Abstract
The international ImMunoGeneTics information system® (IMGT) (http://imgt.cines.fr), created in 1989, by the Laboratoire d'ImmunoGénétique Moléculaire LIGM (Université Montpellier II and CNRS) at Montpellier, France, is a high-quality integrated knowledge resource specializing in the immunoglobulins (IGs), T cell receptors (TRs), major histocompatibility complex (MHC) of human and other vertebrates, and related proteins of the immune systems (RPI) that belong to the immunoglobulin superfamily (IgSF) and to the MHC superfamily (MhcSF). IMGT includes several sequence databases (IMGT/LIGM-DB, IMGT/PRIMER-DB, IMGT/PROTEIN-DB and IMGT/MHC-DB), one genome database (IMGT/GENE-DB) and one three-dimensional (3D) structure database (IMGT/3Dstructure-DB), Web resources comprising 8000 HTML pages (IMGT Marie-Paule page), and interactive tools. IMGT data are expertly annotated according to the rules of the IMGT Scientific chart, based on the IMGT-ONTOLOGY concepts. IMGT tools are particularly useful for the analysis of the IG and TR repertoires in normal physiological and pathological situations. IMGT is used in medical research (autoimmune diseases, infectious diseases, AIDS, leukemias, lymphomas, myelomas), veterinary research, biotechnology related to antibody engineering (phage displays, combinatorial libraries, chimeric, humanized and human antibodies), diagnostics (clonalities, detection and follow up of residual diseases) and therapeutical approaches (graft, immunotherapy and vaccinology). IMGT is freely available at http://imgt.cines.fr.
INTRODUCTION
The international ImMunoGeneTics information system® (IMGT) (http://imgt.cines.fr) (1), created in 1989 by the Laboratoire d'ImmunoGénétique Moléculaire LIGM (Université Montpellier II and CNRS) at Montpellier, France, is a high-quality integrated knowledge resource, specializing in the immunoglobulins (IGs), T cell receptors (TRs), major histocompatibility complex (MHC) of human and other vertebrates, and related proteins of the immune systems (RPI) that belong to the immunoglobulin superfamily (IgSF) and to the MHC superfamily (MhcSF). IMGT is a widely used international reference in immunogenetics and immunoinformatics, and as such, provides a common access to standardized data from genome, proteome, genetics and three-dimensional (3D) structures (1–4).
The accuracy, consistency and integration of the IMGT data, as well as the coherence between the different IMGT components (databases, tools and web resources) are based on IMGT-ONTOLOGY (5), the first ontology in the domain, which provides a semantic specification of the terms to be used in immunogenetics and immunoinformatics, and thus, allows the management of immunogenetics knowledge for all vertebrate species. IMGT-ONTOLOGY comprises five main concepts: IDENTIFICATION, CLASSIFICATION, DESCRIPTION, NUMEROTATION and OBTENTION (3–5). Standardized keywords, standardized labels and sequence annotation rules, standardized IG and TR gene nomenclature, the IMGT unique numbering, and standardized origin/methodology were defined, respectively, based on these five main concepts.
IMGT-ONTOLOGY concepts have been formalized, for the biologists and IMGT users, in the IMGT Scientific chart and, for the computing scientists, in IMGT-ML which uses XML (eXtensible Markup Language) schemata. The IMGT Scientific chart is constituted by controlled vocabulary and annotation rules for data and knowledge management of the IG, TR, MHC and RPI of all vertebrate species. All IMGT data are expertly annotated according to the IMGT Scientific chart rules. IMGT-ML is the formalization of IMGT-ONTOLOGY using XML schemata for interoperability with other information systems.
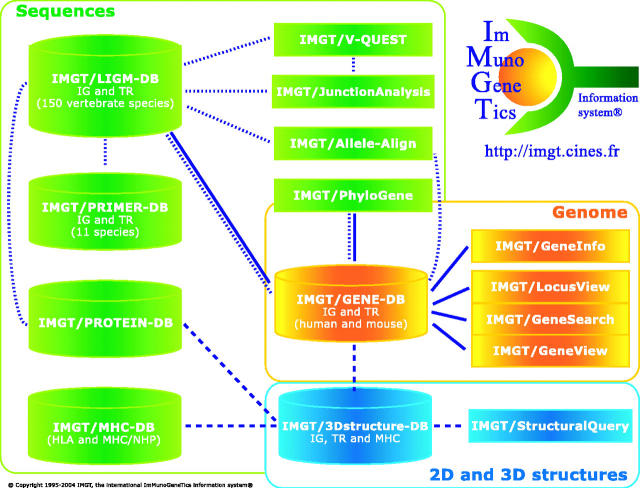
The IMGT information system consists of databases, tools and Web resources summarized in Figure 1. Databases include sequence databases (IMGT/LIGM-DB, IMGT/MHC-DB, IMGT/PRIMER-DB and IMGT/PROTEIN-DB), one genome database (IMGT/GENE-DB) and one 3D structure database (IMGT/3Dstructure-DB). Interactive tools are provided for sequence analysis (IMGT/V-QUEST, IMGT/Junction Analysis, IMGT/Allele-Align and IMGT/PhyloGene), genome analysis (IMGT/LocusView, IMGT/GeneView, IMGT/GeneSearch, IMGT/CloneSearch and IMGT/GeneInfo) and 3D structure analysis (IMGT/StructuralQuery). Web resources (‘IMGT Marie-Paule page’) comprise 8000 HTML pages of synthesis [IMGT Repertoire (for IG and TR, MHC, RPI)], knowledge [IMGT Scientific chart, IMGT Education (Aide-mémoire, Tutorials, Questions and answers, IMGT Lexique, the IMGT Medical page, the IMGT Veterinary page, and the IMGT Biotechnology page), IMGT Index], and external links [IMGT Bloc-notes (The IMGT Immunoinformatics page, Interesting links, etc.) and Other accesses (SRS, BLAST, etc.)].
Figure 1.
The IMGT international ImMunoGeneTics information system® (http://imgt.cines.fr). Genomics, genetics and structural components (databases and tools) are in yellow, green and blue, respectively. The IMGT Repertoire and other Web resources are not shown. Interactions in the genomics, genetics and structural approaches are represented with continuous, dotted and broken lines, respectively.
The IMGT components (databases, tools and IMGT Repertoire web resources) have been developed according to three main biological approaches. The IMGT genomics approach is gene-centered and mainly orientated toward the study of the genes within their loci and on the chromosomes. The IMGT genetics approach refers to the study of the genes in relationship with their sequence polymorphisms and mutations, their expression, their specificity and their evolution. The IMGT structural approach refers to the study of the 2D and 3D structures of the IG, TR, MHC and RPI, and to the antigen- or ligand-binding characteristics in relationship with the protein functions, polymorphisms and evolution. IMGT-Choreography, based on the Web service architecture paradigm, will enable significant biological and clinical requests involving every part of the IMGT information system.
IMGT GENOMICS COMPONENTS
IMGT genome database
IMGT/GENE-DB is the comprehensive IMGT genome database, created by LIGM (Montpellier, France) on the Web since January 2003 (6). All the human and mouse IG and TR genes are available in IMGT/GENE-DB. The human IMGT gene names (7,8) were approved by the Human Genome Organisation (HUGO) Nomenclature Committee (HGNC) in 1999 (9), and entered in IMGT/GENE-DB, Genome DataBase GDB (Canada), LocusLink at NCBI (USA) and GeneCards. Reciprocal links exist between IMGT/GENE-DB and the generalist nomenclature (HGNC Genew) and genome databases (GDB, LocusLink and Entrez Gene at NCBI, and GeneCards). The mouse IG and TR gene names with IMGT reference sequences were provided by IMGT to HGNC and to the Mouse Genome Database (MGD) in July 2002. Queries in IMGT/GENE-DB can be performed according to IG and TR gene classification criteria (7,8) and IMGT reference sequences have been defined for each allele of each gene based on one or, whenever possible, several of the following criteria: germline sequence, first sequence published, longest sequence and mapped sequence. IMGT/GENE-DB interacts dynamically with IMGT/LIGM-DB to download and display gene-related sequence data. In November 2004, IMGT/GENE-DB contained 1375 genes and 2204 alleles (673 IG and TR genes and 1028 alleles from Homo sapiens, and 702 IG and TR genes and 996 alleles from Mus musculus, Mus cookii, Mus pahari, Mus spretus, Mus saxicola and Mus minutoides).
IMGT genome analysis tools and Web resources
Tools
The IMGT genome analysis tools comprise IMGT/LocusView, IMGT/GeneView, IMGT/GeneSearch, IMGT/CloneSearch and IMGT/GeneInfo. IMGT/LocusView and IMGT/GeneView manage the locus organization and the gene location and provide the display of physical maps for the human IG, TR and MHC loci and for the mouse TRA/TRD locus. IMGT/GeneSearch and IMGT/CloneSearch allow retrieval of information concerning genes and clones analysed in IMGT/LocusView. IMGT/GeneInfo provides and displays information on the potential TR rearrangements in human and mouse (10).
Genome Web resources
The genomic Web resources are compiled in the IMGT Repertoire ‘Locus and genes’ section that includes Chromosomal localizations, Locus representations, Locus description, Gene exon/intron organization, Gene exon/intron splicing sites, Gene tables, Potential germline repertoires, the complete lists of human and mouse IG and TR genes, and the correspondences between nomenclatures (7,8). The IMGT Repertoire ‘Probes and RFLP’ section provides data on gene insertion/deletion.
IMGT GENETICS COMPONENTS
IMGT sequence databases
IMGT/LIGM-DB
The comprehensive IMGT database of IG and TR nucleotide sequences from human and other vertebrate species, with translation for fully annotated sequences. It was created in 1989 by LIGM (Montpellier, France), and is on the Web since July 1995 (1–4,11) (http://www3.oup.co.uk/nar/database/summary/504). In November 2004, IMGT/LIGM-DB contained more than 87 700 sequences of 150 vertebrate species. The unique source of data for IMGT/LIGM-DB is EMBL (12), which shares data with the other two generalist databases GenBank and DNA DataBank of Japan (DDBJ). Based on expert analysis, specific detailed annotations are added to IMGT flat files. The Web interface allows searches according to immunogenetic specific criteria and is easy to use without any knowledge in a computing language. Selection is displayed at the top of the resulting sequence pages, so the users can check their own queries. Users have the ability to modify their request or to consult the results with a choice of nine possibilities (3). IMGT/LIGM-DB gene and allele name assignment and sequence annotations are performed according to the IMGT Scientific chart rules. These annotations allow retrieval of data from IMGT/LIGM-DB for queries in other IMGT databases or tools. As an example, the IMGT/LIGM-DB accession numbers of the cDNA expressed sequences for each human and mouse IG and TR gene are available, with direct links to IMGT/LIGM-DB, in the IMGT/GENE-DB entries. IMGT/LIGM-DB data are also distributed by anonymous FTP servers at CINES (ftp://ftp.cines.fr/IMGT/) and EBI (ftp://ftp.ebi.ac.uk/pub/databases/imgt/) and from many Sequence Retrieval System (SRS) sites. IMGT/LIGM-DB can be searched by BLAST or FASTA on different servers (EBI, IGH, INFOBIOGEN, Institut Pasteur, etc.).
IMGT/Automat for IMGT/LIGM-DB annotations
IMGT/Automat is an integrated internal IMGT Java tool that automatically performs the annotation of rearranged cDNA sequences that represent half of the IMGT/LIGM-DB content. The annotation procedure includes the IDENTIFICATION of the sequences, the CLASSIFICATION of the IG and TR genes and alleles, and the DESCRIPTION of all IG and TR specific and constitutive motifs within the nucleotide sequences. Accuracy and reliability of the annotation are mainly estimated by the program itself with the evaluation of the alignment scores, the deduced sequence functionality, and the coherence of the characterized and delimited IG and TR motifs. More than 7500 human and mouse IG and TR cDNA sequences have been automatically annotated by the IMGT/Automat tool, with annotations being as reliable and accurate as those provided by a human annotator.
Other IMGT sequences databases
IMGT/PRIMER-DB (13) (http://www3.oup.co.uk/nar/database/summary/505) is the IMGT oligonucleotide primer database for IG and TR, created by LIGM, Montpellier in collaboration with EUROGENTEC SA, Belgium, on the Web since February 2002. In November 2004, IMGT/PRIMER-DB contained 1827 entries. IMGT/PRIMER-DB provides standardized information on oligonucleotides (or Primers) and combinations of primers (Sets, Couples) for IG and TR. These primers are useful for combinatorial library constructions, scFv, phage display or microarray technologies. The IMGT primer cards are linked to the IMGT/LIGM-DB flat files, and to the IMGT Repertoire (IMGT Colliers de Perles and Alignments of alleles of the IMGT/LIGM-DB reference sequence used for the primer description). IMGT/PROTEIN-DB is a new database related to IG and TR amino acid sequences. The database will be available on the IMGT website in 2005. IMGT/MHC-DB comprises databases hosted at the EBI and includes a database of human MHC allele sequences or IMGT/MHC-HLA (IMGT/HLA), developed by Cancer Research (UK) and maintained by ANRI (London, UK), on the Web since December 1998, and a database of MHC sequences from non-human primates IMGT/MHC-NHP, curated by BPRC (The Netherlands) on the Web since April 2002 (14).
IMGT sequence analysis tools and genetics Web resources
IMGT/V-QUEST
IMGT/V-QUEST (V-QUEry and STandardization) is an integrated software for IG and TR (15), used for the identification of the V, D and J genes and of their mutations. This user-friendly tool analyses an input IG or TR germline or rearranged variable nucleotide sequence. The IMGT/V-QUEST results comprise the identification of the V, D and J genes and alleles and the nucleotide alignments by comparison with sequences from the IMGT reference directory, the FR-IMGT and CDR-IMGT delimitations based on the IMGT unique numbering, the translation of the input sequence, the display of nucleotide and amino acid mutations compared to the closest IMGT reference sequence, the identification of the JUNCTION and results from IMGT/JunctionAnalysis (default option), and the V-REGION IMGT Collier de Perles.
IMGT/JunctionAnalysis
IMGT/JunctionAnalysis (16) is a tool, complementary to IMGT/V-QUEST, which provides a thorough analysis of the V-J and V-D-J junctions that confer the antigen receptor specificity to IG and TR rearranged genes. IMGT/JunctionAnalysis identifies the D-GENEs and alleles involved in the IGH, TRB and TRD V-D-J rearrangements by comparison with the IMGT reference directory, and delimits precisely the P, N and D regions. Several hundreds of junction sequences can be analysed simultaneously.
IMGT/Allele-Align
IMGT/Allele-Align is used for the detection of polymorphisms. It allows the comparison of two alleles, highlighting the nucleotide and amino acid differences.
IMGT/PhyloGene
IMGT/PhyloGene (17) is an easy to use tool for phylogenetic analysis of IG and TR variable region (V-REGION) and constant domain (C-DOMAIN) sequences. This tool is particularly useful in developmental and comparative immunology. The users can analyse their own sequences by comparing with the IMGT standardised reference sequences for human and mouse IG and TR.
Genetics Web resources
The genetics Web resources are compiled in the IMGT Repertoire ‘Proteins and alleles’ section which includes Protein displays, Alignments of alleles, Tables of alleles, Allotypes, Isotypes, etc.
IMGT STRUCTURAL COMPONENTS
IMGT structural database
IMGT/3Dstructure-DB is the IMGT 3D structure database for IG, TR, MHC and RPI, created by LIGM, and on the Web since November 2001 (18). In November 2004, IMGT/3Dstructure-DB contained 809 atomic coordinate files. IMGT/3Dstructure-DB comprises IG, TR, MHC and RPI with known 3D structures. Coordinate files extracted from the Protein Data Bank (PDB) (http://www.rcsb.org/pdb/) (19) are renumbered according to the standardized IMGT unique numbering (20,21). The IMGT/3Dstructure-DB cards provide IMGT annotations [assignment of IMGT genes and alleles, IMGT chain and domain labels, IMGT Colliers de Perles (22) on one layer and two layers], downloadable renumbered IMGT/3Dstructure-DB flat files, vizualization tools and external links. The IMGT/3Dstructure-DB residue cards provide detailed information on the inter- and intra-domain contacts of each residue position. An IMGT/3Dstructure-DB card provides receptor and chain description, IMGT gene and allele names, domain delimitations, amino acid positions according to the IMGT unique numbering. Structural and functional domains of the IG and TR chains comprise the variable domain or V-DOMAIN (9-strand β-sandwich) that corresponds to the V-J-REGION or V-D-J-REGION and is encoded by two or three genes (7,8), the constant domain or C-DOMAIN (7-strand β-sandwich), and, for the MHC chains, the groove domain or G-DOMAIN (4 β-strand and one α-helix). The IMGT unique numbering has been extended to the V-LIKE-DOMAINs (20) and C-LIKE-DOMAINs (21) of IgSF proteins other than IG and TR, and to the G-LIKE-DOMAINs of MhcSF proteins other than MHC.
IMGT structural analysis tool and Web resources
IMGT/StructuralQuery
The IMGT/StructuralQuery tool (18) analyses the interactions of the residues of the antigen receptors (IG and TR), MHC, RPI, antigens and ligands. The contacts are described per domain (intra- and inter-domain contacts) and annotated in terms of IMGT labels (chains, domains), positions (IMGT unique numbering), backbone or side-chain implication. IMGT/StructuralQuery allows retrieval of the IMGT/3Dstructure-DB entries, based on specific structural characteristics, such as φ and ψ angles, accessible surface area (ASA), amino acid type, distance in angstrom between amino acids and CDR-IMGT lengths. It is currently available for the V-DOMAINs.
Structural Web resources
The structural Web resources are compiled in the IMGT Repertoire ‘2D and 3D structures’ section that includes 2D representations or IMGT Colliers de Perles (22), 3D representations, FR-IMGT and CDR-IMGT lengths, amino acid chemical characteristics profiles (23), etc.
IMGT-CHOREOGRAPHY
IMGT-Choreography (24) is based on the Web service architecture paradigm (see W3C; http://www.w3.org/). Its goal is to orchestrate dynamic procedure calls between IMGT databases querying and analysis tools. Conversations between Web services are expressed using the sole IMGT-ML language both for queries and result fetches. This ensures semantic consistency between exchanged messages as IMGT-ML (available at IMGT Index>IMGT-ML) is an XML schema formalization of the IMGT-ONTOLOGY concepts. IMGT Web services are developed using the JAVA programming language and deployed using the Apache Axis (http://ws.apache.org/axis/) Web services development framework. Composition and chaining of IMGT Web services through IMGT-Choreography will enable processing of complex significant biological and clinical requests involving every part of the IMGT information system.
CONCLUSION
Since July 1995, IMGT has been available on the Web at the IMGT Home page http://imgt.cines.fr (Montpellier, France). IMGT has an exceptional response with more than 140 000 requests a month. IMGT is the international reference for immunogenetics and immunoinformatics and provides a common access to all standardized data that include nucleotide and protein sequences, oligonucleotide primers, gene maps, genetic polymorphisms, specificities, 2D and 3D structures, based on IMGT-ONTOLOGY. The information is of much value to clinicians and biological scientists in general. IMGT databases and tools are extensively queried and used by scientists, from both academic and industrial laboratories, who are equally distributed between the United States (one-third), Europe (one-third) and the remaining World (one-third). IMGT is used in very diverse domains: (i) fundamental research and medical research (repertoire analysis of the IG antibody sites and of the TR recognition sites in normal and pathological situations such as autoimmune diseases, infectious diseases, AIDS, leukemias, lymphomas, myelomas); (ii) veterinary research (IG and TR repertoires in farm and wild life species); (iii) genome diversity and genome evolution studies of the adaptive immune responses; (iv) structural evolution of the IgSF and MhcSF proteins; (v) biotechnology related to antibody engineering (single chain Fragment variable (scFv), phage displays, combinatorial libraries, chimeric, humanized and human antibodies); (vi) diagnostics (clonalities, detection and follow up of residual diseases); and (vii) therapeutical approaches (grafts, immunotherapy, vaccinology). Owing to its high quality and data distribution based on IMGT-ONTOLOGY, IMGT has an important role to play in the development of immunogenetics Web services. The design of IMGT-Choreography and the creation of dynamic interactions between the IMGT databases and tools, using Web services and IMGT-ML, represent novel and major developments of IMGT, the international reference in immunogenetics and immunoinformatics.
CITING IMGT
Users are requested to cite this article and quote the IMGT home page URL, http://imgt.cines.fr.
Acknowledgments
ACKNOWLEDGEMENTS
E.D. is holder of a doctoral grant from the Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche (MENESR). Q.K. was the recipient of a doctoral grant from the MENESR and is currently supported by a grant from the Association pour la Recherche sur le Cancer (ARC). O.C. is supported in the frame of the BIOSTIC-LR programme. IMGT is a registered CNRS mark. IMGT is an RIO platform since 2001 (CNRS, INSERM, CEA, INRA). IMGT was funded in part by the BIOMED1 (BIOCT930038), Biotechnology BIOTECH2 (BIO4CT960037) and 5th PCRDT Quality of Life and Management of Living Resources (QLG2-2000-01287) programmes of the European Union and received subventions from ARC and from the Génopole-Montpellier-Languedoc-Roussillon. IMGT is currently supported by the Centre National de la Recherche Scientifique (CNRS), the MENESR (Université Montpellier II Plan Pluri-Formation, BIOSTIC-LR2004 Région Languedoc-Roussillon and ACI-IMPBIO IMP82-2004).
REFERENCES
- 1.Lefranc M.P. (2003) IMGT, the international ImMunoGeneTics database®. Nucleic Acids Res., 31, 307–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lefranc M.P. (2003) IMGT® databases, web resources and tools for immunoglobulin and T cell receptor sequence analysis, http://imgt.cines.fr. Leukemia, 17, 260–266. [DOI] [PubMed] [Google Scholar]
- 3.Lefranc M.-P., Giudicelli,V., Ginestoux,C., Bosc,N., Folch,G., Guiraudou,D., Jabado-Michaloud,J., Magris,S., Scaviner,D., Thouvenin,V., Combres,K., Girod,D., Jeanjean,S., Protat,C., Yousfi Monod,M., Duprat,E., Kaas,Q., Pommié,C., Chaume,D. and Lefranc,G. (2004) IMGT-ONTOLOGY for Immunogenetics and Immunoinformatics. In Silico Biol., 4, 17–29. [PubMed] [Google Scholar]
- 4.Lefranc M.-P. (2004) IMGT-ONTOLOGY and IMGT databases, tools and web resources for immunogenetics and immunoinformatics. Mol. Immunol., 40, 647–659. [DOI] [PubMed] [Google Scholar]
- 5.Giudicelli V. and Lefranc,M.-P. (1999) Ontology for Immunogenetics: the IMGT-ONTOLOGY. Bioinformatics, 12, 1047–1054. [DOI] [PubMed] [Google Scholar]
- 6.Giudicelli V., Chaume,D. and Lefranc,M.-P. (2005) IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res., 33, D256–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lefranc M.-P. and Lefranc,G. (2001) The Immunoglobulin FactsBook. Academic Press, London, UK, 458 pages. [Google Scholar]
- 8.Lefranc M.-P. and Lefranc,G. (2001) The T cell receptor FactsBook. Academic Press, London, UK, 398 pages. [Google Scholar]
- 9.Wain H.M., Bruford,E.A., Lovering,R.C., Lush,M.J., Wright,M.W. and Povey,S. (2002) Guidelines for human gene nomenclature. Genomics, 79, 464–470. [DOI] [PubMed] [Google Scholar]
- 10.Baum T.P., Pasqual,N., Thuderoz,F., Hierle,V., Chaume,D., Lefranc,M.-P., Jouvin-Marche,E., Marche,P.,N. and Demongeot,J. (2004) IMGT/GeneInfo: enhancing V(D)J recombination database accessibility. Nucleic Acids Res., 32, D51–D54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaume D., Giudicelli,V. and Lefranc,M.-P. (2004) IMGT/LIGM-DB. In Galperin, M.Y. (ed.), The Molecular Biology Database Collection: 2004 update, Database listing. Nucleic Acids Res., 32, D3–D22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulikova T., Aldebert,P., Althorpe,N., Baker,W., Bates,K., Browne,P., van den Broek,A., Cochrane,G., Duggan,K., Eberhardt,R., Faruque,N., Garcia-Pastor,M., Harte,N., Kanz,C., Leinonen,R., Lin,Q., Lombard,V., Lopez,R., Mancuso,R., McHale,M., Nardone,F., Silventoinen,V., Stoehr,P., Stoesser,G., Tuli,M.A., Tzouvara,K., Vaughan,R., Wu,D., Zhu,W. and Apweiler,R. (2004) The EMBL Nucleotide Sequence Database. Nucleic Acids Res., 32, D27–D30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Folch G., Bertrand,J., Lemaitre,M. and Lefranc,M.-P. (2004) IMGT/PRIMER-DB. In: Galperin M.Y. (ed.), The Molecular Biology Database Collection: 2004 update. Database listing. Nucleic Acids Res., 32, D3–D22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson J., Waller,M.J., Parham,P., de Groot,N., Bontrop,R., Kennedy,L.J., Stoehr,P. and Marsh,S.G. (2003) IMGT/HLA and IMGT/MHC sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res., 31, 311–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giudicelli V., Chaume,D. and Lefranc,M.-P. (2004) IMGT/V-QUEST, an integrated software program for immunoglobulin and T cell receptor V-J and V-D-J rearrangement analysis. Nucleic Acids Res., 32,W435–W440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.YousfiMonod M., Giudicelli,V., Chaume,D. and Lefranc,M.-P. (2004) IMGT/JunctionAnalysis: the first tool for the analysis of the immunoglobulin and T cell receptor complex V-J and V-D-J JUNCTIONs. Bioinformatics, 20, I379–I385. [DOI] [PubMed] [Google Scholar]
- 17.Elemento O. and Lefranc,M.-P. (2003) IMGT/PhyloGene: an on-line tool for comparative analysis of immunoglobulin and T cell receptor genes. Dev. Comp. Immunol., 27, 763–779. [DOI] [PubMed] [Google Scholar]
- 18.Kaas Q., Ruiz,M. and Lefranc,M.-P. (2004) IMGT/3Dstructure-DB and IMGT/StructuralQuery, a database and a tool for immunoglobulin, T cell receptor and MHC structural data. Nucleic Acids Res., 32, D208–D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lefranc M.-P., Pommié,C., Ruiz,M., Giudicelli,V., Foulquier,E., Truong,L., Thouvenin-Contet,V. and Lefranc,G. (2003) IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol., 27, 55–77. [DOI] [PubMed] [Google Scholar]
- 21.Lefranc M.-P., Pommié,C., Kaas,Q., Duprat,E., Bosc,N., Guiraudou,D., Jean,C., Ruiz,M., Da Piedade,I., Rouard,M., Foulquier,E., Thouvenin,V. and Lefranc,G. (2005) IMGT unique numbering for immunoglobulin and T cell receptor constant domains and Ig superfamily C-like domains. Dev. Comp. Immunol., 29, 185–203. [DOI] [PubMed] [Google Scholar]
- 22.Ruiz M. and Lefranc,M.-P. (2002) IMGT gene identification and Colliers de Perles of human immunoglobulins with known 3D structures. Immunogenetics, 53, 857–883. [DOI] [PubMed] [Google Scholar]
- 23.Pommié C., Sabatier,S., Lefranc,G. and Lefranc,M.-P. (2004) IMGT standardized criteria for statistical analysis of immunoglobulin V-REGION amino acid properties. J. Mol. Recognit., 17, 17–32. [DOI] [PubMed] [Google Scholar]
- 24.Lefranc M.-P., Clément,O., Kaas,Q., Duprat,E., Chastellan,P., Coelho,I., Combres,K., Ginestoux,C., Giudicelli,V., Chaume,D. and Lefranc,G. (2004) IMGT-Choreography for Immunogenetics and Immunoinformatics. In Silico Biol. (in press). [PubMed] [Google Scholar]