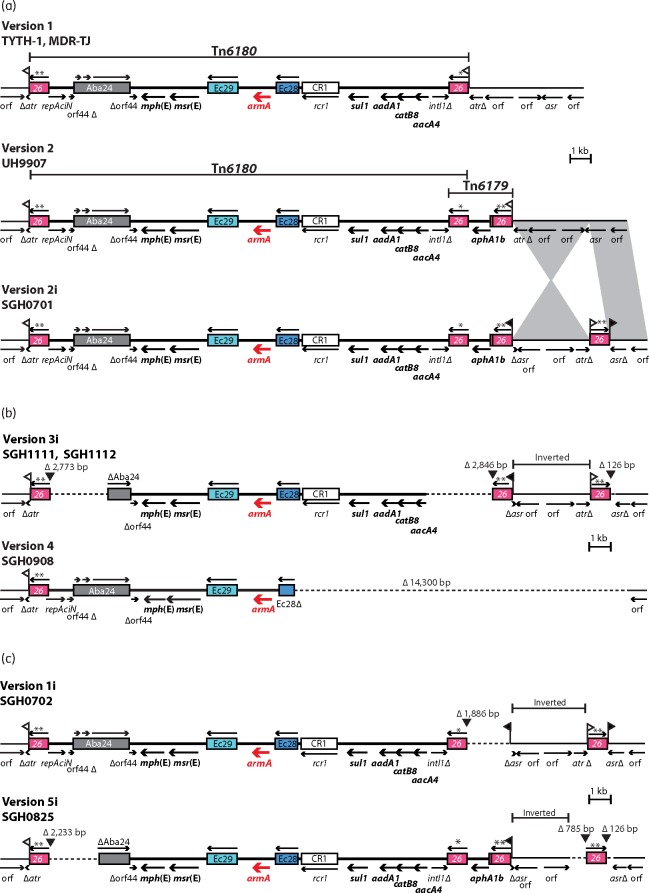
Figure 2.
AbGRI3 structures. (a) Comparison of version 2i with published versions of AbGRI3. (b) AbGRI3 variant forms in the sequenced isolates from Singapore. (c) Variants characterized by mapping in the unsequenced isolates from Singapore. Thick lines represent sequence internal to the resistance island while thin lines indicate chromosomal sequence. Arrows below show the extent and orientation of the genes and antibiotic resistance genes are in bold. IS and CR elements are depicted as boxes with their numbers inside. Arrows above the IS show the orientation of the tnp transposase gene. An asterisk indicates the IS26 is different to the standard sequence of IS26 at three positions [G(459)A, G(613)A and G(614)A], and two asterisks indicates two differences [G(613)A and G(614)A]. Flags indicate the 8 bp TSDs. In (a), grey shading highlights regions of chromosomal sequence with >99.9% nucleotide identity. Version 2 was drawn to scale from GenBank accession number AY0H00000000. In (b) and (c), dashed lines show sequence that is missing and the size of the deletion is indicated along with a black triangle showing the predicted origin of the deletion. The region of inverted sequence is indicated above the line. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
AbGRI3 in SGH0701
SGH0701 contained a 17.5 kb and a 1.1 kb contig, matching the internal sequence of Tn6180 and Tn6179, respectively, and flanked by fragments of IS26. Chromosomal contigs 133 and 3.1 kb in size, each contained part of atr directly adjacent to a fragment of IS26 and the boundaries between IS26 and the chromosome-derived sequence were the same as in AbGRI3 version 1 (found in MDR-TJ and TYTH-1) and version 2 (found in UH9907) (Figure 2a). The repAciN end of Tn6180 was linked to the atr end of the 133 kb contig and the other end was connected to Tn6179. While it was possible to link the other side of Tn6179 to the remainder of atr in the 3.1 kb contig, it is not clear if this is the actual arrangement or if the 3.1 kb segment is inverted as shown in Figure 2(a) (see below).
The 3.1 kb chromosomal contig, which matches atr at one end, is bounded by IS26 at both ends. The other end matches a part of an aspartate racemase gene (corresponding to ABK1_1287 in 1656-2, GenBank accession number CP001921) or asr for simplicity. The remainder of asr was found adjacent to an IS26 outer end on another 142 kb chromosomal contig. Therefore, an additional copy of IS26 interrupts the asr gene in SGH0701. It appears that an inversion of 3.1 kb of the chromosome has been caused by the replicative intramolecular transposition20 of the IS26 at the right end of AbGRI3 to a site in asr, creating an 8 bp target site duplication (TSD) and duplicating the IS26 involved, and this configuration is shown as version 2i, where ‘i’ indicates the adjacent inversion in Figure 2(a). The sequence of the instigating IS26 and the duplicated copy both differ from the standard sequence of IS26 by the same two base pairs (IS26** in Figures 1 and 2), which is consistent with this mechanism (Figure 2a).
However, we were unable to assign unambiguously the orientation of the 3.1 kb segment. Using PCR, Tn6179 could be linked to both ends of the 3.1 kb contig (primer RH831 with RH2005 or RH2015), and both ends of the 3.1 kb contig could also be joined to the asr end of the 142 kb contig (primer RH2006 with RH2005 or RH2015). These two orientations were confirmed by sequencing the amplicons. As the IS26 at the ends of the 3.1 kb contig are in opposite orientation, the 3.1 kb segment could be flipping due to either homologous recombination or the action of IS26. To attempt to determine the major direction of the 3.1 kb segment, boiled DNA preparations for 10 single colonies of SGH0701 were checked for amplification of both orientations using long extension times that should allow polymerization to proceed through IS26 and to prevent PCR artefacts (see the Materials and methods section). Though the re-inverted orientation could still be detected in each single colony, the inverted orientation yielded brighter products.
AbGRI3 in SGH1111 and SGH1112
SGH1111 and SGH1112, both ST208 (Oxford) and both from 2011, had the same AbGRI3 structure, named version 3i. The IS26 at the left end of Tn6180 has caused a deletion that extends into ISAba24 (776/2421 bp remaining) (Figure 2b). The IS26 on the right of Tn6179 has removed aphA1b, the remainder of the intI1 gene, the attI1 site and 7 bp of the aacA4 gene cassette, leaving the aacA4 reading frame intact. Like version 2i, version 3i has an additional copy of IS26 downstream of the resistance island and the 3.1 kb chromosomal sequence in-between can be detected in both orientations. However, part of asr is missing as another deletion, likely caused by the adjacent IS26, has removed 126 bp of this gene (Figure 2b).
AbGRI3 in SGH0908
Version 4 of AbGRI3 in SGH0908 contains the standard boundary on the left but an unusual recombination event has caused the loss of 14 300 bp relative to version 2i (11 586 bp relative to version 1) (Figure 2b). The deletion extends from 696 bp into ISEc28 (201 bp lost) to chromosomal sequence 3112 bp to the right of the resistance island (Figure 2b) removing the invertible segment. There is no clear evidence to explain how this event occurred but an alignment revealed that there was a 4 of 5 bp identity [5'-AA(T)AGT-3'] between the end of the ISEc28 fragment and the chromosomal sequence that could have facilitated the recombination.
Predicted structures of AbGRI3 in the unsequenced SGH isolates
Mapping strategies used to assemble AbGRI3 in the sequenced isolates were applied to predict the structures of AbGRI3 in the remaining armA-positive isolates (Table 1). SGH0402 and SGH0403 from 2004 also contained AbGRI3 version 2i. A further six isolates had AbGRI3 version 4. Two additional AbGRI3 variants were found, each in a single isolate (Figure 2c). SGH0702 contains version 1i, which like version 1 has only Tn6180 (Figure 2). The simplest explanation for this configuration is that the IS26 shared by Tn6180 and Tn6179 caused a deletion that removes the Tn6179 translocatable unit (TU), which is a circular fragment of DNA with a single copy of IS26, as has been described elsewhere.32,33 Version 5i, found in SGH0825, had a smaller deletion at the left end of Tn6180 relative to version 4, retaining 1316 of the 2421 bp ISAba24 (Figure 2b and c). Tn6179 was also present but deletions in the chromosomal DNA on each side of the additional IS26 were detected. The left side has lost 785 bp while the right has lost 126 bp and this deletion is also in version 3i. In both variant 1i and 5i, where the additional IS26 interrupting the asr gene is present, the chromosomal segment in-between could be detected in both orientations. The complete structure of AbGRI3 in SGH0602 could not be determined using the PCR mapping strategy. The left junction of Tn6180 is intact, and the sequence up to armA is present but no amplicon was produced for the remainder of the mapping PCRs.
AbGRI3 in publicly available sequences
The finished genomes of MDR-TJ and TYTH-1 were the first found to contain a form of AbGRI3, which was comprised of just Tn6180 (AbGRI3-1). Since then, the content of the AbGRI3 islands in NCGM_2377 and YU-61234 has been described. Other completed genomes were analysed in this study. Most included both Tn6180 and Tn6179 (AbGRI3-2), though others had only Tn6180 or a deletion derivative of this transposon (Table 4). An IS26 interrupting asr was not present in any of the sequences. Interestingly, PKAB07 (GenBank accession number CP006963) has the same deletion present as AbGRI3-4 and this version of AbGRI3 is also in isolate A072 (GenBank accession number KT354507).35
Table 4.
Structures of AbGRI3 present in finished GC2 genomes
| Isolatea | Year of isolation | GenBank accession number | Chromosome on the left | AbGRI3 | Chromosome on the right |
|---|---|---|---|---|---|
| MDR-ZJ06 | 2006 | CP001937 | + | IS26b | – (Δ28147 bp) |
| MDR-TJ | <2011 | CP003500 | + | Tn6180 | + |
| TYTH-1 | 2008 | CP003856 | + | Tn6180 | + |
| BJAB07104 | 2007 | CP003846 | + | IS26b | + |
| BJAB0868 | 2008 | CP003849 | + | IS26b | + |
| XH860 | 2009 | CP014538 | + | IS26 | – (Δ3338 bp) |
| XH856 | 2010 | CP014541 | + | Tn6180, Tn6179 | + |
| ORAB01 | 2012 | CP015483 | + | Tn6180, Tn6179 | + |
| KPN10P02143 | 2012 | CP013924 | – (Δ 2850 bp) | Tn6180, Tn6179 | + |
| NCGM_237 | 2012 | AP013357 | – (Δ2814 bp) | Tn6180 | – (Δ40215 bp) |
| PKAB07 | 2014 | CP006963 | + | ΔTn6180c | – (3937 bp) |
| XH386 | 2014 | CP010779 | + | Tn6180, Tn6179 | + |
| YU-R612 | 2014 | CP014215 | – (Δ2850 bp) | two copies of ΔTn6180d | + |
The finished genomes with clearly discernible AbGRI3 regions have been included.
These genomes include a Tn6180-Tn6179 TU form said to be a plasmid.
Two copies of IS26 bound the left end of Tn6180 and a deletion identical to version 4 of AbGRI3 is present.
First copy of ΔTn6180 contains the first 308 bp of mph(E) to 26 bp of intI1 and the second copy of ΔTn6180 contains same bases of mph(E) but instead has 364 bp of intI1.
A circular form of AbGRI3-2 (Tn6180 and Tn6179) has been reported previously as a plasmid in three genomes, MDR-ZJ06,10 BJAB0868 and BJAB07104,9 though pMDR-ZJ06 carries the cassette array (aacC1-orfP-orfQ-aadA1) normally seen in the integron of AbGRI2. In the chromosome of BJAB0868 and BJAB07104, an IS26 is present in the location of AbGRI3, i.e. interrupting the atr gene, and it is flanked by the same 8 bp TSD. MDR-ZJ06 also has a suitably located IS26 but 22.8 kb of adjacent chromosomal sequence is missing. As it has been shown that repAciN is not a functional replication initiation protein,22 it seems unlikely that this form could exist stably as a plasmid. Hence, it is likely that the proposed plasmid forms instead belong in the chromosome, as AbGRI3.
Can TUs be detected?
Recently, the AbGRI3-2 structure that includes both Tn6180 and Tn6179 was found in the isolate A071 (GenBank accession number KT317075) and named Tn6279.35 This study also reported that circular TU forms of AbGRI3-2 (Tn6180 and Tn6179 with only two copies of IS26) as well as just AbGRI3-1 (Tn6180 with one copy of IS26) could be detected by PCR in this isolate. However, using DNA from SGH0701, which carries AbGRI3-2, and PCR conditions that allow polymerization through IS26, thereby avoiding PCR artefacts, no amplicons were detected for the circular forms of AbGRI3-2 (RH2002-RH831) or AbGRI3-1 (RH2002-RH2003). Hence, the possibility that the earlier observations were due to artefacts caused by short PCR extension times or other factors in the case of A071 should be investigated.
Discussion
In several sequenced GC2 genomes, the armA gene is found in AbGRI3, the third resistance island typically found in this group. We have shown previously that ancestral forms of AbGRI1 and AbGRI2 were acquired by a GC2 isolate prior to 1982.20 The AbGRI3 island appears to have been acquired in the early 2000s by a GC2 isolate already carrying these two resistance islands, probably in Asia. The earliest isolate with the armA gene recovered at SGH was from 2004 and it contained AbGRI3-2. However, in most of the SGH isolates, a 3.1 kb segment of chromosome adjacent to the Tn6179 end has been inverted and an IS26 has been duplicated in the process. This arrangement is unique to the isolates from SGH and could prove useful in tracking their spread. A surprising finding was that the 3.1 kb segment could be detected in both orientations using PCR, and further experiments such as cloning the fragment or PacBio sequencing will be required to determine if the segment is actually inverting frequently.
The presence of an 8 bp TSD surrounding AbGRI3 demonstrates that, like AbGRI2,20,36 this island entered the chromosome of a GC2 isolate through the action of IS26. What is less clear is if both Tn6180 and Tn6179 entered together or individually.33 Based on the sequences of the IS26 at the outer boundaries of AbGRI3-2, a likely option is that a TU derived from both Tn6180 and Tn6179 inserted into the atr gene by a Tnp26-mediated replicative transposition event, duplicating an IS26** and producing the TSD.
The AbGRI3 region has continued to evolve in situ. As for AbGRI2,20,21 this variation has mainly occurred through deletions adjacent to copies of IS26. The other versions of AbGRI3 found in the remaining SGH isolates are all smaller than AbGRI3-2i. They have clearly evolved from AbGRI3-2i, by various deletions of sequence adjacent to copies of IS26, as version 1i, 3i and 5i all contain the adjacent chromosomal inversion. Whether AbGRI3-4 is also derived from AbGRI3-2i cannot be discerned, as the inverted chromosomal segment is not present in AbGRI3-4 as its characteristic deletion extends past this region of the chromosome. This deletion has been detected in isolates from Singapore, India (PKAB07) and Sweden (A072) suggesting that the sub-lineage containing AbGRI3-4 has dispersed quite successfully.
Tracking the movement of ESKAPE organisms within and between hospitals or countries is key to effective infection control and the simple set of PCRs we devised to map AbGRI3 variants should assist in distinguishing A. baumannii carrying the armA gene.
Conclusions
Five different forms of AbGRI3 were identified in this study. As each variant of AbGRI3 defines a sub-lineage of the GC2 clone, the characterization of the AbGRI3 forms can assist detailed tracking of GC2 isolates locally and globally.
Supplementary Material
Acknowledgements
We thank Dr Tse Hsien Koh, from the Department of Pathology at SGH, for helping to provide the bacterial isolates used in this study. We thank Dr Johanna Kenyon, from the Queensland University of Technology, for the identification of the KL type.
Funding
Funding for this study was received from the National Health and Medical Research Council (NHMRC) grant 1079616. K. E. H. was supported by NHMRC Fellowship 1061409 and G. A. B. is supported by an Australian Postgraduate Award.
Transparency declarations
None to declare.
Supplementary data
Table S1 is available as Supplementary data at JAC Online.
References
- 1.Doi Y, Arakawa Y.. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin Infect Dis 2007; 45: 88–94. [DOI] [PubMed] [Google Scholar]
- 2.Galimand M, Courvalin P, Lambert T.. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob Agents Chemother 2003; 47: 2565–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee H, Yong D, Yum JH. et al. Dissemination of 16S rRNA methylase-mediated highly amikacin-resistant isolates of Klebsiella pneumoniae and Acinetobacter baumannii in Korea. Diagn Microbiol Infect Dis 2006; 56: 305–12. [DOI] [PubMed] [Google Scholar]
- 4.Doi Y, Adams JM, Yamane K. et al. Identification of 16S rRNA methylase-producing Acinetobacter baumannii clinical strains in North America. Antimicrob Agents Chemother 2007; 51: 4209–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wright MS, Haft DH, Harkins DM. et al. New insights into dissemination and variation of the health care-associated pathogen Acinetobacter baumannii from genomic analysis. MBio 2014; 5: e00963-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan AP, Sutton G, DePew J. et al. A novel method of consensus pan-chromosome assembly and large-scale comparative analysis reveal the highly flexible pan-genome of Acinetobacter baumannii. Genome Biol 2015; 16: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tada T, Miyoshi-Akiyama T, Shimada K. et al. Dissemination of 16S rRNA methylase ArmA-producing Acinetobacter baumannii and emergence of OXA-72 carbapenemase coproducers in Japan. Antimicrob Agents Chemother 2014; 58: 2916–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang H, Yang ZL, Wu XM. et al. Complete genome sequence of Acinetobacter baumannii MDR-TJ and insights into its mechanism of antibiotic resistance. J Antimicrob Chemother 2012; 67: 2825–32. [DOI] [PubMed] [Google Scholar]
- 9.Zhu L, Yan Z, Zhang Z. et al. Complete genome analysis of three Acinetobacter baumannii clinical isolates in China for insight into the diversification of drug resistance elements. PLoS One 2013; 8: e66584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou H, Zhang T, Yu D. et al. Genomic analysis of the multidrug-resistant Acinetobacter baumannii strain MDR-ZJ06 widely spread in China. Antimicrob Agents Chemother 2011; 55: 4506–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lean SS, Yeo CC, Suhaili Z. et al. Whole-genome analysis of an extensively drug-resistant clinical isolate of Acinetobacter baumannii AC12: insights into the mechanisms of resistance of an ST195 clone from Malaysia. Int J Antimicrob Agents 2015; 45: 178–82. [DOI] [PubMed] [Google Scholar]
- 12.Shrestha S, Tada T, Miyoshi-Akiyama T. et al. Molecular epidemiology of multidrug-resistant Acinetobacter baumannii isolates in a university hospital in Nepal reveals the emergence of a novel epidemic clonal lineage. Int J Antimicrob Agents 2015; 46: 526–31. [DOI] [PubMed] [Google Scholar]
- 13.Saranathan R, Tomar A, Sudhakar P. et al. Draft genome sequence of a multidrug-resistant Acinetobacter baumannii PKAB07 clinical strain from India belonging to sequence type 195. Genome Announc 2014; 2: e00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milan A, Furlanis L, Cian F. et al. Epidemic dissemination of a carbapenem-resistant Acinetobacter baumannii clone carrying armA two years after its first isolation in an Italian hospital. Microb Drug Resist 2016; 22: 668–74. [DOI] [PubMed] [Google Scholar]
- 15.Karah N, Haldorsen B, Hermansen NO. et al. Emergence of OXA-carbapenemase- and 16S rRNA methylase-producing international clones of Acinetobacter baumannii in Norway. J Med Microbiol 2011; 60: 515–21. [DOI] [PubMed] [Google Scholar]
- 16.Seputiene V, Povilonis J, Suziedeliene E.. Novel variants of AbaR resistance islands with a common backbone in Acinetobacter baumannii isolates of European clone II. Antimicrob Agents Chemother 2012; 56: 1969–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nigro SJ, Hall RM.. Tn6167, an antibiotic resistance island in an Australian carbapenem-resistant Acinetobacter baumannii GC2, ST92 isolate. J Antimicrob Chemother 2012; 67: 1342–6. [DOI] [PubMed] [Google Scholar]
- 18.Post V, Hall RM.. AbaR5, a large multiple-antibiotic resistance region found in Acinetobacter baumannii. Antimicrob Agents Chemother 2009; 53: 2667–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nigro SJ, Farrugia DN, Paulsen IT. et al. A novel family of genomic resistance islands, AbGRI2, contributing to aminoglycoside resistance in Acinetobacter baumannii isolates belonging to global clone 2. J Antimicrob Chemother 2013; 68: 554–7. [DOI] [PubMed] [Google Scholar]
- 20.Blackwell GA, Nigro SJ, Hall RM.. Evolution of AbGRI2-0, the progenitor of the AbGRI2 resistance island in global clone 2 of Acinetobacter baumannii. Antimicrob Agents Chemother 2015; 60: 1421–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nigro SJ, Hall RM.. Loss and gain of aminoglycoside resistance in global clone 2 Acinetobacter baumannii Australia via modification of genomic resistance islands and acquisition of plasmids. J Antimicrob Chemother 2016; 71: 2432–40. [DOI] [PubMed] [Google Scholar]
- 22.Mierzejewska J, Kulinska A, Jagura-Burdzy G.. Functional analysis of replication and stability regions of broad-host-range conjugative plasmid CTX-M3 from the IncL/M incompatibility group. Plasmid 2007; 57: 95–107. [DOI] [PubMed] [Google Scholar]
- 23.Turton JF, Gabriel SN, Valderrey C. et al. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin Microbiol Infect 2007; 13: 807–15. [DOI] [PubMed] [Google Scholar]
- 24.Nigro SJ, Post V, Hall RM.. Aminoglycoside resistance in multiply antibiotic-resistant Acinetobacter baumannii belonging to global clone 2 from Australian hospitals. J Antimicrob Chemother 2011; 66: 1504–9. [DOI] [PubMed] [Google Scholar]
- 25.Bailey JK, Pinyon JL, Anantham S. et al. Commensal Escherichia coli of healthy humans: a reservoir for antibiotic-resistance determinants. J Med Microbiol 2010; 59: 1331–9. [DOI] [PubMed] [Google Scholar]
- 26.Hamidian M, Hancock DP, Hall RM.. Horizontal transfer of an ISAba125-activated ampC gene between Acinetobacter baumannii strains leading to cephalosporin resistance. J Antimicrob Chemother 2013; 68: 244–5. [DOI] [PubMed] [Google Scholar]
- 27.Woodford N, Ellington MJ, Coelho JM. et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents 2006; 27: 351–3. [DOI] [PubMed] [Google Scholar]
- 28.Hamidian M, Hall RM.. AbaR4 replaces AbaR3 in a carbapenem-resistant Acinetobacter baumannii isolate belonging to global clone 1 from an Australian hospital. J Antimicrob Chemother 2011; 66: 2484–91. [DOI] [PubMed] [Google Scholar]
- 29.Camacho C, Coulouris G, Avagyan V. et al. BLAST+: architecture and applications. BMC Bioinformatics 2009; 10: 421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magiorakos AP, Srinivasan A, Carey RB. et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18: 268–81. [DOI] [PubMed] [Google Scholar]
- 31.Kenyon JJ, Hall RM.. Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii genomes. PLoS One 2013; 8: e62160.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harmer CJ, Hall RM.. IS26-Mediated formation of transposons carrying antibiotic resistance genes. mSphere 2016; 1: e00038-16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harmer CJ, Moran RA, Hall RM.. Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. MBio 2014; 5: e01801-14.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee Y, D'Souza R, Yong D. et al. Prediction of putative resistance islands in a carbapenem-resistant Acinetobacter baumannii global clone 2 clinical isolate. Ann Lab Med 2016; 36: 320–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karah N, Dwibedi CK, Sjostrom K. et al. Novel aminoglycoside resistance transposons and transposon-derived circular forms detected in carbapenem-resistant Acinetobacter baumannii clinical isolates. Antimicrob Agents Chemother 2016; 60: 1801–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blackwell GA, Hamidian M, Hall RM.. IncM plasmid R1215 is the source of chromosomally located regions containing multiple antibiotic resistance genes in the globally disseminated Acinetobacter baumannii GC1 and GC2 clones. mSphere 2016; 1: e00117-16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.