Abstract
PDB-Ligand (http://www.idrtech.com/PDB-Ligand/) is a three-dimensional structure database of small molecular ligands that are bound to larger biomolecules deposited in the Protein Data Bank (PDB). It is also a database tool that allows one to browse, classify, superimpose and visualize these structures. As of May 2004, there are about 4870 types of small molecular ligands, experimentally determined as a complex with protein or DNA in the PDB. The proteins that a given ligand binds are often homologous and present the same binding structure to the ligand. However, there are also many instances wherein a given ligand binds to two or more unrelated proteins, or to the same or homologous protein in different binding environments. PDB-Ligand serves as an interactive structural analysis and clustering tool for all the ligand-binding structures in the PDB. PDB-Ligand also provides an easier way to obtain a number of different structure alignments of many related ligand-binding structures based on a simple and flexible ligand clustering method. PDB-Ligand will be a good resource for both a better interpretation of ligand-binding structures and the development of better scoring functions to be used in many drug discovery applications.
INTRODUCTION
Understanding the interaction between protein and small molecular ligand is very important in post-genomics life science because many important proteins require small molecular ligands or cofactors such as ATP or NAD, in order to function properly. In addition, there is a huge need to design small molecular inhibitors for new drug discovery, based on the analysis of protein–ligand interaction.
The first step for understanding protein–ligand interaction would be to analyze the known protein–ligand complex structures in the Protein Data Bank (PDB) (1) (http://www.rcsb.org/). When analyzing protein–ligand structures, it is often necessary to cluster related ligand-binding structures, according to the ligand conformation, the three-dimensional (3D) ligand-binding structures, and the relative position and orientation of any important residues at the ligand-binding sites.
There are already many protein cluster databases. Protein structure classification databases such as SCOP (2), FSSP (3) and CATH (4) are based on the clustering of the whole 3D structures of protein domains. Other databases such as Pfam (5), Swiss-Model (6) and CDD (7) are primarily based on sequence similarities. With the structural genomics initiatives, these databases have been greatly expanded in size and the structure and function of many experimentally undetermined proteins are now readily inferred using these databases. However, these databases are more focused on the protein structure and function rather than on the structures of ligand or ligand-binding sites. These ligand-binding structures are probably more important in many post-genomics applications such as small molecular inhibitor design for new drug discovery.
There are also many web-based databases of the ligand-binding structure of PDB, including PDBSum (8), Relibase (9) (http://relibase.ebi.ac.uk/), Hic-Up (http://xray.bmc.uu.se/hicup/) and PLD (10). Although these ligand databases provide very useful information on the ligand–protein binding structures, they cannot easily be used to compare or to classify the ligand-binding structures in 3D. Therefore, there is a need for a convenient tool to analyze and classify the ligand-binding structures based on the clustering of the relevant 3D-structures using all the PDB data.
PDB-Ligand is a 3D ligand-binding structure database, derived from the PDB. It is also a database tool that can be used to build such a database and for conveniently browsing through these databases. One novel feature of PDB-Ligand is that it allows an interactive clustering of ligand-binding structures based on user-specific clustering criteria such as root-mean-square deviation (RMSD) using flexible combinations of the atoms at the ligand-binding sites.
DATABASE CONTENTS AND FEATURES
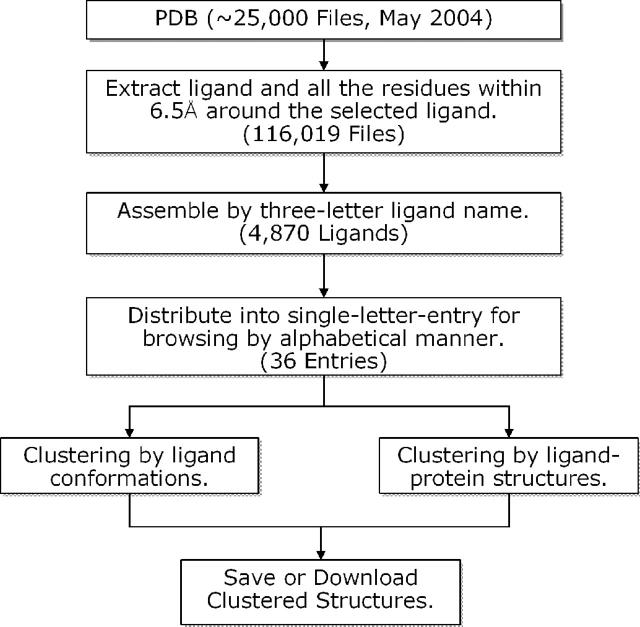
Figure 1 shows the scheme used in PDB-Ligand database construction. Currently, PDB-Ligand holds 4870 different types of ligands, extracted from 116,019 ligand-binding structures derived from about 25 000 PDB entries. In PDB-Ligand, a ligand-binding structure is defined by the ligands and all the residues and other atoms that are within 6.5 Å around the ligand. Thus, every ligand-binding structure in PDB-Ligand database is surrounded by the residues of the protein, DNA, RNA, solvent or even other ligands. PDB-Ligand uses Chime Plug-in (http://www.mdli.com) as a web-based molecular graphics interface for visualization. It also provides a URL-link to the original PDB file for each ligand-binding structure so that one can easily view the whole ligand–protein structure with other related ligand-binding structures.
Figure 1.
Scheme used in the PDB-Ligand database construction.
One of the most useful features of PDB-Ligand is the interactive clustering of ligand-binding structures, based on the RMSD between different ligand-binding structures. When analyzing the ligand-binding structures for many biologically important ligands such as ATP or FAD, one wants to know how many are in a similar binding environment, and how similar they are in 3D conformation. PDB-Ligand database and its clustering tool allow fast structural classification of the similar ligand-binding structures from all the ligand-binding structures in the PDB. The structure-based clustering feature of PDB-Ligand may be more effectively used with other ligand-binding analysis tools such as LIGPLOT (11) and LPC (12), or with other ligand databases such as Relibase (9) and Ligand-Depot (http://ligand-depot-i.rutgers.edu/).
In addition, PDB-Ligand allows more flexible clustering based on both the ligand and the protein residues at the ligand-binding sites. This feature is useful, for example, when analyzing the same ligand-binding structures of a structurally related protein family.
CLUSTERING AND STRUCTURE ALIGNMENT
Since PDB-Ligand aims to be a 3D ligand-binding structure database with an interactive clustering feature, it only uses the ligand and the residues within 6.5 Å around the ligand in RMSD-based clustering by structure–structure alignment. Thus, using only the selected number of atoms at the ligand-binding site can greatly speed up the structure-alignment operation for RMSD calculations, while including all the important residues at the ligand-binding sites.
In PDB-Ligand, the clustering of ligand-binding structures is based on the RMSD value between all the corresponding atoms in the ligand after 3D structure superposition by Kabsch method (13). By default, all atoms of the ligand are considered in the superimposition for clustering. Therefore, in this case, every ligand in each cluster will have an overall structural similarity defined by the RMSD cut-off value (default is 0.5 Å).
However, if the ligand shows several different binding modes, it is more important to consider a part of ligand atoms and/or any critical residues at the ligand-binding site in the clustering process. In order to provide users with more convenient atom-selection, PDB-Ligand uses ‘copy-and-paste’ mechanism, based on chime script utilities (e.g. see E. Martz, http://www.umass.edu/microbio/chime/). For example, if a user selects main chain atoms of the residues at the ligand-binding site in the graphics window, these atoms are listed in the chime-log window, then they can be used in the clustering by ‘copy-and-paste’ into the selected atoms window. The user can copy any set of atoms shown in this window and paste them into the ‘Selected Atoms’ window. These atoms are then used to compute the superposition matrix. The simplicity and flexibility of the atom selection mechanism allow the users to perform a more precise clustering of ligand-binding structures. Currently, all the atoms in ligand and protein main chain atoms (N, CA, C and O), are allowed in the clustering.
As a clustering method, a simple greedy algorithm similar to that used by Hobohm and Sander (14) is used. In PDB-Ligand, a reference ligand-binding structure is always the one at the top of the list. Based on a given RMSD cut-off, all the structures similar to the reference structure are clustered together and removed from the list. The clustering is complete if no structure remains in the list.
AN EXAMPLE: ATP-BINDING STRUCTURES
In the current release of PDB-Ligand, there are 321 ATP-binding structures derived from 161 PDB entries. The ATP is the 46th most abundant ligand. If these 321 ATP-binding structures are clustered using 0.5 Å RMSD cut-off, we obtain 165 clusters (see Table 1). It means that there are 165 different conformations of ATP, each one of which is different from all others, at least, by 0.5 Å in RMSD. If 1.0 Å RMSD is used, we obtain 91 different structural clusters for ATP.
Table 1. Examples of the structure-based clustering of selected ligands in PDB-Ligand database.
| Ligand name | Description | Number of models/PDB entries | Number of clusters, classified by RMSD cut-off valuesa | |||||
|---|---|---|---|---|---|---|---|---|
| 0.5 Å | 1.0 Å | 1.5 Å | 2.0 Å | 2.5 Å | 3.0 Å | |||
| NAG | N-acetyl-d-glucosamine | 5227/967 | 223 | 20 | 8 | 5 | 5 | 5 |
| NAD | Nicotinamide-adenine–dinucleotide | 831/306 | 171 | 67 | 48 | 27 | 12 | 8 |
| ADP | Adenosine-5′-diphosphate | 623/292 | 176 | 64 | 25 | 10 | 7 | 6 |
| FAD | Flavin-adenine dinucleotide | 599/320 | 105 | 35 | 25 | 16 | 12 | 8 |
| ATP | Adenosine-5′-triphosphate | 321/161 | 165 | 91 | 32 | 10 | 5 | 3 |
| NAP | NADP, nicotinamide-adenine-dinucleotide phosphate | 278/150 | 110 | 53 | 29 | 16 | 12 | 5 |
| NDP | NADPH | 222/103 | 91 | 45 | 27 | 19 | 11 | 6 |
| AMP | Adenosine monophosphate | 217/92 | 52 | 17 | 4 | 2 | 1 | 1 |
| GDP | Guanosine-5′-diphosphate | 198/130 | 57 | 24 | 8 | 4 | 2 | 1 |
| GTP | Guanosine-5′-triphosphate | 135/46 | 38 | 25 | 17 | 8 | 3 | 2 |
| HEM | Protoporphyrin Ix containing Fe | 2320/1112 | 357 | 22 | 9 | 8 | 6 | 5 |
| HEC | Heme C | 389/110 | 131 | 38 | 4 | 1 | 1 | 1 |
| FMN | Flavin mononucleotide | 295/181 | 46 | 18 | 6 | 3 | 2 | 1 |
| DLE | d-Leucine | 177/22 | 50 | 12 | 1 | 1 | 1 | 1 |
| COA | Coenzyme A | 122/63 | 68 | 51 | 30 | 30 | 17 | 9 |
| ACO | Acetyl coenzyme A | 56/24 | 34 | 18 | 14 | 9 | 7 | 3 |
| PTR | O-phosphotyrosine | 127/79 | 50 | 21 | 3 | 1 | 1 | 1 |
In PDB-Ligand database, most abundant ligands are usually trivial ones such as MSE (selenomethionine), MG (magnesium ion), SO4 (Sulfate ion), etc. (see statistics at PDB-Ligand, http://www.idrtech.com/PDB-Ligand/).
aRMSD cut-off values are based on all ligand atoms, thus each cluster will have similar ligand conformations (see text).
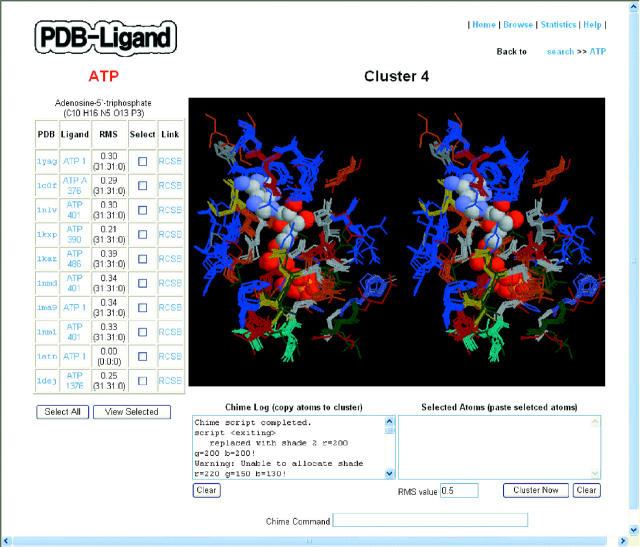
Figure 2 shows a sample cluster of ATP-binding structures using 0.5 Å RMSD cut-off. One can easily see in this figure the common 3D structure of the amino acids surrounding the ligand. Interestingly, based on SCOP 1.65 protein family classification (2), the ATP-binding structures shown in Figure 2 are classified as Actin/Hsp70 protein family. This result may be useful for the users who want to investigate further the ATP-binding structures of such protein family.
Figure 2.
A typical ATP-ligand cluster, obtained using default options. The 10 ATP-binding structures from 10 PDB entries are displayed in the ligand-information table on the left-hand side. The superimposed ATP-binding structures are shown in the chime-window in stereo view. ATP is shown in CPK model and the surrounding amino acids as connected line segments in different colors depending on the residue type. The chime utilities were used for the graphics rendering.
FUTURE DIRECTIONS AND APPLICATIONS
The ligand-binding structures in PDB-Ligand will be updated, at least, every four months. In addition, the methods and algorithms for ligand-binding structure clustering will be improved for speed and convenience. Substructure search among ligand structures will also be included in the future. This feature will be useful in analyzing binding structures of various functional groups in many important ligands. Protein sequence and structure information based on the clustered ligand-binding structures will be also useful because it provides more complete information about ligand-binding structures. We also believe that the methods and the strategies used in PDB-Ligand, based on the clustering of ligand-binding structures, will be very useful in many applications for new drug discovery. For an example, based on the classification of similar ligand-binding structures, we have a plan to derive more accurate scoring functions for ligand-docking, virtual screening and lead-optimization for specific target proteins.
AVAILABILITY
PDB-Ligand is freely accessible through the URL at http://www.idrtech.com/PDB-Ligand/.
Acknowledgments
ACKNOWLEDGEMENTS
We thank B. K. Lee for useful discussions and for valuable suggestions on the manuscript. We also thank H. C. Shin, S. M. Kim, C. K. Han, J. H. Yoon, Y. H. In and N. D. Kim for useful discussions and comments. We also thank M. R. Roh and Y. W. Kim for maintaining the website. This study was supported by a grant of Korean Health 21 R&D Project, Ministry of Health and Welfare, Republic of Korea (Grant ID: 03-PJ2-PG4-BD02-0001).
REFERENCES
- 1.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreeva A., Howorth,D., Brenner,S.E., Hubbard,T.J.P., Chothia,C. and Murzin,A.G. (2004) SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res., 32, D226–D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holm L. and Sander,C. (1996) Mapping the protein universe. Science, 273, 595–602. [DOI] [PubMed] [Google Scholar]
- 4.Pearl F.M.G., Lee,D., Bray,J.E., Sillitoe,I., Todd,A.E., Harrison,A.P., Thornton,J.M. and Orengo,C.A. (2000) Assigning genomic sequences to CATH. Nucleic Acids Res., 28, 277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bateman A., Coin,L., Durbin,R., Finn,R.D., Hollich,V., Griffiths-Jones,S., Khanna,A., Marshall,M., Moxon,S., Sonnhammer,E.L.L. et al. (2004) The Pfam protein families database. Nucleic Acids Res., 32, D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kopp J. and Schwede,T. (2004) The SWISS-MODEL Repository of annotated three-dimensional protein structure homology models. Nucleic Acids Res., 32, D230–D234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchler-Bauer A., Anderson,J.B., DeWeese-Scott,C., Fedorova,N.D., Geer,L.Y., He,S., Hurwitz,D.I., Jackson,J.D., Jacobs,A.R., Lanczycki,C.J. et al. (2003) CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res., 31, 383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laskowski R.A. (2001) PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res., 29, 221–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendlich M., Bergner,A., Gunther,J. and Klebe,G. (2003) Relibase—design and development of a database for comprehensive analysis of protein–ligand interactions. J. Mol. Biol., 326, 607–620. [DOI] [PubMed] [Google Scholar]
- 10.Puvanendrampillai D. and Mitchell,J.B.O. (2003) Protein Ligand Database (PLD): additional understanding of the nature and specificity of protein–ligand complexes. Bioinformatics, 19, 1856–1857. [DOI] [PubMed] [Google Scholar]
- 11.Wallace A.C., Laskowski,R.A. and Thornton,J.M. (1995) LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng., 8, 127–134. [DOI] [PubMed] [Google Scholar]
- 12.Sobolev V., Sorokine,A., Prilusky,J., Abola,E.E. and Edelman,M. (1999) Automated analysis of interatomic contacts in proteins. Bioinformatics, 15, 327–332. [DOI] [PubMed] [Google Scholar]
- 13.Kabsch W. (1978) A discussion of the solution for the best rotation to relate two sets of vectors. Acta Crystallogr., A34, 827–828. [Google Scholar]
- 14.Hobohm J. and Sander,C. (1994) Enlarged representative set of protein structures. Protein Sci., 3, 522–524. [DOI] [PMC free article] [PubMed] [Google Scholar]