Abstract
To efficiently utilize the growing body of available protein localization data, we have developed Organelle DB, a web-accessible database cataloging more than 25 000 proteins from nearly 60 organelles, subcellular structures and protein complexes in 154 organisms spanning the eukaryotic kingdom. Organelle DB is the first on-line resource devoted to the identification and presentation of eukaryotic proteins localized to organelles and subcellular structures. As such, Organelle DB is a strong resource of data from the human proteome as well as from the major model organisms Saccharomyces cerevisiae, Arabidopsis thaliana, Drosophila melanogaster, Caenorhabditis elegans and Mus musculus. In particular, Organelle DB is a central repository of yeast data, incorporating results—and actual fluorescent images—from ongoing large-scale studies of protein localization in S.cerevisiae. Each protein in Organelle DB is presented with its sequence and, as available, a detailed description of its function; functions were extracted from relevant model organism databases, and links to these databases are provided within Organelle DB. To facilitate data interoperability, we have annotated all protein localizations using vocabulary from the Gene Ontology consortium. We also welcome new data for inclusion in Organelle DB, which may be freely accessed at http://organelledb.lsi.umich.edu.
INTRODUCTION
With an expanding volume of protein data at our disposal, we are now approaching the point at which we can begin to consider protein function within the eukaryotic cell from a more holistic perspective. In particular, we need to bear in mind the obvious fact that proteins do not function in isolation but, rather, at a subcellular level, in organelles or as components of multi-part complexes (1). It is this subcellular unit, and not the individual protein—nor possibly the entire proteome—that represents the most enlightening and promising avenue for immediate studies of eukaryotic cell function. As a result, we need to define as completely as possible the constituent proteins comprising these organelles and eukaryotic protein complexes.
Through a large and varied complement of proteomic methodologies, our understanding of organelle composition and protein localization is advancing rapidly. For example, in the simple eukaryote Saccharomyces cerevisiae, localization data are now available for approximately 5000 proteins (∼90% of the yeast proteome)—thanks in large part to several recent efforts in which genome-wide collections of epitope-tagged genes and reporter fusions were constructed and employed for large-scale studies of yeast protein localization (2–4). Similar strategies are now being utilized in higher eukaryotes with promising early results (5,6). In addition, several groups have effectively coupled subcellular fractionation biochemistry with mass spectrometry as part of a blossoming sub-discipline termed subcellular proteomics (7). In subcellular proteomics, traditional biochemical fractionation techniques are used to isolate distinct subcellular compartments, and peptide mass spectrometry is subsequently utilized to resolve these complex mixtures into constituent proteins (8).
Despite the increasing popularity of large-scale protein localization projects, the majority of localization data available at present have been generated piecemeal from independent small-scale studies. It is thus particularly vital that these findings be collected and presented in central databases so that the typical researcher may easily access and utilize this knowledgebase in its entirety. Accordingly, we present Organelle DB—a web-accessible database cataloging the protein constituents of 57 organelles, subcellular structures and protein complexes from 154 organisms across the eukaryota. That is, Organelle DB provides a cross-species listing of eukaryotic proteins localized to a given organelle, subcellular structure or protein complex. At present, Organelle DB encompasses greater than 25 000 proteins; our data set is drawn both from our own studies in yeast as well as from published data deposited in either the Gene Ontology (GO) database (9), in SWISS-PROT (10) or in one of the major model organism sites. We have taken particular care to incorporate accurate gene names, amino acid sequences and detailed functional data for each protein when available. In order to facilitate automated data processing and interoperability, each localization entry in Organelle DB has been annotated using controlled vocabulary from the GO consortium (9). We envision Organelle DB as a repository for primary protein localization data (i.e. fluorescent micrographs, etc.); Organelle DB already includes approximately 1500 fluorescent micrographs of yeast cells stained with antibodies directed against epitope-tagged proteins, and we welcome localization data submissions for all eukaryotic organisms. In total, Organelle DB constitutes a singular resource consolidating our knowledge of the proteins comprising eukaryotic organelles and subcellular structures.
DESIGN AND IMPLEMENTATION
Organelle DB has been developed and implemented using the MySQL relational database system and the PHP server-side scripting language. Our server is run on the Unix operating system. We have populated Organelle DB partly by warehousing the GO database (9) and extracting from GO a listing of relevant proteins and cellular compartments. In addition, we have extracted protein localization data deposited in each major model organism database [i.e. the Saccharomyces Genome Database (SGD) (11), the Drosphila melanogaster database FlyBase (12), the Caenorhabdits elegans database WormBase (13), the Mouse Genome Database MGD (14), the Arabidopsis Information Resource TAIR (15) and SWISS-PROT (10)]. Yeast protein localization data sets from ongoing studies are being manually compiled for entry into Organelle DB, as are the results from several mass spectrometry-based studies of eukaryotic organelles. All data sets are entered into Organelle DB via web page forms maintaining a controlled vocabulary consistent with the GO project.
DATA CONTENT
Organelles, subcellular structures and protein complexes
Organelle DB houses data for proteins localized to the organelles, subcellular complexes and structures listed in Figure 1. In total, 57 GO terms have been used to classify the protein localization data maintained in Organelle DB; note that many terms in GO encompass sub-terms, such that more than 57 terms are actually housed in Organelle DB (9). For simplicity of presentation, these terms have been grouped into six broad categories: endoplasmic reticulum, nucleus, membrane protein, mitochondrion, protein complex and others. Note that these groupings are often necessarily heterogeneous in content. For example, the ‘protein complex’ category contains a number of distinct protein complexes that could be sub-grouped within other organelles; these proteins have been grouped separately to provide users with the most specific and detailed information available. If desired, users may easily download data sets from Organelle DB and re-sort the files as desired (see Data Access).
Figure 1.
Listing of GO terms used in Organelle DB. Specific GO terms have been grouped into six broad categories (indicated in boxes): Nucleus, Mitochondrion, Endoplasmic Reticulum, Membrane Proteins, Protein Complexes and Others. The number of protein records encompassed in each category is indicated. Note that many of the GO terms listed here encompass a large family of sub-terms as well (e.g. Mitochondrion, GO: 0005739); these sub-terms may be accessed from the GO website (http://www.geneontology.org/).
Organisms
Organelle DB is a cross-species resource: a sampling of eukaryotic organisms represented in our database is presented in Table 1. The data sets in Organelle DB are drawn largely from studies of the major model organisms (i.e. S.cerevisiae, Arabidopsis thaliana, D.melanogaster, C.elegans and Mus musculus). In addition, we have developed records for nearly 4000 human proteins, largely from the data deposited in SWISS-PROT (10). A listing of protein localization records per major organism is listed in Table 2. In particular, Organelle DB is a principal repository for yeast protein localization data; published data sets from S.cerevisiae have been supplemented with unpublished protein localization results from our laboratory. To establish our site as a timely channel for data dissemination, we will actively incorporate relevant localization data into Organelle DB in advance of publication.
Table 1. Organisms represented in Organelle DB.
| Organism | Genus/species | Localized proteins |
|---|---|---|
| Budding yeast | Saccharomyces cerevisiae | 3858 |
| Mouse-ear cress plant | Arabidopsis thaliana | 1988 |
| Fruit fly | Drosophila melanogaster | 2654 |
| Nematode | Caenorhabditis elegans | 534 |
| Mouse | Mus musculus | 2600 |
| Human | Homo sapiens | 3989 |
| Others (148 in total) | N/A | 9472 |
Table 2. Protein localization records in Organelle DB.
| Organism | Nucleus | Mitochondrion | ER | Membrane protein | Protein complex | All |
|---|---|---|---|---|---|---|
| S.cerevisiae | 2225 | 818 | 477 | 772 | 594 | 5294 |
| Plants (A.thaliana) | 1021 | 7783 | 147 | 528 | 628 | 10 402 |
| D.melanogaster | 1337 | 488 | 132 | 820 | 529 | 3576 |
| C.elegans | 116 | 360 | 9 | 70 | 8 | 582 |
| M.musculus | 1221 | 268 | 155 | 966 | 195 | 3089 |
| H.sapiens | 1651 | 328 | 226 | 1821 | 242 | 4704 |
Note that records do not correspond exactly with proteins; one protein may have more than one record if it has been found within more than one organelle.
Protein data
Each protein record in Organelle DB has been annotated with functional and sequence data; a representative protein report is presented in Figure 2. We have taken particular care to utilize accurate gene/protein names for each record in Organelle DB, conforming to the established naming conventions in place for each organism. When possible, both a systematic name and a standard name have been provided for each protein. The systematic name for a given gene is typically derived from its physical location within the organism's genome, while the standard name often reflects the function or observed phenotype associated with a gene. Also, as available, each protein record is invested with a brief description of the gene or protein product. In most cases, this description of protein function has been extracted from the protein's corresponding model organism database. An additional ‘comment’ field has been added to some records, reflecting any known mutant phenotypes associated with the gene.
Figure 2.

Sample query output and protein report from Organelle DB. (A) Output from a query for yeast nuclear proteins. Search results are grouped according to constituent sub-categories and organisms; a table of chromosomal proteins is shown here. (B) By clicking on an individual protein from the Results table, users may view a detailed protein report for that gene containing all fields for which we have available data. A sample protein report for YBL003C is shown here.
Proteins in Organelle DB have been further annotated with biological data from the GO project. The GO consortium has developed a controlled vocabulary to describe the ‘cellular component’ to which a protein localizes, that protein's ‘molecular function’ and its ‘biological process’ (9). The ‘cellular component’ field directly describes the localization of a given protein, and a large number of proteins in Organelle DB have been classified according to this cellular component data set extracted from GO. The terms ‘molecular function’ and ‘biological process’ are fairly intuitive. By the GO vocabulary, the general biological process to which a protein contributes is indicated by the ‘biological process’ term, while the specific function or activity of a given protein within this broader biological process is indicated by the ‘molecular function’ term. For example, a mitogen-activated protein kinase (MAPK) would be indicated as a kinase by its ‘molecular function’, but may contribute to one of many ‘biological processes’ (e.g. response to mating factor pheromone or external conditions of high osmolarity).
In presenting ‘cellular component’ data from the GO project, we do occasionally encounter instances in which the GO annotation is at odds with data deposited in one of the model organism sites. In such cases, we have chosen to present the subcellular localization presented within the model organism database. Accordingly, the particular source of a given protein localization record is indicated within each protein data report in the ‘Data Source’ field.
The amino acid sequence and length of each localized protein is also presented within Organelle DB. Note that for many proteins from higher eukaryotes, multiple splice variants and isoforms may exist. Whenever possible, we have included all such sequences for a given gene as output in our standard report; as an example, please see our report for the C.elegans gene clr-1.
In Organelle DB, localization data sets for the budding yeast S.cerevisiae were generated largely as part of a proteome-scale analysis during which yeast genes were epitope-tagged (through a variety of approaches) for subsequent localization by indirect immunofluorescence (2,16). Fluorescent micrographs from this study have been incorporated into Organelle DB, providing a visual library of yeast protein localization. Two images are provided per protein: the left-most image is of yeast cells stained with the DNA-binding dye DAPI, highlighting the nucleus and mitochondria, while the right-most image is of the same cells stained with monoclonal antibody directed against the epitope-tagged protein. Please refer the Organelle DB report of the yeast gene YNL052W for an example; YNL052W (or COX5A) encodes subunit Va of cytochrome c oxidase at the mitochondrial inner membrane. As similar large-scale studies are underway in our laboratory, additional images are currently being incorporated into Organelle DB. We further welcome any raw data describing protein localization in any eukaryote for inclusion in our database.
The availability of experimentally derived protein localization data in yeast raises an important point: not all protein localization records in Organelle DB are experimentally verified. In considering the validity of a given record, examine the data source for that result. Obviously, as large-scale studies of protein localization proceed in many organisms, some predicted localization records will be experimentally verified and others contradicted. We will endeavor to maintain Organelle DB consistent with these emerging results.
DATA RETRIEVAL
Data sets in Organelle DB may be accessed through a variety of search options. From the Quick Search features available on our homepage, users may specifically view all proteins localized to a given organelle, subcellular structure or protein complex. Similar options are provided on the Quick Search form such that users may alternatively browse records related to a single organism or gene/protein. Choices for these searches have been kept purposefully broad for the sake of simplicity; detailed subcategories of organelles, protein complexes and organisms may be directly accessed from our Advanced Search forms. These Advanced Search options offer a full list of organelles and organisms contained within Organelle DB; for example, through our Advanced Search, users may select an organelle (e.g. endoplasmic reticulum) and further select a subcategory of that organelle (e.g. integral to endoplasmic reticulum membrane). In addition, users may specify a specific organelle and organism, thereby limiting the output to only those organelle-localized proteins from the indicated organism. By entering an organelle, organism and gene name, users will limit the search to only a specific protein; note that, in some cases, a single gene name is shared among many orthologues within several different organisms. Regardless of the specific options employed, each search returns a list of proteins with a brief functional description (when functional information is available). Users may click upon any individual protein to view a full report containing all data fields described in Figure 2.
Users may also download data sets from Organelle DB in bulk. Specifically, all data in Organelle DB may be downloaded as a tab-delimited text file; a separate file is provided with GO annotation for each protein. In addition, we provide a file of amino acid sequences (in FASTA format) for all protein entries. Note that multiple sequences are available for certain proteins in Organelle DB. These protein sequences can be correlated to a single protein entry (in the tab-delimited text file described above) through the Accession ID field. The results from individual searches may also be downloaded as a flat file by clicking on the ‘Download in Flat File’ button from our advanced search form.
SIGNIFICANCE
Organelle DB is a singular consolidation of protein localization data specifically describing the protein composition of eukaryotic organelles, subcellular structures and select protein complexes. As such, it represents a data resource through which users may pose unique questions regarding the function and evolution of organelles and cell structures. Furthermore, Organelle DB provides a highly useful data set that strongly complements the existing protein interaction studies as a tool for proteomics and functional genomics.
Organelle function
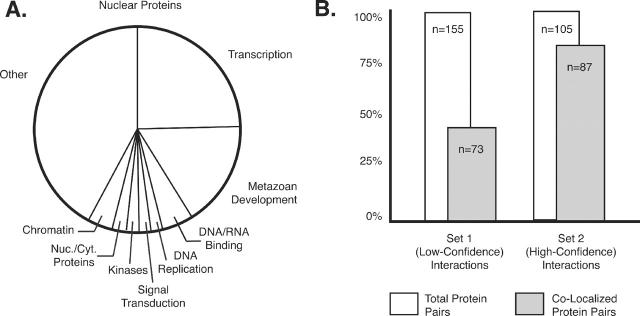
Through the functional descriptions and GO terms incorporated in Organelle DB, we can address some fundamental questions regarding the cellular functions carried out in an organelle. For example, consider the nucleus (Figure 3A). In total, Organelle DB houses records for 7546 nuclear proteins in more than 100 different organisms. As expected, the majority of these proteins carry out functions related to the performance, fidelity and maintenance of transcription. Also, a large fraction of nuclear proteins (∼16%) act to organize and coordinate processes related to development in metazoans. However, the total nuclear protein complement is far from homogenous. The nucleus is an obvious endpoint for many cellular signaling pathways, and a fair number of kinases and phosphatases have been localized to the nucleus. Typically perceived as cytoplasmic proteins, kinases frequently shuttle between the cytoplasm and nucleus. The presence of these nuclear-localized kinases underscore the importance of this translocation as a regulatory mechanism and further suggests the utility of proteome-wide protein localization data.
Figure 3.
Applications of Organelle DB. (A) Organelle DB is a useful tool by which we can investigate organelle function. The indicated pie chart illustrates functions (as drawn from GO) associated with nuclear-localized proteins in Organelle DB. Multiple functions may be associated with a single protein; the figure indicates only the most common functions associated with nuclear proteins. (B) The localization data in Organelle DB can be effectively integrated with various data sets (e.g. two-hybrid data). Set 1 refers to a sampling of 155 randomly selected protein interactions identified in either of the landmark yeast two-hybrid studies of Ito et al. (20) or Uetz et al. (21) but not in both studies; hence, this is a low-confidence data set and is more likely to contain false positive interactions. Set 2 consists of a sampling of 105 interactions identified in both studies; hence, this is a higher confidence data set less likely to contain false positives. Total protein–protein interactions are indicated by the white bars; interactions consisting of two proteins localized to the same cell compartment are represented by the gray bars. Note that the high-confidence two-hybrid data set consists largely of proteins sharing identical localization patterns, highlighting the utility of localization data in screening two-hybrid results for genuine protein interactions.
The cross-species complement of organelle proteins contained in Organelle DB will ultimately facilitate evolutionary analysis of the organelle as a functional unit. At present, a truly comprehensive catalog of proteins within any single organelle is unavailable for any organism. Advancing protein localization studies, however, promise to rectify that situation within the coming years. Once such data sets are available, we can begin to consider the minimal protein complement necessary for a functioning organelle. Even in its current state, the data sets in Organelle DB bear witness to the increased numbers of proteins localized within a given organelle as we trace upwards from lower to higher eukaryotes. The mitochondrion provides a strong example. In yeast, 768 proteins have been localized to the mitochondrion (as presented in Organelle DB), while recent estimates suggest that mammalian mitochondria may possess upwards of 1200 proteins (17). As the mitochondrion is more comprehensively defined in each organism, we may be able to derive a minimal mitochondrion by comparing the orthologues between the yeast and mammalian forms of this organelle.
Integrating localization data
The localization data set housed in Organelle DB is extremely valuable as a means by which protein–protein interaction data may be validated. Protein interactions are typically identified through two-hybrid studies or co-immunoprecipitation techniques; however, both these techniques are prone to the identification of potential false positive interactions, particularly when performed in on a large-scale (18–21). As two interacting proteins must necessarily localize to the same cell compartment, putative protein partners can be screened for genuine interactions by integrating interaction data with localization data. In Figure 3B, we have identified two sets of putative interacting yeast proteins randomly drawn from two large-scale two-hybrid studies in S.cerevisiae (20,21): Set 1 consists of low-confidence interactions identified in one or the other study (but not in both studies), while Set 2 contains high-confidence interactions identified independently in both studies. We have integrated these interaction data sets with localization data from Organelle DB. Note that the high-confidence data set consists largely (83%) of protein pairs localized to the same cell compartment; however, only 47% of the two-hybrid interactions from the low-confidence grouping consisted of co-localized protein pairs. We, therefore, expect the data sets in Organelle DB to be effective as a tool in filtering protein–protein interaction data, and, more generally, useful as a complement to other orthologous functional data sets.
DATABASE ACCESS
Organelle DB may be freely accessed through the Life Sciences Institute/University of Michigan at http://organelledb.lsi.umich.edu. User support may be obtained from the Organelle DB staff by contacting anujk@umich.edu. Please direct all technical concerns and questions to this address as well. When referencing Organelle DB, please cite this article.
Acknowledgments
ACKNOWLEDGEMENTS
We thank John Herlocher and Jim Zajkowski for their technical expertise and support. We also thank Dr Hosagrahar Jagadish for terrific assistance in the establishment of this project.
REFERENCES
- 1.Kumar A. and Snyder,M. (2002) Protein complexes take the bait. Nature, 415, 123–124. [DOI] [PubMed] [Google Scholar]
- 2.Kumar A., Agarwal,S., Heyman,J.A., Matson,S., Heidtman,M., Piccirillo,S., Umansky,L., Drawid,A., Jansen,R., Liu,Y. et al. (2002) Subcellular localization of the yeast proteome. Genes Dev., 16, 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross-Macdonald P., Coelho,P.S., Roemer,T., Agarwal,S., Kumar,A., Jansen,R., Cheung,K.H., Sheehan,A., Symoniatis,D., Umansky,L. et al. (1999) Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature, 402, 413–418. [DOI] [PubMed] [Google Scholar]
- 4.Huh W.K., Falvo,J.V., Gerke,L.C., Carroll,A.S., Howson,R.W., Weissman,J.S. and O'Shea,E.K. (2003) Global analysis of protein localization in budding yeast. Nature, 425, 686–691. [DOI] [PubMed] [Google Scholar]
- 5.Sutherland H.G., Mumford,G.K., Newton,K., Ford,L.V., Farrall,R., Dellaire,G., Caceres,J.F. and Bickmore,W.A. (2001) Large-scale identification of mammalian proteins localized to nuclear sub-compartments. Hum. Mol. Genet., 10, 1995–2011. [DOI] [PubMed] [Google Scholar]
- 6.Simpson J.C., Wellenreuther,R., Poustka,A., Pepperkok,R. and Wiemann,S. (2000) Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep., 1, 287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dreger M. (2003) Subcellular proteomics. Mass Spectrom. Rev., 22, 27–56. [DOI] [PubMed] [Google Scholar]
- 8.Andersen J.S., Lyon,C.E., Fox,A.H., Leung,A.K., Lam,Y.W., Steen,H., Mann,M. and Lamond,A.I. (2002) Directed proteomic analysis of the human nucleolus. Curr. Biol., 12, 1–11. [DOI] [PubMed] [Google Scholar]
- 9.Harris M.A., Clark,J., Ireland,A., Lomax,J., Ashburner,M., Foulger,R., Eilbeck,K., Lewis,S., Marshall,B., Mungall,C. et al. (2004) The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res., 32, D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M.C., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O'Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christie K.R., Weng,S., Balakrishnan,R., Costanzo,M.C., Dolinski,K., Dwight,S.S., Engel,S.R., Feierbach,B., Fisk,D.G., Hirschman,J.E. et al. (2004) Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res., 32, D311–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flybase Consortium (2003) The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res., 31, 172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris T.W., Chen,N., Cunningham,F., Tello-Ruiz,M., Antoshechkin,I., Bastiani,C., Bieri,T., Blasiar,D., Bradnam,K., Chan,J. et al. (2004) WormBase: a multi-species resource for nematode biology and genomics. Nucleic Acids Res., 32, D411–D417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bult C.J., Blake,J.A., Richardson,J.E., Kadin,J.A., Eppig,J.T., Baldarelli,R.M., Barsanti,K., Baya,M., Beal,J.S., Boddy,W.J. et al. (2004) The Mouse Genome Database (MGD): integrating biology with the genome. Nucleic Acids Res., 32, D476–D481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhee S.Y., Beavis,W., Berardini,T.Z., Chen,G., Dixon,D., Doyle,A., Garcia-Hernandez,M., Huala,E., Lander,G., Montoya,M. et al. (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res., 31, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar A., DesEtages,S., Coelho,P., Roeder,G. and Snyder,M. (2000) High-throughput methods for the large-scale analysis of gene function by transposon tagging. Methods Enzymol., 328, 550–574. [DOI] [PubMed] [Google Scholar]
- 17.Mootha V.K., Bunkenborg,J., Olsen,J.V., Hjerrild,M., Wisniewski,J.R., Stahl,E., Bolouri,M.S., Ray,H.N., Sihag,S., Kamal,M. et al. (2003) Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell, 115, 629–640. [DOI] [PubMed] [Google Scholar]
- 18.Ho Y., Gruhler,A., Heilbut,A., Bader,G.D., Moore,L., Adams,S.L., Millar,A., Taylor,P., Bennett,K., Boutilier,K. et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature, 415, 180–183. [DOI] [PubMed] [Google Scholar]
- 19.Gavin A.C., Bosche,M., Krause,R., Grandi,P., Marzioch,M., Bauer,A., Schultz,J., Rick,J.M., Michon,A.M., Cruciat,C.M. et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- 20.Ito T., Chiba,T., Ozawa,R., Yoshida,M., Hattori,M. and Sakaki,Y. (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA, 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uetz P., Giot,L., Cagney,G., Mansfield,T.A., Judson,R.S., Knight,J.R., Lockshon,D., Narayan,V., Srinivasan,M., Pochart,P. et al. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]