Abstract
WormBase (http://www.wormbase.org), the model organism database for information about Caenorhabditis elegans and related nematodes, continues to expand in breadth and depth. Over the past year, WormBase has added multiple large-scale datasets including SAGE, interactome, 3D protein structure datasets and NCBI KOGs. To accommodate this growth, the International WormBase Consortium has improved the user interface by adding new features to aid in navigation, visualization of large-scale datasets, advanced searching and data mining. Internally, we have restructured the database models to rationalize the representation of genes and to prepare the system to accept the genome sequences of three additional Caenorhabditis species over the coming year.
DESCRIPTION
WormBase is the model organism database for the biology and genomics of Caenorhabditis elegans and Caenorhabditis briggsae. It is a rapidly evolving resource, which is driven by the fact that C.elegans is widely used as a model organism for a variety of biomedical research topics, including development, neuroscience, apoptosis and aging (1–4), and an increasingly wide range of high-throughput data is available for it. The genome sequence of C.elegans (5) has boosted genome-wide research projects including ORFeome (6), RNA interference (RNAi) (7), microarray (8), interactome (genome-wide protein–protein interactions) (9), serial analysis of gene expression (SAGE) (10,11) and other gene expression profiling techniques (11). These large-scale datasets have enormously enriched WormBase content (2,3). More recently, the availability of the whole C.briggsae genome sequence (12), in addition to that of C.elegans, has established WormBase as a platform for comparative genomics among the Caenorhabditides genus (13).
The International WormBase Consortium, consisting of over 30 scientists from four institutions (http://wormbase.org/about/people.html), collects and annotates both large- and small-scale datasets from C.elegans, C.briggsae and related nematodes, organizes them in a single public database, and makes them available for browsing and downloading on the WormBase website. In addition to acquiring directly deposited data by liaison with the research community, the consortium reviews and extracts data from the complete Caenorhabditis published literature. New releases of the database are made available every two weeks, ensuring that new and updated datasets are available to the community on a timely basis. This paper reviews recent progress in WormBase content and improvements in the user interface, explains how WormBase is evolving and discusses different methods of accessing the data. The paper closes with a discussion of new features planned for the coming year.
RECENT ADDITIONS TO WormBase CONTENTS
Over the past year we have greatly increased the sizes of some existing datasets. For example, there is a 5-fold increase in microarray data points and a dramatic 13-fold increase in microarray experiments, from 8 experiments (reported in 2 papers) to 113 experiments (reported in 15 papers). The number of RNAi experiments producing a non-wild-type phenotype has also more than doubled over the past year.
We continue to refine C.elegans gene models on the basis of new data appearing in the literature, from new sequence data in the public nucleotide databases (GenBank/EMBL/DDBJ), and from personal communications from the Worm community. Most curation activity involves refining the structure of existing gene models. However, we also continue to remove gene predictions that are no longer valid (e.g. very short open reading frames) and we continually add new gene predictions where appropriate (usually corresponding to new isoforms of an existing gene). Despite large numbers of genes being created and removed, the total gene count (for protein-coding genes) has seen only a small net increase (+22 genes) over the year. In contrast to this, the proportion of protein-coding genes that are now confirmed by transcript data (i.e. where every coding exon has transcript support) has increased by 20% (from 4663 to 5569) over the same period. This is due to the availability of more transcript data [particularly expressed sequence tags (ESTs)] and the work of curators to refine gene models to better fit the available transcript data. We have also greatly improved the methods by which transcripts are mapped onto the genome and connected to gene models.
Over the same period, WormBase has added several new large-scale experimental and theoretical datasets. Notable additions include large-scale SAGE datasets (10,11), the interactome dataset (9), 3D structural data and the National Center for Biotechnology Information (NCBI) KOGs (14) set of predicted orthologous groups. Recently, the newly developed technique trans-spliced exon coupled RNA end determination (TEC-RED) has been used to assay the 5′ ends of expressed genes in C.elegans (15) and the dataset is being curated and entered into WormBase.
Genome-wide SAGE
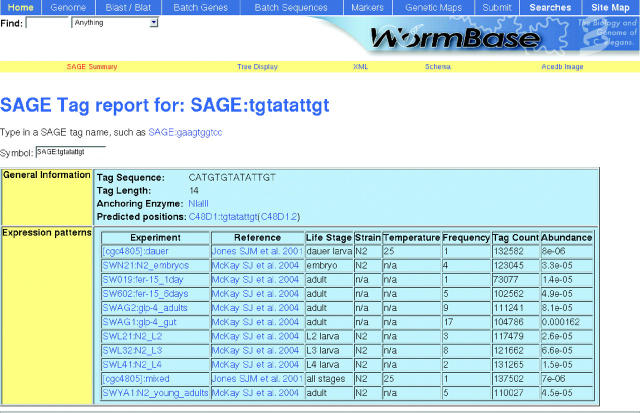
SAGE (10,11) is a sensitive technique for assaying genome-wide gene expression levels that provides a good complement to microarray-based techniques. As of release WS123, WormBase incorporates the results of 12 SAGE libraries, two of which have been published previously (10). The 12 libraries cover various developmental stages (11) from embryo to adult and touch 20 417 genes (coding sequences, WS129) corresponding to 91.9% of all genes annotated in the C.elegans genome in WormBase (22 213 including alternatively spliced coding sequences, WS129). SAGE tags corresponding to a gene can be found at the bottom of the WormBase gene page (e.g. http://www.wormbase.org/db/gene/gene?name=ced-3#Reagents) and are linked to information detailing the SAGE tag's abundance at various life stages in a new SAGE report page (Figure 1).
Figure 1.
SAGE report page.
Interactome
Dissecting a protein's interaction network is often a key to understanding its biological role. WormBase includes the results of the ‘Interactome Project’, a large-scale screen based on the yeast two-hybrid (Y2H) technique (9). In the current dataset, baits are biased towards genes either homologous to human genes, of multicellular functions (genes with homologues in multicellular organisms including Drosophila melanogaster, Homo sapiens and Arabidopsis thaliana but not in Saccharomyces cerevisiae), or having a known role in mitosis and meiosis. Currently, WormBase includes 5534 interactions covering 15% of the C.elegans proteome. Users can view these interactions from the gene summary page.
Protein three-dimensional structures
This small but important dataset is from the Northeast Structural Genomics Consortium (http://www.nesg.org), which aims to produce 340 C.elegans targets. The primary targets of the Consortium focus on proteins of eukaryotic model organisms including S.cerevisiae and D.melanogaster in addition to C.elegans. Currently, structures for six proteins have been deposited in the Protein Data Bank (PDB) (http://www.rcsb.org/pdb/) (16). Detailed information about the status for these 340 C.elegans targets have been included in the WormBase and will be regularly updated.
NCBI KOGs
KOGs are a eukaryote-specific version of the Conserved Orthologous Groups originally devised at the NCBI for microbial genomes (14). KOGs are defined by a triangle of reciprocal best BLASTP hits between domains of eukaryote proteins from highly divergent species (14). Over the last year, WormBase has incorporated these KOG annotations, together with other homology groups (14). Currently, WormBase carries 4852 KOGs, which includes the product of 9427 C.elegans protein-coding genes (i.e. 48% of all predicted protein-coding genes in WS129).
INTERNAL DATA MODEL CHANGES AND NEW IDENTIFIERS
The backend database of WormBase is ACeDB (http://www.acedb.org) (4). During the last year, we have changed the way that a number of data types are represented in the database. These changes to the database schema do not affect usual users. However, advanced users who write scripts to access WormBase need to be aware of them. Significant model changes include the introduction of a unified Gene class (http://wormbase.org/db/misc/model?class=Gene), which holds all relevant information about a gene. Previously, such information was scattered among several interrelated classes. At the same time we have introduced CDS and Transcript classes to manage better the relationships between spliced transcripts and their products, and have significantly improved the derivation of transcript structures from cDNA and EST sequences.
Alongside these changes we have introduced stable anonymous identifiers for genes, of the form WBGene00006741, and for papers, of the form WBPaper0005637, in the same form as the person identifiers of the form WBPerson241. These identifiers track the various names that have been used for the corresponding entity and should be used where possible for database cross-referencing. The website supports URLs of the form http://www.wormbase.org/db/get?name=WBGene00006741;class=Gene. Questions about data models can be directed to wormbase-help@wormbase.org.
USER INTERFACE ENHANCEMENTS
Enhancements to WormBase genome browser
The genome browser is a central component of WormBase that allows users to visualize gene model structures and their supporting evidence, as well as other features such as single nucleotide polymorphisms (SNPs), repetitive elements and experimental reagents. Over the last year, the browser has been enhanced in several ways: (i) scalable vector graphics (SVG) support. WormBase genome browser images have been widely used in presentations and publication illustrations (2,3,17), but their bitmapped nature leads to image degradation when printed at high resolution. We have recently added a facility that allows WormBase users to download specified genome browser images as SVG files (http://www.w3.org/TR/SVG/), which can be displayed, edited and printed at high resolution using SVG compatible software such as Adobe Illustrator 10. (ii) Feature highlighting. To assist location and visualization of features of interest, WormBase now highlights with a yellow background the feature that users have found in a search. This change is especially useful when users browse in large window size with multiple tracks turned on. (iii) Untranslated regions (UTRs). Both the internal data model and the visual display have now been modified to show the untranslated sections of transcripts, as well as internal splices that occur within the 5′- or 3′-UTRs. (iv) More feature tracks, including SNPs, SAGE tags, operon, poly(A) sites and predicted signal sequences. (v) DAS support. The genome browser may now be used as a viewer for Distributed Annotation System (DAS) (18) tracks, allowing users to superimpose their own annotations on WormBase tracks.
EST alignment page and protein alignment page
WormBase now maintains nucleotide-level alignments of ESTs, cDNAs and other sequences both within and between species. For example, the alignment between the C.elegans and C.briggsae genomes can be viewed both in a low-resolution view that emphasizes the relationship among a group of colinear genes (http://www.wormbase.org/db/seq/ebsyn?name=cb25.fpc0143:1..8000), or in a high-resolution text alignment view that shows differences in individual nucleotides. ESTs and cDNAs from C.elegans and other nematodes can be viewed in a multiple alignment view that highlights misalignments and gaps (http://www.wormbase.org/db/seq/aligner?name=WBGene00000423;class=Gene).
At the protein level, WormBase maintains a list of best BLAST matches to longest protein products from other important species including human (H.sapiens), mouse (Mus musculus), rat (Rattus norvegicus), fly (D.melanogaster), yeast (S.cerevisiae) and C.briggsae, which together can provide insights into the function of the related genes. All BLAST results are hyperlinked to a relevant entry in the respective model organism database or to Swiss-Prot/TrEMBL as appropriate. The multiple alignment display highlights conserved amino acid residues using a color code based on the chemical properties of the residues (Figure 2).
Figure 2.
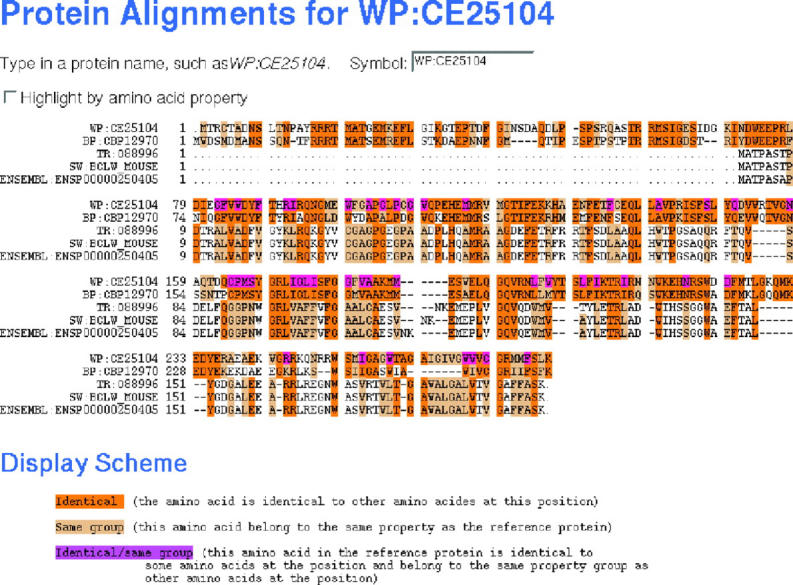
Protein alignment page.
WormBase site map and WormBase glossary
Over the past year, we have added a WormBase site map (http://wormbase.org/db/misc/site_map) to provide an overview of the increasing number of web pages. Users can access this map directly from the navigation banner at the top of every WormBase page. The site map page lists all WormBase pages and provides users with different views. For example, users can choose ‘Detailed View’ to get brief overviews for individual pages before browsing the pages. And ‘Alphabetical View’ lists search pages in alphabetical order. Recently, WormBase has established a glossary page (http://dev.wormbase.org/db/misc/glossary) that lists definitions of common terms used throughout the site.
WormBase AS A PLATFORM FOR DATA MINING
As biologists come to make more sophisticated use of large-scale datasets, there is an increasing need for a resource that is more than a point-and-click repository but provides data analysis and mining tools as well. This section briefly describes existing and recently introduced features that make WormBase suitable for data mining.
WormBase accessing and retrieving
There are five different methods for accessing WormBase, each one suitable for a different set of purposes. Users can choose the most appropriate access methods according to their experience and needs.
Website browsing. This is an one-item-at-a-time approach. WormBase users typically enter WormBase from the front page, searching the gene (or other items) of interests in the search box. Alternatively, users can open the WormBase site map by clicking on a link in the top navigational banner and enter a specific web page for searching, either by sequence (BLAST or BLAT) or by text. Once the users find their item of interest, they can browse related web pages by following links. The advantage of working with WormBase this way is that the users can get detailed views and information about the items of interest.
Batch retrieval. WormBase users increasingly need to obtain customized batch reports. To address this need, WormBase provides two web search pages: ‘Batch Genes’ and ‘Batch Sequences’ (2). The Batch Genes page allows users to retrieve all biologically interesting gene data fields, ranging from external database IDs, to protein motifs, GO terms, genomic positions, phenotypes and underlying DNA and protein sequences. This page gives users the option to download the results in plain text or the HTML format, and provides a variety of ways to select the set of genes of interest. The Batch Sequences page is ideal for retrieving sequence-based data such as UTRs, introns, putative promoter elements and so on. For example, this facility can be used to generate sequence files consisting of a specific length of upstream sequence from a selected set of protein-coding genes. Both pages can be readily accessed from the top navigational banner. The benefit of this method of searching is that it returns results for a large number of items (genes).
Query language searching. For users who are comfortable with the ACeDB database query languages and familiar with WormBase database models, query language searches represent a quick and versatile method of searching WormBase. Two query language search pages are available: one for the WormBase Query Language, the original ACeDB query language, and another for AQL, the new-style ACeDB Query Language that is more similar to SQL. These pages can be accessed from the WormBase ‘Site Map’ page. For users who are not familiar with the ACeDB query languages, the search pages provide instructions and example queries. The major benefit is that users can formulate sophisticated ad hoc queries.
Bulk downloads. Users can download whole gene sets or even the whole database itself. WormBase provides a number of database extracts on its FTP site, including coordinates of genes and other features, protein sequences, gene splicing data and genetic mapping information. The entire genome and its annotations are available in a tabular format that can be loaded into and queried with a variety of relational databases including MySQL, PostgreSQL and Oracle. A table is provided for each release that links PCR products such as are used for microarrays and RNAi experiments to currently annotated genes. WormBase also provides the entire database in the ACeDB format. The advantage of this method is that users do not have to rely on the Internet for data retrieval, so that their data processing is not limited by Internet access. Problems associated with this method are that users need to be very familiar with the nature of the datasets and the database models.
Scripting. For more advanced users who know script programming, WormBase provides an open-access server ‘aceserver’ (at http://aceserver.cshl.org) for direct access to the backend WormBase database (19). The WormBase data mining instruction page provides researchers with details about how to connect to these databases using Perl (http://www.perl.org) application programming interface, AcePerl (http://stein.cshl.org/AcePerl), together with a scripts repository of reusable Perl scripts. Users can run these scripts on their local machines and use them as templates to customize scripts of their own. The biggest advantage of this is users can query, format and process the search results to the extent they desire. An obvious drawback is that users need to acquire some programming skills. However, this is becoming increasingly popular with advanced users.
Specialized data mining tools
As a sequence analysis platform, WormBase has made a large number of sequence analysis tools available to users. These tools include BLAST (20), BLAT (21), ePCR (22), coordinate mapper, EST aligner and protein aligner. In the past year, two new data mining tools, Textpresso (http://www.textpresso.org) (23), a literature search tool, and CisOrtho (24), a comparative cis-elements search tool have also been added to WormBase. Textpresso is a full text search engine, which gives researchers the ability to search the body of all WormBase literature holdings, which includes a substantial percentage of the C.elegans and C.briggsae literature. Currently, the Textpresso database holds 19 985 curated documents, 4420 of which have full texts. These documents come from four major sources: (i) CGC papers. These are scientific journal articles maintained by the Caenorhabditis Genetics Center (http://biosci.umn.edu/CGC/CGChomepage.htm); (ii) Worm Meetings abstracts; (iii) Worm Breeders Gazette abstracts; and (iv) Miscellaneous. These are various other abstracts containing data about C.elegans and C.briggsae. Another useful feature of Textpresso is that it returns the sentences that contain the key words, with links to WormBase paper pages and PubMed pages.
CisOrtho (24) works by starting from a consensus binding site that is represented as a weight matrix. It identifies potential sites in a pre-filtered genome and then further filters by assessing conservation of the putative site in the genome of a related species, a process called phylogenetic footprinting. CisOrtho can be accessed at http://www.wormbase.org/cisortho/.
DATABASE FREEZES
In the past, the WormBase fortnightly update policy presented a problem to researchers who published results based on mining WormBase because by the time their results were published the version of WormBase they based their analysis on had been superseded. To assist in making such research citable and reproducible, we have adopted a new policy in which every tenth WormBase release becomes a frozen release. Frozen releases are available in perpetuity on specially designated WormBase sites named http://ws100.wormbase.org, http://ws110.wormbase.org and so on. The first freeze was http://ws100.wormbase.org, released on May 10, 2003. The most recent freeze is http://ws130.wormbase.org, released on August 16, 2004. Researchers are encouraged to perform large-scale analyses on a frozen release and to cite the release number in their publications. Pointers to all freezes are displayed on the WormBase live site front page.
COLLABORATIONS WITH OTHER MODEL ORGANISM DATABASES
WormBase is a part of the GMOD project (25,26), a broad collaboration among the model organism databases to develop common vocabularies, data models, software tools and user interfaces applicable across all model organism community databases. As part of this project, WormBase provides sequence-similarity-based links between its gene pages and the gene pages of FlyBase (27), The Saccharomyces Genome Database (28,29), Ensembl (29) and Reactome (http://www.reactome.org). Links to RGD (30) and MGD (31) are planned.
Recently, the GMOD project has developed a common representation of genomic sequence features known as the Sequence Ontology (http://song.sourceforge.net), which facilitates exchange of genomic annotations among the various MODs and encourages the use of common analytic and visualization tools. GMOD participants are already using common software packages on their websites for visualizing genome annotations, drawing genetic maps and searching the literature, and this convergence will be enhanced in the near future as the MODs move towards a unified gene page.
FUTURE DIRECTIONS
WormBase has evolved from ACeDB (http://www.acedb.org), to a database which encompass literature curation and biology of C.elegans (4), and recently to a database housing the biology and genomic data of multiple nematode species (2,3). WormBase is still a work in progress. On the user interface front, future enhancements include WormMart, which is based on BioMart, an advanced query and report generation system first developed for use with Ensembl (32). On the data front, we are looking forward to the genome sequencing and annotation of three more nematode species (http://genome.gov/page.cfm?pageID=10002154), bringing up to five the number of Caenorhabditis genomes maintained by WormBase. During 2005, WormBase plans to introduce a browser for nematode intermediate metabolism and higher-order biological pathways. The pathway browser and the underlying dataset will be developed in collaboration with the Reactome and MetaCyc (http://metacyc.org/) (33) projects. Together these will provide an unparalleled resource for dissecting functional elements in the Caenorhabditis genomes and provide valuable insights into the evolution and biological adaptations of these organisms.
The WormBase Consortium will continue to address issues raised by WormBase users, maintaining both a simple and friendly user interface while adding further search and research tools to enable WormBase's evolution from a data repository into a resource for all biologists to use in order to maximize the value of model organism research in C.elegans and its relatives.
As always, we welcome comments, questions, corrections and data submissions (wormbase-help@wormbase.org).
Acknowledgments
ACKNOWLEDGEMENTS
P.W.S. is an Investigator with the Howard Hughes Medical Institute. We thank Sheldon McKay and Kris Gunsalus for critical reading of the manuscript. WormBase is supported by grant P41-HG02223 from the US National Human Genome Research Institute and the British Medical Research Council.
REFERENCES
- 1.Riddle D.L., Blumenthal,T., Meyer,B.J. and Priess,J.R. (1997) C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- 2.Harris T.W., Chen,N., Cunningham,F., Tello-Ruiz,M., Antoshechkin,I., Bastiani,C., Bieri,T., Blasiar,D., Bradnam,K., Chan,J. et al. (2004) WormBase: a multi-species resource for nematode biology and genomics. Nucleic Acids Res., 32 (Database issue), D411–D417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris T.W., Lee,R., Schwarz,E., Bradnam,K., Lawson,D., Chen,W., Blasier,D., Kenny,E., Cunningham,F., Kishore,R. et al. (2003) WormBase: a cross-species database for comparative genomics. Nucleic Acids Res., 31, 133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stein L., Sternberg,P., Durbin,R., Thierry-Mieg,J. and Spieth,J. (2001) WormBase: network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res., 29, 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The C.elegans Sequencing Consortium (1998) Genome sequence of the nematode C.elegans: a platform for investigating biology. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 6.Chen N., Lawson,D., Bradnam,K. and Harris,T.W. (2004) WormBase as an integrated platform for the C. elegans ORFeome. Genome Res. 14, 2155–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamath R.S., Fraser,A.G., Dong,Y., Poulin,G., Durbin,R., Gotta,M., Kanapin,A., Le Bot,N., Moreno,S., Sohrmann,M. et al. (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature, 421, 231–237. [DOI] [PubMed] [Google Scholar]
- 8.Kim S.K., Lund,J., Kiraly,M., Duke,K., Jiang,M., Stuart,J.M., Eizinger,A., Wylie,B.N. and Davidson,G.S. (2001) A gene expression map for Caenorhabditis elegans. Science, 293, 2087–2092. [DOI] [PubMed] [Google Scholar]
- 9.Li S., Armstrong,C.M., Bertin,N., Ge,H., Milstein,S., Boxem,M., Vidalain,P.O., Han,J.D., Chesneau,A., Hao,T. et al. (2004) A map of the interactome network of the metazoan C. elegans. Science, 303, 540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones S.J., Riddle,D.L., Pouzyrev,A.T., Velculescu,V.E., Hillier,L., Eddy,S.R., Stricklin,S.L., Baillie,D.L., Waterston,R. and Marra,M.A. (2001) Changes in gene expression associated with developmental arrest and longevity in Caenorhabditis elegans. Genome Res., 11, 1346–1352. [DOI] [PubMed] [Google Scholar]
- 11.McKay S.J., Johnsen,R., Khattra,J., Asano,J., Baillie,D.L., Chan,S., Dube,N., Fang,L., Goszczynski,B., Ha,E. et al. (2004) Cold Spring Harbor Symposia on Quantitative Biology. Cold Spring Harbor, NY, Vol. 68, pp. 159–170. [DOI] [PubMed] [Google Scholar]
- 12.Stein L.D., Bao,Z., Blasiar,D., Blumenthal,T., Brent,M.R., Chen,N., Chinwalla,A., Clarke,L., Clee,C., Coghlan,A. et al. (2003) The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol., 1, E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta B.P. and Sternberg,P.W. (2003) The draft genome sequence of the nematode Caenorhabditis briggsae, a companion to C. elegans. Genome Biol., 4, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tatusov R.L., Fedorova,N.D., Jackson,J.D., Jacobs,A.R., Kiryutin,B., Koonin,E.V., Krylov,D.M., Mazumder,R., Mekhedov,S.L., Nikolskaya,A.N. et al. (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics, 4, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang B.J., Muller,H.M. and Sternberg,P.W. (2004) Genome annotation by high-throughput 5′ RNA end determination. Proc. Natl Acad. Sci. USA, 101, 1650–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berman H.M., Battistuz,T., Bhat,T.N., Bluhm,W.F., Bourne,P.E., Burkhardt,K., Feng,Z., Gilliland,G.L., Iype,L., Jain,S. et al. (2002) The Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr., 58, 899–907. [DOI] [PubMed] [Google Scholar]
- 17.Stajich J.E., Block,D., Boulez,K., Brenner,S.E., Chervitz,S.A., Dagdigian,C., Fuellen,G., Gilbert,J.G., Korf,I., Lapp,H. et al. (2002) The Bioperl toolkit: Perl modules for the life sciences. Genome Res., 12, 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dowell R.D., Jokerst,R.M., Day,A., Eddy,S.R. and Stein,L. (2001) The distributed annotation system. BMC Bioinformatics, 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stein L.D. and Thierry-Mieg,J. (1998) Scriptable access to the Caenorhabditis elegans genome sequence and other ACEDB databases. Genome Res., 8, 1308–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez R., Silventoinen,V., Robinson,S., Kibria,A. and Gish,W. (2003) WU-Blast2 server at the European Bioinformatics Institute. Nucleic Acids Res., 31, 3795–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kent W.J. (2002) BLAT—the BLAST-like alignment tool. Genome Res., 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuler G.D. (1997) Sequence mapping by electronic PCR. Genome Res., 7, 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller H.M., Kenny,E. and Sternberg,P. (2004) Testpresso: an ontology-based information retrieval and extraction system for C. elegans literature. PLoS Biol., 2, e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bigelow H.R., Wenick,A.S., Wong,A. and Hobert,O. (2004) CisOrtho: a program pipeline for genome-wide identification of transcription factor target genes using phylogenetic footprinting. BMC Bioinformatics, 5, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis S.E., Searle,S.M., Harris,N., Gibson,M., Lyer,V., Richter,J., Wiel,C., Bayraktaroglir,L., Birney,E., Crosby,M.A. et al. (2002) Apollo: a sequence annotation editor. Genome Biol., 3, RESEARCH0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stein L.D., Mungall,C., Shu,S., Caudy,M., Mangone,M., Day,A., Nickerson,E., Stajich,J.E., Harris,T.W., Arva,A. et al. (2002) The generic genome browser: a building block for a model organism system database. Genome Res., 12, 1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.FlyBase (2003) The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res., 31, 172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christie K.R., Weng,S., Balakrishnan,R., Costanzo,M.C., Dolinski,K., Dwight,S.S., Engel,S.R., Feierbach,B., Fisk,D.G., Hirschman,J.E. et al. (2004) Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res., 32 (Database issue), D311–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birney E., Andrews,D., Bevan,P., Caccamo,M., Cameron,G., Chen,Y., Clarke,L., Coates,G., Cox,T., Cuff,J. et al. (2004) Ensembl 2004. Nucleic Acids Res., 32 (Database issue), D468–D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Twigger S., Lu,J., Shimoyama,M., Chen,D., Pasko,D., Long,H., Ginster,J., Chen,C.F., Nigam,R., Kwitek,A. et al. (2002) Rat Genome Database (RGD): mapping disease onto the genome. Nucleic Acids Res., 30, 125–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bult C.J., Blake,J.A., Richardson,J.E., Kadin,J.A., Eppig,J.T., Baldarelli,R.M., Barsanti,K., Baya,M., Beal,J.S., Boddy,W.J. et al. (2004) The Mouse Genome Database (MGD): integrating biology with the genome. Nucleic Acids Res., 32 (Database issue), D476–D481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kasprzyk A., Keefe,D., Smedley,D., London,D., Spooner,W., Melsopp,C., Hammond,M., Rocca-Serra,P., Cox,T. and Birney,E. (2004) EnsMart: a generic system for fast and flexible access to biological data. Genome Res., 14, 160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krieger C.J., Zhang,P., Mueller,L.A., Wang,A., Paley,S., Arnaud,M., Pick,J., Rhee,S.Y. and Karp,P.D. (2004) MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res., 32 (Database issue), D438–D442. [DOI] [PMC free article] [PubMed] [Google Scholar]