Abstract
The Protein Data Bank (PDB) is the central worldwide repository for three-dimensional (3D) structure data of biological macromolecules. The Research Collaboratory for Structural Bioinformatics (RCSB) has completely redesigned its resource for the distribution and query of 3D structure data. The re-engineered site is currently in public beta test at http://pdbbeta.rcsb.org. The new site expands the functionality of the existing site by providing structure data in greater detail and uniformity, improved query and enhanced analysis tools. A new key feature is the integration and searchability of data from over 20 other sources covering genomic, proteomic and disease relationships. The current capabilities of the re-engineered site, which will become the RCSB production site at http://www.pdb.org in late 2005, are described.
INTRODUCTION
The production version of the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) (http://www.pdb.org) is mirrored at seven sites around the world and has been described previously (1,2). In order to improve the accessibility of the PDB's structure data, display the increased level of detail and improved consistency resulting from the RCSB PDB data uniformity project (3,4), and take advantage of advances in database and Web/Internet technologies, the RCSB PDB has re-engineered its database and redesigned the associated website. This site is now available for beta testing at http://pdbbeta.rcsb.org and will henceforth be referred to as PDB Beta. The following features of PDB Beta have been introduced: software architecture, database content and schema, data integration from other sources, and query and analysis capabilities expanded from those reported previously (5).
CONTENT
Software architecture, database content and schema
Using an Enterprise Java framework, the PDB Beta has been redesigned and it is composed of three tiers: an underlying relational database, a presentation tier designed in collaboration with users and an object-relational J2EE middle tier based on Hibernate. The PDB Beta has been tested with both MySQL and IBM DB2 relational database tiers. The current system uses MySQL, which enables unlimited distribution of the primary and secondary data in the RCSB PDB database. Moreover, the current system makes extensive use of freely distributable Java components (Table 1).
Table 1. Software components used by PDB Beta.
| Component | Function |
|---|---|
| Lucene (jakarta.apache.org/lucene/docs) | Text indexing system |
| Jazzy (jazzy.sourceforge.net) | The Java open source spell checker |
| JfreeChart (www.jfree.org/jfreechart) | Java class library for generating charts |
| iText (sourceforge.net/projects/itext/) | A Java PDF library to generate documents in the PDF and/or HTML |
| OpenBabel (sourceforge.net/projects/openbabel) | A program and library to interconvert between many file formats used in molecular modeling and computational chemistry |
| RoboHelp (www.macromedia.com/software/robohelp) | Text indexing and help system |
| Hibernate (www.hibernate.org) | Object/relational persistence and query service for Java |
| Jboss (www.jboss.org/) | Open source J2EE application server |
| MySQL (www.mysql.com) | Open source relational database |
| SimpleViewer (mbt.sdsc.edu/apps/SimpleViewer) | 3D visualization applet based on MBT |
| WebMol (18) | 3D visualization applet |
| KiNG (Kinemage, Next Generation; kinemage.biochem.duke.edu/software/king.php) | Interactive system for 3D visualization |
| Jalview (www.jalview.org) | Multiple alignment editor written in Java |
| MarvinView (www.chemaxon.com/marvin) | Applet for visualizing 2D chemical structure |
The database uses an mmCIF-based (6) schema derived from the PDB Exchange Dictionary (7). Data are loaded from either XML (8) or mmCIF data files with their associated remediated and extended content. Data file parser/structure loaders are SQL-92 compliant and therefore independent of the backend database. Data files are parsed and loaded weekly at the same time as the current production site is updated. The design of this system along with its local weekly data updates will facilitate local distribution and use.
Data integration from other sources
From a user's perspective, macromolecular structure does not exist in isolation, but is generally associated with an inquiry that might include relationships to genomic and proteomic sequence, biological function, cellular location and disease. While the focus of the RCSB PDB remains on fully exposing the features of macromolecular structures, a wider spectrum of inquiry is now possible. This is achieved through the weekly collection and integration (warehousing) of external data. For example, data from the Gene Ontology (GO) (9), Enzyme Commission (http://www.chem.qmul.ac.uk/iubmb/enzyme/), KEGG Pathways (10,11) and NCBI resources (including LocusLink, OMIM, SNP and BookShelf) (12) are mapped onto structures and loaded into the database. This is achieved as follows.
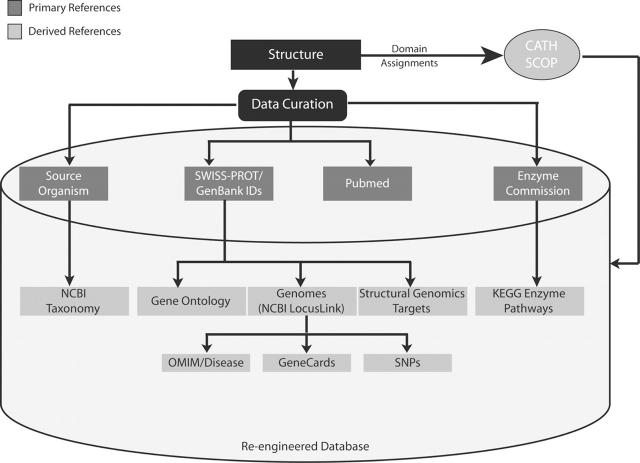
Our ongoing data uniformity efforts enable accurate assignment of external database references to structures in the RCSB PDB database; these include identifiers from Swiss-Prot (13), GenBank, PubMed, EC numbers and the taxonomy of the source organism. These references are used to locate information in a further set of databases. For example, Swiss-Prot identifiers were used to assign GO terms from the Gene Ontology Consortium to structures. Swiss-Prot and GenBank identifiers were also used to obtain genome information: gene name, chromosome location, structural genomics targets (14) and OMIM numbers for structures. Figure 1 summarizes the rich and varied linkages that have been established between structure data in the RCSB PDB database and data from external biological databases. Note that many of these data are related to structure through a one-to-many relationship since a structure consists of one or more components, such as multiple polypeptide chains. The representation of structures as a number of constituent components, each with external data assignments, is an ongoing effort at the RCSB PDB.
Figure 1.
Primary and secondary references assigned to structures. The primary references are assigned during structure annotation/data curation. Secondary references are collected from external databases using the primary reference identifiers and accession numbers. This is rerun on a weekly basis for new and all existing structures and stored in the database.
Data loaders written in Java access the external databases, parse the files and load relevant derived information into the database. In some instances additional external information is retrieved at query run time. For example, KEGG pathways associated with a given EC number are retrieved by issuing a Web service call to the KEGG database at query run time. Under an agreement with the US National Library of Medicine, PubMed identifiers for the primary citation associated with a structure are used to load the PubMed abstracts into the RCSB PDB database. In doing so, abstracts can be searched by keyword(s) as an alternative means to find structures of interest.
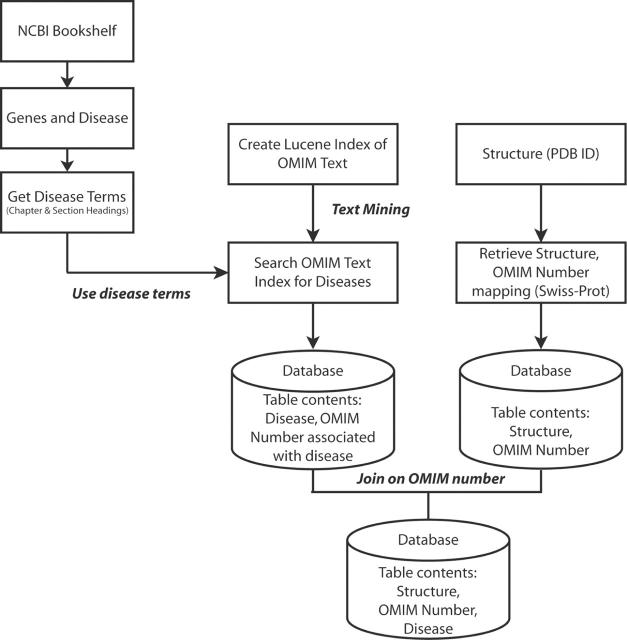
As a final example of how linkages between incorporated data were generated, consider the relationship between structure and disease (Figure 2). The OMIM text was searched for disease terms obtained from the chapter and section headings in the online book Genes and Disease from the NCBI Bookshelf (http://www.ncbi.nlm.nih.gov/books/bv.fcgi?call=bv.View..ShowSection&rid=gnd.preface.91). The disease term and the OMIM numbers returned for the term were loaded into a relational table, as was a mapping of structures to OMIM numbers from the Swiss-Prot site. The two tables were joined using OMIM numbers and the disease term was thus mapped onto structures. These relationships have enabled the implementation of a hierarchical disease tree suitable for browsing. For example, users can locate all the PDB structures identified as being associated with cancer and drill down to find only those associated with breast cancer.
Figure 2.
Procedure to create structure-OMIM-disease mapping. Disease terms listed in the NCBI book Genes and Disease are used to mine the OMIM text for OMIM numbers associated with the terms. This Disease-OMIM mapping is loaded into the database. Performing a join on the Disease-OMIM table created above and the OMIM-Structure mapping table results in Disease-Structure mapping.
Query and analysis
Browsing
A major feature of PDB Beta is the ability to browse database content. Much of the data now integrated with structure is hierarchical and lends itself to display via tree browsers. When each node in a browser tree is moused over, the number of associated structures at that branch is displayed. This gives the user a sense of the size of the result set, even before a query is made. As some data that are browsed are not strictly hierarchical, concessions are made. An example of this concession is a protein chain that has been associated with multiple GO molecular functions. This protein chain would therefore appear multiple times in the browser tree even though it is only associated with a single structure. Browsers are also useful in reverse; knowing the location of a structure in a tree reveals its place in the hierarchy. For example, entering homo sapiens in the taxonomy browser will locate the structures for which this has been identified as a source organism, and highlight where humans are located in the tree of life, at least according to the NCBI classification scheme. The results of browsing can be used as a starting point for query refinement using the SearchFields interface outlined below.
Searching
The PDB Beta has retained the search capability of the RCSB PDB production site but has provided, based on user feedback, more intuitive interfaces and added additional Website navigation tools. Sequence searching functionality via BLAST (15) has also been added.
SearchLite is based on the Lucene text indexing and search engine, and uses the content of the mmCIF versions of the PDB structures which provides indexes of terms not available in the original PDB files. For example, at the time of submission it may not have been known that the structure was associated with apoptosis. When the term is later added to the list of Swiss-Prot keywords, it will be accessible through SearchLite even though there was no reference in the original PDB file. SearchLite can be considered as an inclusive rather than an exclusive search engine, since it produces results that may require further query refinement. This feature is also available on the current production site.
StatusSearch continues to provide information on deposited but unreleased structures. Sequence data are available ahead of a structure's release for some entries, which is useful for theoretical modeling and avoiding duplication of effort.
SearchFields, an interface for performing advanced queries, has been enhanced to include the full extent of the experimental information that is collected and much of the integrated information outlined above. Particular attention has been paid to NMR structures. A new SearchFields option is to search for structures with specific NMR experimental parameters like refinement method, selection criteria, spectrometer details and sample conditions.
Searches performed during a session are recorded and can be recalled and rerun or modified and rerun. Since the result of one query may trigger a new line of inquiry, we have extended the notion of query by example in which a result from one query can be used as a search term in a subsequent query. For example, a search for the structure with the PDB identifier 1AEW displays the Structure Explorer page for the iron storage molecule ferritin (16). According to the GO term for the single polypeptide chain in the asymmetric unit of this structure, it is assigned a molecular function of ‘ferric iron binding’. Clicking on this term on the Structure Explorer page will reveal all other structures in the PDB associated with the same molecular function as defined by the same GO term.
A histogram feature in the early stages of development is applied to quantitative data and is accessible from the results display. For example, an X-ray structure is reported with a resolution of 2.5 Å. A novice user may be interested in how that resolution compares with the contents of the complete database. An icon next to the resolution on the structure page can be selected to present a graphical distribution of the resolutions of all the structures in the PDB. Specific ranges can be expanded and the structures in a selected resolution range displayed in a report format.
The PDB Beta is also composed of ∼1000 curated Web pages, which have been made more accessible through site searching and indexing. All features are in the process of being better documented and made accessible through a context-sensitive help system based on the RoboHelp tool.
Results reporting
Query results are presented on the Structure Explorer page for a single structure or in the Query Results Browser for a set of structures. The Structure Explorer page presents data similar to the format of a scientific paper. This page can be printed as a PDF. Table 2 highlights the new features that are included beyond those available from the current production site. Of the new reports, the Materials and Methods section is customized based on the experiment type. Structure Explorer pages list crystallization, diffraction and refinement information for X-ray structures; and NMR experiment, refinement and ensemble information for NMR entries.
Table 2. New results reported from PDB Beta.
| Summary reports | Features |
|---|---|
| Structure Explorer | Navigation breadcrumbs; Print PDF; Toggle asymmetric and biological unit images; Ligand and ligand–structure interaction viewer; CATH, SCOP (19) and GO terms listing and search feature; Ensemble and refinement information for NMR structures |
| Materials and Methods | Reports customized for X-ray and NMR structures |
| Biology and Chemistry | Detailed information including taxonomy, genome and locus, SNPs, enzyme pathways, disease and function |
| Structural Features | Detailed chemical bond information |
Molecular viewers
Recognizing the difficulties that users may have installing the existing molecular viewers, four new, general purpose molecular viewers have been added to the PDB Beta: KiNG, Jmol, SimpleViewer and WebMol applets. KiNG, Jmol and WebMol require Java-enabled browsers; SimpleViewer requires the installation of Java3D to provide high-quality rendering. SimpleViewer is an example of an application built from the Molecular Biology Toolkit (MBT) (http://mbt.sdsc.edu). The concept behind MBT is to deliver simple context-sensitive molecular graphics applications at the appropriate point in a query. So for example, a ligand viewer has been implemented that provides a detailed view of the interaction between a macromolecule and a ligand (HET group in original PDB terminology), but is not designed to be a general-purpose molecular viewer. A SNPviewer, indicating where non-synonymous single nucleotide polymorphisms (SNPs) are mapped onto structures, is another example of the use of a context-sensitive viewer that has been added to the PDB Beta.
Distribution system
The RCSB production and PDB Beta sites distribute data files in the PDB, mmCIF and XML formats. Data for sequences and complete structural descriptions are available in uncompressed as well as various compressed formats. The PDB data can also be obtained using CORBA and Web services. A CORBA server may be established using C++ (http://deposit.pdb.org/mmcif/FILM/) or the Java OpenMMS software (http://openmms.sdsc.edu) (17). Web services, which are currently in the early implementation stage, will allow users to use XML and SOAP to perform queries and retrieve results programmatically from the PDB Beta. The PDB Beta Web Services Definition Language (WSDL) is available at http://pdbbeta.rcsb.org/jboss-net/services/pdbWebService?wsdl.
CONCLUSION
While the RCSB PDB's primary mandate continues to be the delivery of high-quality structure data in a timely manner, our services are being expanded. Recognizing that structure exists as a point on a spectrum of biological inquiry, more integrated access to structure data is being provided. Using PDB Beta locally to manage private copies of the PDB data on a laptop or larger computer, and a web interface that can be customized for individual user preferences, either locally or on the RCSB's PDB servers are examples of forthcoming deliverables. Comments and suggestions are always welcome by sending email to betafeedback@rcsb.org. Upon completion of the beta testing, anticipated to be in late 2005 the re-engineered site will be available as the PDB production site (http://www.pdb.org).
Acknowledgments
ACKNOWLEDGEMENTS
The RCSB PDB is operated by Rutgers, The State University of New Jersey; the San Diego Supercomputer Center (SDSC) at the University of California San Diego (UCSD); and the Center for Advanced Research in Biotechnology (CARB/UMBI/NIST)—three members of the Research Collaboratory for Structural Bioinformatics. This work is supported by grants from National Science Foundation (NSF), National Institute of General Medical Sciences (NIGMS), Office of Science, Department of Energy (DOE), National Library of Medicine (NLM), National Cancer Institute (NCI), National Center for Research Resources (NCRR), National Institute of Biomedical Imaging and Bioengineering (NIBIB) and National Institute of Neurological Disorders and Stroke (NINDS). The RCSB PDB is a member of wwwPDB. We wish to thank the many users who have provided input into the development of the re-engineered RCSB PDB.
REFERENCES
- 1.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein F.C., Koetzle,T.F., Williams,G.J.B., Meyer,E.F.,Jr, Brice,M.D., Rodgers,J.R., Kennard,O., Shimanouchi,T. and Tasumi,M. (1977) Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol., 112, 535–542. [DOI] [PubMed] [Google Scholar]
- 3.Westbrook J., Feng,Z., Jain,S., Bhat,T.N., Thanki,N., Ravichandran,V., Gilliland,G.L., Bluhm,W., Weissig,H., Greer,D.S. et al. (2002) The Protein Data Bank: unifying the archive. Nucleic Acids Res., 30, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhat T.N., Bourne,P., Feng,Z., Gilliland,G., Jain,S., Ravichandran,V., Schneider,B., Schneider,K., Thanki,N., Weissig,H. et al. (2001) The PDB data uniformity project. Nucleic Acids Res., 29, 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourne P.E., Addess,K.J., Bluhm,W.F., Chen,L., Deshpande,N., Feng,Z., Fleri,W., Green,R., Merino-Ott,J.C., Townsend-Merino,W., Weissig,H., Westbrook,J. and Berman,H.M. (2004) The distribution and query systems of the RCSB Protein Data Bank. Nucleic Acids Res., 32, D223–D225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourne P.E., Berman,H.M., Watenpaugh,K., Westbrook,J.D. and Fitzgerald,P.M.D. (1997) The macromolecular Crystallographic Information File (mmCIF). Methods Enzymol., 277, 571–590. [DOI] [PubMed] [Google Scholar]
- 7.Westbrook J., Henrick,K., Ulrich,E.L. and Berman,H.M. (2004) Definition and exchange of crystallographic data. In International Tables for Crystallography. Kluwer Academic Publishers, Dordrecht, The Netherlands, Vol. G (in press). [Google Scholar]
- 8.Westbrook J., Ito,N., Nakamura,H., Henrick,K. and Berman,H.M. (2004) PDBML: the representation of archival macromolecular structure data in XML. Bioinformatics, doi:10.1093/bioinformatics/bti082. [DOI] [PubMed] [Google Scholar]
- 9.The Gene Ontology Consortium (2000) Gene Ontology: tool for the unification of biology. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanehisa M. (1997) A database for post-genome analysis. Trends Genet., 13, 375–376. [DOI] [PubMed] [Google Scholar]
- 11.Kanehisa M. and Goto,S. (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res., 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wheeler D.L., Church,D.M., Edgar,R., Federhen,S., Helmberg,W., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Sequeira,E. et al. (2004) Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res., 32, D35–D40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L., Oughtred,R., Berman,H.M. and Westbrook,J. (2004) TargetDB: a target registration database for structural genomics projects. Bioinformatics, 20, 2860–2862. [DOI] [PubMed] [Google Scholar]
- 15.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 16.Hempstead P.D., Yewdall,S.J., Fernie,A.R., Lawson,D.M., Artymiuk,P.J., Rice,D.W., Ford,G.C. and Harrison,P.M. (1997) Comparison of the three-dimensional structures of recombinant human H and horse L ferritins at high resolution. J. Mol. Biol., 268, 424–448. [DOI] [PubMed] [Google Scholar]
- 17.Greer D.S., Westbrook,J.D. and Bourne,P.E. (2002) An ontology driven architecture for derived representations of macromolecular structure. Bioinformatics, 18, 1280–1281. [DOI] [PubMed] [Google Scholar]
- 18.Walther D. (1997) WebMol—a Java-based PDB viewer. Trends Biochem. Sci., 22, 274–275. [DOI] [PubMed] [Google Scholar]
- 19.Conte L.L., Brenner,S.E., Hubbard,T.J., Chothia,C. and Murzin,A.G. (2002) SCOP database in 2002: refinements accommodate structural genomics. Nucleic Acids Res., 30, 264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]