Abstract
Breast cancers are solid tumors frequently characterized by regions with low oxygen concentrations. Cellular adaptations to hypoxia are mainly determined by “hypoxia inducible factors” that mediate transcriptional modifications involved in drug resistance and tumor progression leading to metastasis and relapse occurrence. In this study, we investigated the prognostic value of hypoxia-related gene expression in breast cancer. A systematic review was conducted to select a set of 45 genes involved in hypoxia signaling pathways and breast tumor progression. Gene expression was quantified by RT-qPCR in a retrospective series of 32 patients with invasive ductal carcinoma. Data were analyzed in relation to classical clinicopathological criteria and relapse occurrence. Coordinated overexpression of selected genes was observed in high-grade and HER2+ tumors. Hierarchical cluster analysis of gene expression significantly segregated relapsed patients (p = 0.008, Chi2 test). All genes (except one) were up-regulated and six markers were significantly expressed in tumors from recurrent patients. The expression of this 6-gene set was used to develop a basic algorithm for identifying recurrent patients according to a risk score of relapse. Analysis of Kaplan-Meier relapse-free survival curves allowed the definition of a threshold score of 2 (p = 0.021, Mantel-Haenszel test). The risk of recurrence was increased by 40% in patients with a high score. In addition to classical prognostic factors, we showed that hypoxic markers have potential prognostic value for outcome and late recurrence prediction, leading to improved treatment decision-making for patients with early-stage invasive breast cancer. It will be necessary to validate the clinical relevance of this prognostic approach through independent studies including larger prospective patient cohorts.
Introduction
Breast cancer is a heterogeneous disease with diverse clinical outcomes. Current therapeutic options, including initial surgery and both adjuvant chemotherapy and endocrine therapy, are effective in the earlier stages of disease and improve clinical outcome. However, despite the proven benefits of these treatments, breast cancer patients still have a risk of relapse after the first 5 years. The clinical outcome of breast cancer is based primarily on clinicopathological criteria such as tumor size, histological grade and the status of estrogen, progesterone and HER2 receptors. These parameters are prognostic markers for early recurrence, but their role in late recurrence is less clear [1, 2]. Prediction of late recurrence at diagnosis could help individualize therapeutic options, thereby preventing unnecessary treatments. Identification of factors predicting long-term relapse-free survival in breast cancer patients has become an important promising field of biomarker research [3].
Accumulating evidence from clinical studies suggests that tumor hypoxia might have an important role for clinical outcome and late recurrence in human cancer, including invasive breast cancer. The hypoxic tumor microenvironment is associated with a poorer prognosis for outcome and survival [4]. Several authors have shown that molecular mechanisms of adaptation to hypoxia make tumor cells more aggressive and more resistant to chemotherapy and radiotherapy, thereby promoting tumor progression [5, 6]. Hypoxic areas arise when the metabolic requirements of cancer cells are higher than the availability of intravascular oxygen content in tumors. Cellular adaptations to hypoxia are mainly mediated by a family of transcription factors called hypoxia inducible factors (HIFs). HIF-1 was the first member of this family and is ubiquitously expressed [7]. These transcription factors are heterodimers composed of an alpha subunit and a beta subunit [8]. Under normoxic conditions, HIF-1 alpha is hydroxylated by a family of dioxygenases known as prolyl-hydroxylases (PHD). Hydroxylated proline residues are recognized by the Von Hippel-Lindau tumor suppressor, leading to polyubiquitination and subsequent proteasomal degradation. Under hypoxic conditions, oxygen levels are not sufficient for the enzymatic activation of PHD. Consequently, HIF-1 alpha is not degraded and is translocated to the nucleus, where it binds to the subunit HIF-1 beta and the transcriptional coactivator p300 [9]. The active transcription complex regulates the expression of multiple genes by binding specific DNA sequences called hypoxia response elements (HRE). Regulation of HIF-1 alpha protein is not limited to hypoxic conditions. Several studies have also revealed oxygen-independent mechanisms that result from genetic alterations such as activation of oncogenes (HER2) and/or loss of tumor suppressor genes (VHL or PTEN). Dysfunctions of the PI3K/AKT and RAS/MAPK signaling pathways are also involved in HIF-1 alpha regulation [10, 11]. Activation of hypoxia-related genes plays an important role in tumor progression because of the involvement of these genes in several cellular processes, including cell differentiation, survival, angiogenesis, migration and metastasis [12].
Thus, assessment of tumor hypoxia appears to be a potential strategy for clinical outcome prediction of solid tumors. However, it remains difficult to perform quantitative measures of tumor hypoxia as well as to determine the relationship between hypoxia and clinical parameters in human cancers. Several measurement methods of tumor oxygenation, including both direct and indirect approaches, have been described. The main direct approach for measuring the partial pressure of oxygen in tumors is based on the polarographic method using oxygen microelectrodes. This method revealed a mean partial oxygen pressure of 28 mmHg in breast tumors and 65 mmHg in normal breast tissue [13]. Indirect methods consist essentially of immunohistochemical measurement in tumor biopsies of the expression of HIF-1 alpha as well as proteins regulated by HIF complexes, such as carbonic anhydrase 9 (CA9) and vascular endothelial growth factor (VEGF). Several previous reports have already associated breast cancer outcomes with levels of HIF-1 alpha or CA9 proteins [14–16]. Other non-invasive techniques, such as molecular imaging, allow the identification of intratumoral hypoxia by analyzing its effect on the metabolism of tumor cells. The low oxygen pressures observed in solid tumors force cells to shift from aerobic to anaerobic glucose metabolism [17]. Positron emission tomography (PET) imaging with 18F-fluorodeoxyglucose (18F-FDG) permits the detection of increased glucose consumption by cancer cells. 18F-FDG uptake correlates with reduced partial pressure of oxygen and increased HIF-1 alpha protein levels in diverse types of tumors [18, 19]. More recently, the analysis of changes induced by hypoxia in the transcriptome has also provided an indirect method with prognostic and predictive values [12]. Several molecular signatures have been constructed from non-specific genetic markers of hypoxic responses and breast cancer. Most of these signatures were generated by differential strategies based on whole-transcriptome analysis or were implemented initially from other solid tumors [20, 21]. Winter et al. defined a molecular signature of 99 genes whose expression in a series of head and neck squamous cell carcinomas clustered with the expression of 10 well-known hypoxia-regulated genes. This signature was shown to be a prognostic factor for relapse-free survival in an independent breast cancer series [20].
These studies highlight the importance of hypoxia-related gene expression for outcome prediction in breast cancer. The quantification of biomarkers involved in both hypoxia signaling pathways and breast cancer development may facilitate the prediction of prognosis according to the molecular profile of tumors. The aim of this study was to generate a molecular signature of tumor hypoxia with potential prognostic significance in breast cancer. We analyzed the expression of 45 well-known hypoxia-regulated genes in a retrospective series of 32 tumor samples from patients with early-stage invasive breast cancer. This set of genes was selected from a systematic review according to objective criteria based on their implication in breast cancer aggressiveness and hypoxia signaling pathways. Gene expression was investigated in relation to clinicopathological data (stage, grade mSBR, HER2 status, and relapse occurrence).
Materials and methods
Patients and clinicopathological data
A retrospective study of a total of 32 patients with previously untreated primary breast cancer was conducted. Patients were diagnosed between 1994 and 1998 and had undergone surgery at the Jean Perrin Comprehensive Cancer Center. Fine-needle aspiration biopsies were performed in patients, and an aliquot of each aspirate was immediately smeared on a slide to serve as a control for the presence of malignant cells and the absence of important stromal and fat contamination. The remaining aspirated material was processed for embedding in a paraffin block for later use in immunophenotyping or stored in liquid nitrogen until total RNA extraction. Tumors samples were conserved in the Biological Resource Center of Jean Perrin Comprehensive Cancer Center, identified under No. BB-0033-00075 (Clermont-Ferrand, France). The clinical history of patients was collected with the help of an oncologist. Tumors were classified histologically according to the World Health Organization criteria as ductal invasive breast carcinoma. Initial staging comprised complete and detailed clinical examination including the International Union Against Cancer TNM (tumor size, nodes, metastases) classification. Histopathological evaluation of tumors was performed using the Scarff-Bloom-Richardson histologic grading system as modified by Le Doussal [22, 23]. Under French law on biomedical research, this is an epidemiological study that does not have to be submitted to an Institutional Review Board. All clinical data and tissue samples were fully anonymized and de-identified before they were accessed by the researchers for this study.
Immunohistochemical studies
Patients were screened for estrogen, progesterone and HER2 receptor status by immunohistochemistry (IHC) on paraffin-embedded tissue sections. Immunostaining was performed with a Nexes automated immunostainer following the manufacturer's guidelines (Ventana, Illkirch, France). Sections were scored semiquantitatively by two pathologists using standard light-microscopic evaluation. A threshold of 10% total stained tumor cells was considered positive for estrogen and progesterone status. Immunohistochemical staining for HER2 was performed using the HercepTest kit (Dako, Carpinteria, CA, USA) and was scored according to the standard scoring system recommended by the manufacturer. Intensity scores of 0 or 1+ were designated as negative for HER2 expression. Scores of 3+ were considered positive and were defined as HER2 overexpression in the presence of complete membrane staining with high intensity. Scores of 2+ were considered equivocal cases, and HER2 fluorescence in situ hybridization (FISH) assay was performed for detection of HER2 amplification using the HER2 FISH pharmDx kit (Dako) according to the manufacturer's instructions. Tumors with amplification of HER2 were considered HER2 positive (3+). Patient and tumor characteristics are summarized in Table 1.
Table 1. Clinical and histopathological characteristics of patients.
| Characteristics | Classification | All patients (n = 32) |
|---|---|---|
| Age | < 50 ≥ 50 |
n = 8 n = 24 |
| Estrogen receptors | Negative Positive |
n = 1 n = 31 |
| Progesterone receptors | Negative Positive |
n = 8 n = 24 |
| Lymph nodes | Negative Positive |
n = 20 n = 12 |
| Tumor stage | 1 2–3 |
n = 7 n = 25 |
| Grade mSBR | 1-2-3 4–5 |
n = 25 n = 7 |
| HER2 status | Negative Positive |
n = 27 n = 5 |
| Recurrence | No Yes |
n = 18 n = 14 |
RNA extraction and reverse transcription
Total RNA was extracted from frozen tumor samples using Trizol reagent according to the manufacturer’s protocol (Invitrogen Life Technologies, Carlsbad, CA, USA). The quality and concentration of the total RNA were assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Foster City, CA, USA). Two micrograms of total RNA were reverse transcribed in a total volume of 20 μl using the High Capacity cDNA kit with RNase inhibitor according to the manufacturer’s instructions (Applied Biosystems, Foster City, CA, USA). The reaction conditions were 25°C for 10 min, 37°C for 120 min and 85°C for 5 min.
Assay design and real-time quantitative PCR
A qualitative review of literature on breast cancer was performed in PubMed/MEDLINE to select 45 genes known to be regulated by hypoxia and involved in breast carcinogenesis. Selection of these genes was performed according to several criteria, including the presence of HRE elements in promoters, ability to be activated by hypoxia and/or hypoxia-mimetic agents such as desferrioxamine or cobalt chloride, and involvement in breast cancer aggressiveness (Table 2). Real-time quantitative PCR analysis was performed using custom-made Taqman low-density arrays (TLDAs), which are 384-well microfluidic cards preloaded with sets of primers and specific probes designed to amplify selected genes (Applied Biosystems, Foster City, CA, USA). Samples of cDNA (50 μl) were mixed with 50 μl of 2X Taqman Universal PCR Master Mix (Applied Biosystems), and a total of 100 μl of reaction mixture was loaded on TLDA cards, followed by centrifugation twice 1 min at 1200 rpm to distribute the samples from the loading port into each well. The cards were sealed, and real-time quantitative PCR amplification was performed using an ABI Prism 7900 HT Sequence Detection System according to the manufacturer's instructions (Applied Biosystems). Relative quantification (RQ) analysis was performed with RQ Manager 1.2 software (Applied Biosystems). A threshold cycle (Ct) value equal to 35 was used as the cutoff for non-expressed genes. The set of genes included two housekeeping genes used as internal controls (RPL32 and 18S). In addition, gene expression stability was determined by the NormFinder program, and optimal reference genes for normalization were identified among the selected genes [24]. The average expression level of all markers was also used to perform data normalization. The RQ of gene expression was determined using the comparative ΔΔCt method based on the equation RQ = 2-ΔΔCt [25]. This method allows the determination of the relative fold change ratio of a target gene between two different groups.
Table 2. List of selected gene expression assays.
| Gene symbol | Assay reference | Gene name |
|---|---|---|
| Endogenous genes | ||
| 18S | Hs99999901_s1 | - |
| RPL32 | Hs00851655_g1 | Ribosomal protein L32 |
| Cell survival, proliferation, differentiation | ||
| BNIP3 | Hs00969291_m1 | BCL2/adenovirus E1B 19 kd-interacting protein 3 |
| BRCA1 | Hs00173233_m1 | Breast cancer 1 |
| CCND1 | Hs00277039_m1 | Cyclin D1 |
| EPO | Hs01071096_g1 | Erythropoietin |
| HER2 | Hs01001595_m1 | Erythroblastic leukemia viral oncogene homolog 2 |
| IGF2 | Hs01005964_g1 | Insulin-like growth factor 2 |
| NDRG1 | Hs00608387_m1 | N-myc downstream regulated gene 1 |
| BNIP3L | Hs00188949_m1 | BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| TGFB3 | Hs00234245_m1 | Transforming growth factor beta |
| TGM2 | Hs00190278_m1 | Transglutaminase 2 |
| Transcription factors and feed back | ||
| CEBPA | Hs00269972_s1 | CCAAT/Enhancer binding protein alpha |
| CITED2 | Hs00366696_m1 | Cbp/p300-interacting transactivator, 2 |
| ETS1 | Hs00901425_m1 | v-ets erythroblastosis virus E26 oncogene homolog 1 |
| FOXO3A | Hs00921424_m1 | Forkhead box O3 |
| NR4A1 | Hs00374230_m1 | Nuclear receptor subfamily 4, group A, member 1 |
| PHD2 | Hs00254392_m1 | HIF-prolyl hydroxylase 2 |
| SNAI1 | Hs00195591_m1 | Snail homolog 1 |
| TWIST1 | Hs00361186_m1 | Twist homolog 1 |
| VHL | Hs00184451_m1 | Von Hippel-Lindau |
| PTEN | Hs00829813_s1 | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase |
| Extracellular matrix, motility | ||
| CTSD | Hs00157201_m1 | Cathepsin D |
| CDH1 | Hs01023895_m1 | E-cadherin |
| KRT19 | Hs00761767_s1 | Keratin 19 |
| CTGF | Hs01026926_g1 | Connective tissue growth factor |
| CXCR4 | Hs00607978_s1 | Chemokine (C-X-C motif) receptor 4 |
| MET | Hs01565582_g1 | The proto-oncogene MET |
| MMP2 | Hs00234422_m1 | Matrix metallopeptidase 2 |
| PLAUR | Hs00182181_m1 | Plasminogen activator, urokinase receptor |
| VIM | Hs00185584_m1 | Vimentin |
| Glucose metabolism, pH | ||
| GPI | Hs00976711_m1 | Glucose phosphate isomerase |
| CA9 | Hs00154208_m1 | Carbonic anhydrase 9 |
| ENO1 | Hs00361415_m1 | Enolase 1 |
| GLUT1 | Hs00892681_m1 | Glucose transporter 1 |
| LDHA | Hs00855332_g1 | Lactate dehydrogenase A |
| NHERF1 | Hs00188594_m1 | Na/H exchanger regulatory factor 1 |
| PGK1 | Hs00943178_g1 | Phosphoglycerate kinase 1 |
| TPI | Hs01593134_gH | Triose-phosphate isomerase |
| Angiogenesis | ||
| COX2 | Hs01573471_m1 | Cyclo-oxygenase 2 |
| EDN1 | Hs00174961_m1 | Endothelin |
| ENG | Hs00164438_m1 | Endoglin |
| LEP | Hs00174877_m1 | Leptin |
| VEGF | Hs00900054_m1 | Vascular endothelial growth factor |
| Drug resistance | ||
| AK3 | Hs00750261_s1 | Adenylate Kinase 3 |
| ABCB1 | Hs01067802_m1 | ATP-binding cassette, sub-family B member 1 |
| ABCG2 | Hs01053790_m1 | ATP-binding cassette, sub-family G member 2 |
Statistical analysis
Different groups of patients were defined according to clinicopathological criteria such as tumor stage, histological grade, HER2 status and occurrence of relapse. For each gene, the average RQ was calculated in each group. The ratio of the average RQ between 2 groups was used to determine the fold induction for the expression of each gene in a group of patients relative to the corresponding control group. A positive fold change of 1 indicated 2-fold up-regulation, and a negative fold change of -1 indicated 2-fold down-regulation. A comparative analysis of gene expression profiles was performed between different groups. A parametric (Student's t-test) or a non-parametric test (Kruskal-Wallis test) was used to identify genes that were significantly differently expressed between groups.
Unsupervised hierarchical clustering analysis based on ΔCt values was performed using the Euclidean distance and Ward’s method based on barycenter calculation. Gene expression profiles were analyzed using all selected genes and differentially expressed genes with statistical significance between the recurrent group and non-recurrent group. Secondary to cluster calculation, the Chi2 test was used to compare the proportion of relapses in the main selected clusters of patients. This approach permits the validation of the relevance of the cluster analysis and the influence of the expression of the genes on relapse risk.
Kaplan-Meier survival curves were constructed for distant or local relapse-free survival, and statistical significance was examined using the Mantel-Haenszel test. Relapse-free survival was defined as the time of diagnosis to the development of distant or local recurrence. The internal consistency of predictive markers of relapse was assessed using Cronbach’s alpha coefficient as a measure of scale reliability. All analyses were performed using the SEM statistical software [26], and a probability value p < 0.05 was considered significant.
Results
Comparative analysis of hypoxia-related gene expression according to clinicopathological data
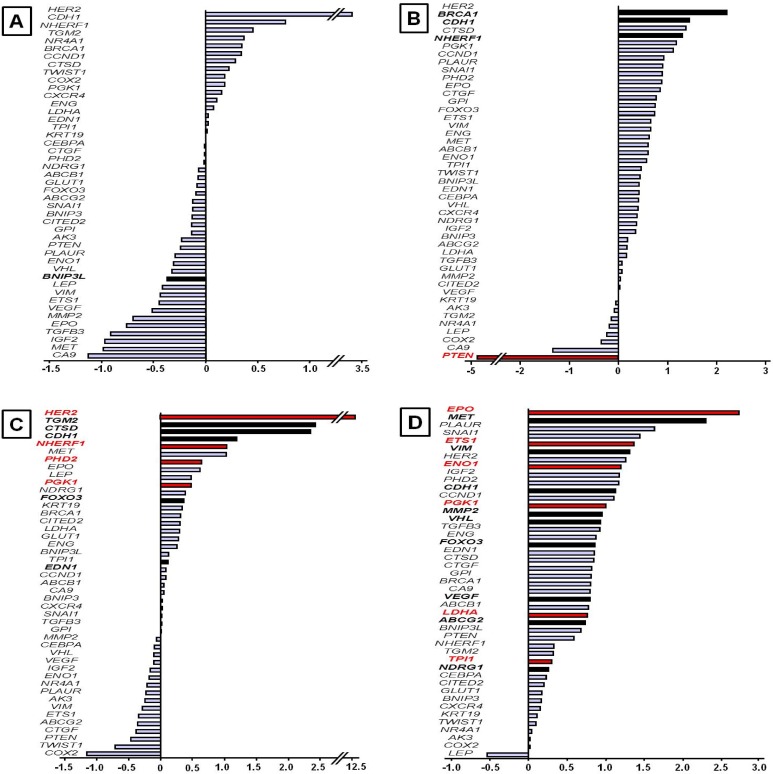
The expression of selected genes was quantified by real-time quantitative PCR using Taqman low-density arrays (Applied Biosystems). The relative quantification (RQ) of each gene was determined in 32 tumor samples from 32 patients with breast carcinoma (S1 Table). Several groups of patients were defined according to tumor stage (tumor stage 2–3 vs tumor stage 1), histological grade (high mSBR grades vs low mSBR grades), HER2 status (HER2+ vs HER2-), and relapse occurrence (recurrent vs non-recurrent patients). The distribution of patients according to these clinicopathological criteria is presented in Table 1. A comparative analysis of gene expression based on the fold induction values was performed between different groups (Fig 1). Breast tumors were staged at diagnosis. The major tumor characteristic used to determine the stage was the tumor size. All patients have been diagnosed with early-stage invasive breast cancer. However, two groups were defined: a group of 7 patients with stage 1 and a group of 25 patients with stage 2 or 3. Only one patient was diagnosed with stage 3. The gene expression profile was randomly distributed, and no genes were significantly differently expressed between stages 2–3 and stage 1 (Fig 1A) (S2 Table). Patients were then divided into two groups based on the mSBR grading system modified by Le Doussal. This modified grading system was built from the nuclear pleomorphism and the mitotic index and retains five prognostic classes instead of three. Le Doussal et al. have demonstrated that mSRB grades 1, 2 and 3 have a lower risk for developing metastasis than mSRB grades 4 and 5. Seven patients were mSBR grade 4 or 5, and 25 patients were mSBR grade 1, 2 or 3. Overall, almost all genes were overexpressed in patients with high mSBR grades compared with low grades. The genes in the high-grade group were overexpressed by approximately 45% compared with the low-grade group. Interestingly, the PTEN gene was 6-fold down-expressed in high grades compared with low-grade tumors (p = 0.022). Insignificant differences (p < 0.10) were observed for BRCA1, CDH1 and NHERF1, which are overexpressed in high-grade tumors, with increases of up to 2.2-, 1.6- and 1.5-fold, respectively (Fig 1B). Only 5 tumors were HER2+ (ICH+ and FISH+), and 27 tumors were HER2-. Overexpression of the majority of genes was observed in the group of patients with HER2+ breast cancer compared with the HER2- group. The genes in the HER+ group were overexpressed by an average of approximately 50% compared with HER- group. The HER2 gene was overexpressed by more than 12-fold in the HER2+ group (p = 0.0007). The NHERF1, PGK1 and PHD2 genes were also significantly overexpressed (p = 0.028, p = 0.032 and p = 0.048, respectively). In addition, the TGM2, CDH1, CTSD, FOXO3A and EDN1 genes were positively correlated with the HER2+ group (P < 0.10) (Fig 1C). A comparative analysis of gene expression profiles between recurrent and non-recurrent patients was also performed. With the exception of the LEP gene, all genes were overexpressed in the relapse group compared with the non-relapse group. The average gene overexpression in the relapse group was approximately 75%. Six genes was significantly overexpressed in the group of patients who relapsed: EPO (p = 0.013), ETS1 (p = 0.022), ENO1 (p = 0.003), PGK1 (p = 0.021), LDHA (p = 0.011) and TPI (p = 0.048). In addition, MET, VIM, CDH1, MMP2, VHL, FOXO3, VEGF, ABCG2 and NDRG1 were associated with recurrent group (p < 0.10) (Fig 1D).
Fig 1. Hypoxia-related gene expression profiles according to clinicopathological data.
Gene expression was determined using quantitative real-time PCR as described in the Materials and Methods. The results are presented as the fold induction of relative quantification by classification in ascending order. A positive fold change of 1 indicated 2-fold up-regulation, and a negative fold change of -1 indicated 2-fold down-regulation. A comparative analysis was performed between (A) high tumor stage vs low tumor stage, (B) high mSBR grades vs low mSBR grades, (C) HER2+ status vs HER2- status, and (D) recurrent patients vs non-recurrent patients. Statistical analysis was performed between groups using Student’s t or Kruskal Wallis tests (red bar: p < 0.05; black bars: p < 0.10).
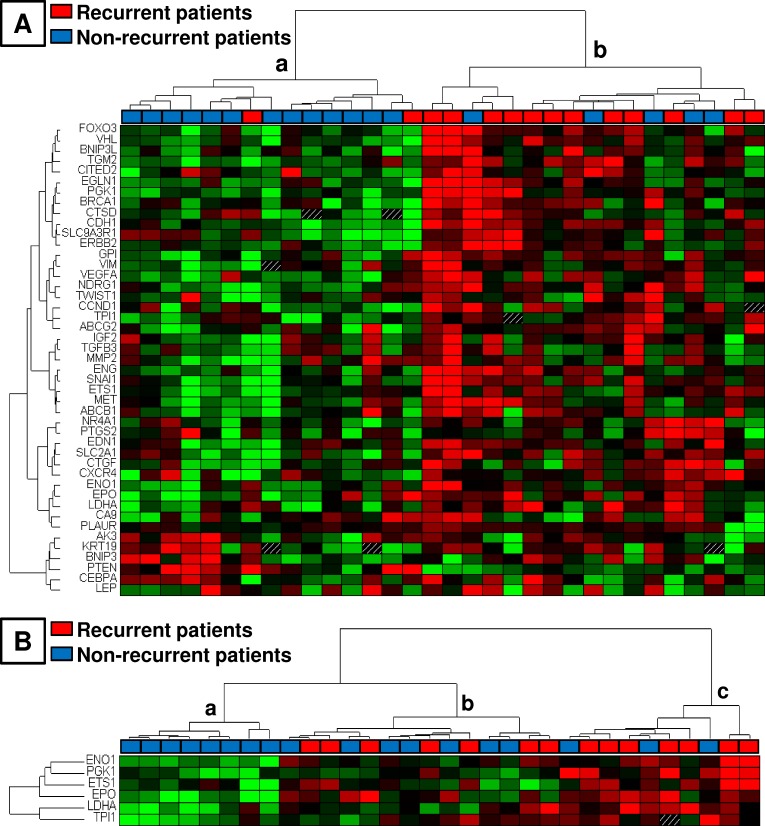
Hierarchical clustering analysis of hypoxia-related gene expression
Data are presented in heat map format combined with hierarchical clustering, thus revealing the distribution of genes according to their expression in each tumor sample (Fig 2). Hierarchical clustering analysis of tumors based on the expression of all selected genes identified two main clusters of patients that were significantly associated with relapse occurrence (p = 0.008, Chi2 test). In cluster a and cluster b, 13% and 70% of patients relapsed, respectively (Fig 2A). The clustering based on the 6 significantly differentially expressed genes between the recurrent group and non-recurrent group (EPO, ETS1, ENO1, PGK1, LDHA and TPI) also significantly segregated patients who had relapsed: 0% of relapse in group a, 50% in b and 70% in c (p = 0.0095). For the comparison of groups b and c together with a, p = 0.03 (Fig 2B).
Fig 2. Profile of hypoxia-related gene expression in 32 tumors from patients with early-stage breast cancer.
Data are presented in heat map format combined with hierarchical clustering using ΔCt values of gene expression. Each row represents a gene, and each column represents a patient. Gene expression is relative to the median of ΔCt values. Genes in red and green indicate expression above and below the median, respectively. (A) Hierarchical cluster analysis using all selected genes. (B) Hierarchical cluster analysis using the 6 differentially expressed genes with statistical significance between the recurrent group and non-recurrent group.
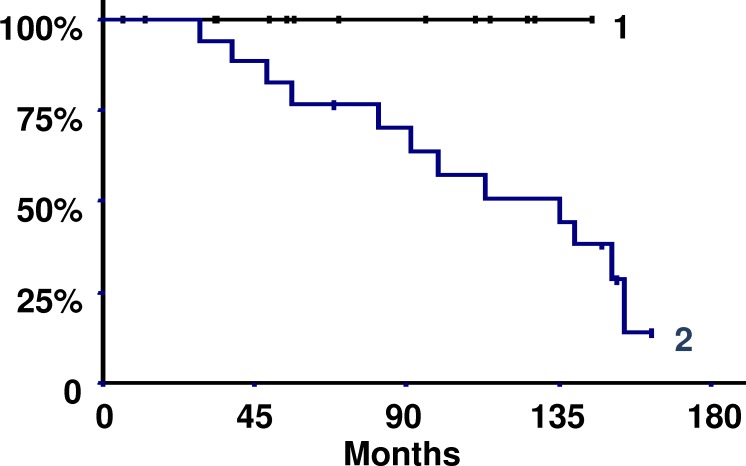
Risk score of relapse
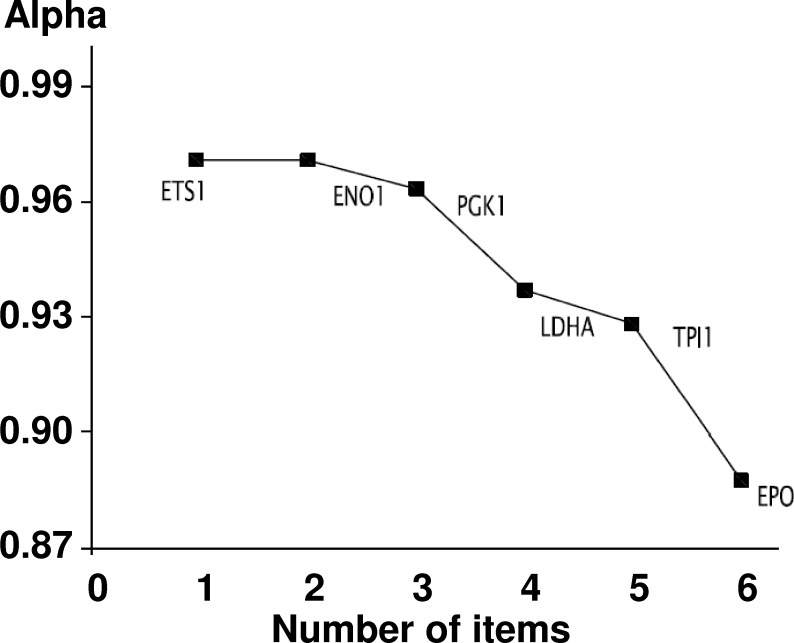
The comparison of gene expression between the relapse group and non-relapse group allowed the identification of six significantly differentially expressed genes: EPO, ETS1, ENO1, PGK1, LDHA and TPI. A basic algorithm was developed to classify patients according to a risk score of relapse. To define this score, the optimum level of each gene significantly expressed in the relapse group was determined by an iterative approach using the difference in relapse-free survival as the main criterion (Table 3). For each one of the six genes, a value of 1 was given if its expression was higher than the optimum thresholds presented in Table 3. The risk score was then calculated by summing the values attributed to each gene. Analysis of the Kaplan-Meier relapse-free survival curves using the Mantel-Haenszel test statistic permitted the definition of a threshold score of 2 (Fig 3). As shown in Fig 3, a threshold score equal to 2 yielded a significant difference between recurrent and non-recurrent patients (p = 0.021). The risk of relapse was multiplied by 1.384 if the score was ≥ 3, which indicated that the risk of relapse was increased by 40%. In the group with a score ≥ 3, the relapse rate was 19% after 5 years and 42% after 10 years; by contrast, the rate was 0% in the other group because no patient belonging to this group had relapsed. In addition, the statistical index of Cronbach's alpha [27] indicated that there was good consistency between all markers (alpha = 0.9) (Fig 4). In summary, the analysis of the expression values of EPO, ETS1, ENO1, PGK1, LDHA and TPI permitted the generation of a risk score of relapse in which a risk score of ≥ 3 indicates a short relapse time and a risk score ≤ 2 indicates a long relapse time.
Table 3. Optimum level of gene expression thresholds discriminating relapse-free survival.
| Gene | Optima |
|---|---|
| EPO | 7.10 |
| ETS1 | 1.81 |
| ENO1 | 1.00 |
| PGK1 | 1.37 |
| LDHA | 1.20 |
| TPI | 1.14 |
Fig 3. Kaplan-Meier relapse-free survival curves according to the risk score of relapse.
Curve 1: 15 patients with score ≤ 2. Curve 2: 17 patients with score ≥ 3. The 14 recurrent patients were in curve 2 (p = 0.021, Mantel-Haenszel test).
Fig 4. Analysis of the internal consistency of the 6 genes differentially expressed between recurrent and non-recurrent patients.
Cronbach’s alpha coefficient was calculated to measure internal consistency (alpha = 0.90).
Discussion
A number of experimental and clinical studies have shown that adaptations of tumor cells to hypoxia are associated with malignant progression and development of resistance to both chemotherapy and radiotherapy [5, 28]. The expression of various hypoxic markers in breast cancer has been linked to a worse prognosis. In a clinical series of breast cancer patients, resistance to endocrine therapy combined with chemotherapy has been associated with overexpression of the HIF-1 alpha and CA9 proteins [29]. Cellular adaptations to hypoxia involve transcriptional modifications responsible for tumor aggressiveness and resistance to treatment [30]. Many approaches have sought to target the cellular response to hypoxia in human cancers [31, 32]. Inhibition of HIF-1 alpha activation appears to be the main approach and may improve the response to chemotherapy. Analysis of hypoxia-related gene expression may be useful in the development of novel therapeutic strategies for breast cancer. Several authors have identified different molecular signatures predicting the clinical outcome of cancer diseases; but these signatures differ greatly and share quite a few genes [21, 33, 34]. The aim of this study was to quantify the expression of well-known hypoxia-related genes in primary tumors from patients with early-stage breast cancer to assess their potential value as prognostic and predictive markers for cancer development and relapse occurrence. All patients included in this study received chemotherapy and endocrine therapy after initial surgery.
Multiple genes have been reported to be regulated by HIF complexes. These hypoxia regulated genes are involved in biological processes allowing tumor progression, such as cell proliferation and differentiation, survival, glucose metabolism, angiogenesis, migration, motility and drug resistance [12, 35]. A qualitative review of relevant literature related to tumor hypoxia and breast cancer enabled the selection of a set of candidate genes. Among these genes, we established a molecular signature composed of 45 genes involved in hypoxia signaling pathways and breast cancer progression. The expression of selected genes was quantified in a retrospective series including 32 tumor samples derived from patients with early-stage invasive ductal carcinoma without treatment at diagnosis. A comparison analysis of gene expression was performed according to clinicopathological features (stage, mSBR grade, HER2 status) and relapse occurrence.
Analysis of gene expression did not appear to be influenced by the clinical stage of tumors previously defined from the tumor node metastasis (TNM) classification. All patients were diagnosed with stage 1 or 2 breast cancer, with the exception of one patient with stage 3. The distribution of gene expression according to clinical stage showed no significant difference between stage 1 and stages 2 and 3. All tumors were less than 5 cm (T1 or T2) and were node-negative or 1 to 3 lymph-node positive (N0 or N1). These clinical criteria defined a relatively homogeneous group with no distant metastasis (M0). A hypoxic microenvironment has been consistently identified as a feature that promotes metastatic processes in breast cancer [36]. HIF factors regulate the transcription of several genes involved in different steps of the metastatic process, including angiogenesis, extracellular matrix modulation, cell migration and adhesion [37]. High proportions of hypoxic cells and increased levels of HIF-1 alpha protein in primary tumors of breast cancer patients indicate increased risk of metastasis and decreased overall survival rates [38]. Bos et al. revealed that high levels of HIF-1 alpha were significantly associated with overall survival in patients with negative lymph node status. However, no correlation was observed between levels of HIF-1 alpha expression and tumor size or lymph node status in a retrospective series of early-stage breast tumors [39]. In agreement with this finding, the expression of the hypoxia-regulated genes selected in this study was not associated with stage in this series of breast cancer patients.
In contrast to clinical stage, overexpression of almost all markers was observed in the group of patients with high-grade tumors. This group was also characterized by a significant decrease in PTEN gene expression. PTEN is a tumor suppressor that encodes a phosphatase involved in downregulation of the PI3K/AKT signaling pathway. The PTEN gene is frequently mutated or inactivated in multiple human cancers, including a large proportion of breast cancers. A number of clinical studies have demonstrated that loss or reduced expression of PTEN is involved in breast cancer progression, poor prognosis and resistance to treatment [40]. PTEN is also a negative regulator of HIF-1 alpha expression [10]. Some mutations or deletions of PTEN induce hyperactivation of the PI3K/AKT signaling pathway and activation of HIF complexes. In vitro studies of PTEN knockout in cancer cell lines have provided evidence for the role of PTEN in the stability and activity of the HIF-1 complex [10]. Tumor hypoxia and loss of PTEN function result in activation of HIF factors, followed by increased transcription of hypoxia-related genes and the development of a more aggressive breast cancer.
A similar analysis was performed in the group of patients with HER2+ breast cancer vs the group of HER2- patients. As expected, patients with HER2+ status harbored strong amplification of the HER2 gene. In addition, significant differences in gene expression were observed for NHERF1 (Na/H exchange regulatory factor), PHD2 (prolyl-hydroxylase 2) and PGK1 (phosphoglycerate kinase 1). The NHERF1 gene encodes a protein capable of interacting with the HER2 receptor [41]. The mechanism of action of NHERF1 in tumor cells has not been elucidated, but it has been reported that NHERF1 plays an important role in cancer development. NHERF1 overexpression is associated with high-grade tumors and increased expression of HIF-1 alpha protein in breast cancer [42]. Transcriptional activation of NHERF1 by hypoxia has also been established in in vitro models, including several breast cancer cell lines [43]. The protein encoded by the PHD2 gene is a dioxygenase that catalyzes the post-translational hydroxylation of HIF-1 alpha protein under normoxia. This enzyme plays a central role in the regulation and stability of HIF complexes. In vitro studies have demonstrated that levels of PHD2 expression are increased in hypoxic conditions. The promoter of PHD2 contains HRE elements, allowing the establishment of a positive feedback loop under hypoxia [44, 45]. In addition, increased levels of PHD2 protein have been correlated with relapse and tumor metastasis [46].
The PGK1 gene was also significantly overexpressed in the group of patients with high mSBR grade as well as in the group of recurrent patients. Indeed, the comparative analysis of gene expression between recurrent patients and non-recurrent patients revealed overexpression of almost all genes. The PGK1, LDHA, TPI, ENO1, EPO and ETS1 markers were significantly overexpressed in the relapse group compared with the non-relapse group. Among these 6 significantly differentially expressed genes, PGK1, ENO1 (enolase), LDHA (lactate dehydrogenase) and TPI (triose phosphate isomerase) are directly involved in glucose metabolism and encode glycolytic enzymes. HIF factors have long been implicated in the regulation of genes involved in glucose metabolism in tumor cells [47]. These genes have HRE elements in their respective promoters and therefore bind HIF complexes [48]. In hypoxia, cancer cells redirect their aerobic metabolism to anaerobic metabolism by activating glycolysis, which becomes the main source of energy. Several other genes targeted by HIF factors are involved in multiple steps of glucose metabolism and are up-regulated under hypoxia. Overexpression of the TPI, PGK1 and ENO1 enzymes has been demonstrated in a series of breast tumors [49]. Expression of LDHA is increased in hypoxic tumor cells, leading to increased ATP production and cell proliferation. This enzyme catalyzes the conversion of pyruvate into lactate under hypoxia. The lactate is absorbed by non-hypoxic tumor cells for use as a respiratory substrate for promoting angiogenesis and metastasis [50]. In several breast cancer cell lines, inactivation of LDHA inhibits cell proliferation and induces apoptosis [51]. The EPO gene encodes erythropoietin, which is a specific stimulator of erythropoiesis [52]. The HIF-1 factor was discovered by the identification of HRE elements in the promoter of EPO [53]. Regulation of EPO by HIF complexes under hypoxic conditions is well documented [54]. EPO is a potent inhibitor of apoptosis caused by ischemia and hypoxia [55]. In erythrocytes, binding of EPO to its receptor (EPOR) results in the activation of multiple signaling pathways responsible for cell proliferation and differentiation [56, 57]. EPO and its receptor are also expressed in other cell types, including endothelial cells and mammary epithelial cells [58]. High mRNA and protein levels of EPO and EPOR have been reported in several cancer cell lines. In vitro studies in breast cancer cell lines have demonstrated that autocrine/paracrine production of EPO and EPOR under hypoxia contributes to cell survival and proliferation. Other authors have shown that the EPO/EPOR axis plays an important role in the regulation of the migration and invasion of breast cancer cells [59]. The ETS1 gene is a proto-oncogene encoding a transcription factor involved in the proliferation of normal breast epithelial cells. This gene is also involved in tumor progression in breast cancers and contributes to aggressive tumor phenotypes by activating the transcription of genes involved in angiogenesis, extracellular matrix remodeling, cell adhesion and invasion [60]. In addition, HRE elements have been identified in the promoter of ETS1, suggesting transcriptional activation under hypoxic conditions [60]. Span et al. demonstrated that increased expression of ETS1 was associated with increased risk of recurrence in a series of invasive breast cancers. In agreement with these previous studies, overexpression of this 6-gene set appears to be involved in tumor progression contributing to the occurrence of relapse.
Overall, the expression of the 45-gene set was associated with aggressive tumors characterized by high grade, HER2+ status and increased recurrence risk. This gene signature reflects the impact of the hypoxic microenvironment on cancer cells. Our findings provide further evidence that hypoxia-related genes are involved in the clinical outcome of breast cancer by activating hypoxia signaling pathways. Although this study is based on a limited number of patients, assessment of hypoxia-related gene expression in breast cancer could have potential prognostic value. In particular, quantification of the expression of EPO, ETS1, PGK1, TPI, LDHA and ENO1 in a primary tumor sample provides information on the risk of recurrence for patients with early-stage invasive breast cancer. The calculation of a score from the expression of this 6-gene set permitted the classification of patients with a low or high risk of relapse. A primary breast tumor with a risk score ≥ 3 has a high risk of recurrence, and a tumor with a risk score ≤ 2 has a low risk of recurrence. Furthermore, hierarchical clustering analysis of gene expression identified two main groups of patients significantly associated with relapse occurrence.
In summary, we have defined a molecular signature specific to hypoxia responses in breast cancer. This gene signature was associated with tumor aggressiveness and the risk of recurrence. The expression of the 6-gene set allowed the calculation of a relapse risk score. In addition to existing clinicopathological parameters, we showed that the assessment of hypoxia-related gene expression using simple real-time PCR assays in frozen breast tumor samples could improve the prediction of recurrence risk in breast cancer. Although this study has some limitations, such as its retrospective nature and the limited number of patients, our results provide additional clinical evidence that hypoxia-related gene expression has prognostic potential. Of course, it will be necessary to validate the clinical relevance of the risk score based on these 6 genes in independent studies including larger prospective patient cohorts. In addition, this risk score provides a prediction of relapse likelihood regardless of treatment type. Thus, it will be interesting to assess the potential value of the risk score of relapse following specific therapies.
Supporting information
(DOCX)
(DOCX)
Acknowledgments
We would like to gratefully acknowledge all staff members of the Biological Resource Center of Jean Perrin Comprehensive Cancer Center for their help in centralizing and managing biological collections.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported in part by grant from the Conseil Régional d’Auvergne (http://www.auvergne.fr). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. There was no additional external funding received for this study. The internal funding was provided by the Centre Jean Perrin (Clermont-Ferrand, France) where the majority of the authors of this article are employed.
References
- 1.Sestak I, Cuzick J. Markers for the identification of late breast cancer recurrence. Breast Cancer Res. 17:10 Epub 2015/04/08. doi: 10.1186/s13058-015-0516-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn SG, Lee HM, Cho SH, Bae SJ, Lee SA, Hwang SH, et al. The difference in prognostic factors between early recurrence and late recurrence in estrogen receptor-positive breast cancer: nodal stage differently impacts early and late recurrence. PLoS One. 8(5):e63510 Epub 2013/05/30. doi: 10.1371/journal.pone.0063510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farr A, Wuerstlein R, Heiduschka A, Singer CF, Harbeck N. Modern Risk Assessment for Individualizing Treatment Concepts in Early-stage Breast Cancer. Rev Obstet Gynecol. 6(3–4):165–73. Epub 2013/01/01. [PMC free article] [PubMed] [Google Scholar]
- 4.Magnon C, Opolon P, Ricard M, Connault E, Ardouin P, Galaup A, et al. Radiation and inhibition of angiogenesis by canstatin synergize to induce HIF-1alpha-mediated tumor apoptotic switch. J Clin Invest. 2007;117(7):1844–55. Epub 2007/06/09. PubMed Central PMCID: PMC1884687. doi: 10.1172/JCI30269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaupel P, Kelleher DK, Hockel M. Oxygen status of malignant tumors: pathogenesis of hypoxia and significance for tumor therapy. Semin Oncol. 2001;28(2 Suppl 8):29–35. Epub 2001/06/08. [DOI] [PubMed] [Google Scholar]
- 6.El Guerrab A, Zegrour R, Nemlin CC, Vigier F, Cayre A, Penault-Llorca F, et al. Differential impact of EGFR-targeted therapies on hypoxia responses: implications for treatment sensitivity in triple-negative metastatic breast cancer. PLoS One. 6(9):e25080 Epub 2011/10/04. doi: 10.1371/journal.pone.0025080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gassmann M, Kvietikova I, Rolfs A, Wenger RH. Oxygen- and dioxin-regulated gene expression in mouse hepatoma cells. Kidney Int. 1997;51(2):567–74. Epub 1997/02/01. [DOI] [PubMed] [Google Scholar]
- 8.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92(12):5510–4. Epub 1995/06/06. PubMed Central PMCID: PMC41725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salceda S, Beck I, Caro J. Absolute requirement of aryl hydrocarbon receptor nuclear translocator protein for gene activation by hypoxia. Arch Biochem Biophys. 1996;334(2):389–94. Epub 1996/10/15. doi: 10.1006/abbi.1996.0469 [DOI] [PubMed] [Google Scholar]
- 10.Zundel W, Schindler C, Haas-Kogan D, Koong A, Kaper F, Chen E, et al. Loss of PTEN facilitates HIF-1-mediated gene expression. Genes Dev. 2000;14(4):391–6. Epub 2000/02/26. PubMed Central PMCID: PMC316386. [PMC free article] [PubMed] [Google Scholar]
- 11.Semenza G. Signal transduction to hypoxia-inducible factor 1. Biochem Pharmacol. 2002;64(5–6):993–8. Epub 2002/09/06. [DOI] [PubMed] [Google Scholar]
- 12.Favaro E, Lord S, Harris AL, Buffa FM. Gene expression and hypoxia in breast cancer. Genome Med. 3(8):55 Epub 2011/08/31. doi: 10.1186/gm271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaupel P, Hockel M, Mayer A. Detection and characterization of tumor hypoxia using pO2 histography. Antioxid Redox Signal. 2007;9(8):1221–35. Epub 2007/06/01. doi: 10.1089/ars.2007.1628 [DOI] [PubMed] [Google Scholar]
- 14.Bos R, Zhong H, Hanrahan CF, Mommers EC, Semenza GL, Pinedo HM, et al. Levels of hypoxia-inducible factor-1 alpha during breast carcinogenesis. J Natl Cancer Inst. 2001;93(4):309–14. Epub 2001/02/22. [DOI] [PubMed] [Google Scholar]
- 15.Chia SK, Wykoff CC, Watson PH, Han C, Leek RD, Pastorek J, et al. Prognostic significance of a novel hypoxia-regulated marker, carbonic anhydrase IX, in invasive breast carcinoma. J Clin Oncol. 2001;19(16):3660–8. Epub 2001/08/16. doi: 10.1200/JCO.2001.19.16.3660 [DOI] [PubMed] [Google Scholar]
- 16.Wong C, Wellman TL, Lounsbury KM. VEGF and HIF-1alpha expression are increased in advanced stages of epithelial ovarian cancer. Gynecol Oncol. 2003;91(3):513–7. Epub 2003/12/17. [DOI] [PubMed] [Google Scholar]
- 17.Chitneni SK, Palmer GM, Zalutsky MR, Dewhirst MW. Molecular imaging of hypoxia. J Nucl Med. 52(2):165–8. Epub 2011/01/15. doi: 10.2967/jnumed.110.075663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robey IF, Lien AD, Welsh SJ, Baggett BK, Gillies RJ. Hypoxia-inducible factor-1alpha and the glycolytic phenotype in tumors. Neoplasia. 2005;7(4):324–30. Epub 2005/06/22. PubMed Central PMCID: PMC1501147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li XF, Du Y, Ma Y, Postel GC, Civelek AC. (18)F-fluorodeoxyglucose uptake and tumor hypoxia: revisit (18)f-fluorodeoxyglucose in oncology application. Transl Oncol. 7(2):240–7. Epub 2014/04/05. doi: 10.1016/j.tranon.2014.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winter SC, Buffa FM, Silva P, Miller C, Valentine HR, Turley H, et al. Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res. 2007;67(7):3441–9. Epub 2007/04/06. doi: 10.1158/0008-5472.CAN-06-3322 [DOI] [PubMed] [Google Scholar]
- 21.Seigneuric R, Starmans MH, Fung G, Krishnapuram B, Nuyten DS, van Erk A, et al. Impact of supervised gene signatures of early hypoxia on patient survival. Radiother Oncol. 2007;83(3):374–82. Epub 2007/05/29. doi: 10.1016/j.radonc.2007.05.002 [DOI] [PubMed] [Google Scholar]
- 22.Elston EW, Ellis IO. Method for grading breast cancer. J Clin Pathol. 1993;46(2):189–90. Epub 1993/02/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Doussal V, Tubiana-Hulin M, Friedman S, Hacene K, Spyratos F, Brunet M. Prognostic value of histologic grade nuclear components of Scarff-Bloom-Richardson (SBR). An improved score modification based on a multivariate analysis of 1262 invasive ductal breast carcinomas. Cancer. 1989;64(9):1914–21. Epub 1989/11/01. [DOI] [PubMed] [Google Scholar]
- 24.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–50. Epub 2004/08/04. doi: 10.1158/0008-5472.CAN-04-0496 [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. Epub 2002/02/16. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 26.Kwiatkowski F, Girard M, Hacene K, Berlie J. [Sem: a suitable statistical software adaptated for research in oncology]. Bull Cancer. 2000;87(10):715–21. Epub 2000/11/21. [PubMed] [Google Scholar]
- 27.Cronbach LJ, Warrington WG. Time-limit tests: estimating their reliability and degree of speeding. Psychometrika. 1951;16(2):167–88. Epub 1951/06/01. [DOI] [PubMed] [Google Scholar]
- 28.Unruh A, Ressel A, Mohamed HG, Johnson RS, Nadrowitz R, Richter E, et al. The hypoxia-inducible factor-1 alpha is a negative factor for tumor therapy. Oncogene. 2003;22(21):3213–20. Epub 2003/05/23. doi: 10.1038/sj.onc.1206385 [DOI] [PubMed] [Google Scholar]
- 29.Generali D, Berruti A, Brizzi MP, Campo L, Bonardi S, Wigfield S, et al. Hypoxia-inducible factor-1alpha expression predicts a poor response to primary chemoendocrine therapy and disease-free survival in primary human breast cancer. Clin Cancer Res. 2006;12(15):4562–8. Epub 2006/08/11. doi: 10.1158/1078-0432.CCR-05-2690 [DOI] [PubMed] [Google Scholar]
- 30.Vaupel P, Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev. 2007;26(2):225–39. Epub 2007/04/19. doi: 10.1007/s10555-007-9055-1 [DOI] [PubMed] [Google Scholar]
- 31.Semenza GL. HIF-1 and tumor progression: pathophysiology and therapeutics. Trends Mol Med. 2002;8(4 Suppl):S62–7. Epub 2002/04/03. [DOI] [PubMed] [Google Scholar]
- 32.Binley K, Askham Z, Martin L, Spearman H, Day D, Kingsman S, et al. Hypoxia-mediated tumour targeting. Gene Ther. 2003;10(7):540–9. Epub 2003/03/21. doi: 10.1038/sj.gt.3301944 [DOI] [PubMed] [Google Scholar]
- 33.Sotiriou C, Piccart MJ. Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nat Rev Cancer. 2007;7(7):545–53. Epub 2007/06/23. doi: 10.1038/nrc2173 [DOI] [PubMed] [Google Scholar]
- 34.Chi JT, Wang Z, Nuyten DS, Rodriguez EH, Schaner ME, Salim A, et al. Gene expression programs in response to hypoxia: cell type specificity and prognostic significance in human cancers. PLoS Med. 2006;3(3):e47 Epub 2006/01/19. PubMed Central PMCID: PMC1334226. doi: 10.1371/journal.pmed.0030047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rankin EB, Giaccia AJ. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 2008;15(4):678–85. Epub 2008/02/09. doi: 10.1038/cdd.2008.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai YP, Wu KJ. Hypoxia-regulated target genes implicated in tumor metastasis. J Biomed Sci. 19:102 Epub 2012/12/18. doi: 10.1186/1423-0127-19-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilkes DM, Semenza GL. Role of hypoxia-inducible factors in breast cancer metastasis. Future Oncol. 9(11):1623–36. Epub 2013/10/26. doi: 10.2217/fon.13.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rundqvist H, Johnson RS. Tumour oxygenation: implications for breast cancer prognosis. J Intern Med. 274(2):105–12. Epub 2013/07/13. doi: 10.1111/joim.12091 [DOI] [PubMed] [Google Scholar]
- 39.Bos R, van der Groep P, Greijer AE, Shvarts A, Meijer S, Pinedo HM, et al. Levels of hypoxia-inducible factor-1alpha independently predict prognosis in patients with lymph node negative breast carcinoma. Cancer. 2003;97(6):1573–81. Epub 2003/03/11. doi: 10.1002/cncr.11246 [DOI] [PubMed] [Google Scholar]
- 40.Campbell RA, Bhat-Nakshatri P, Patel NM, Constantinidou D, Ali S, Nakshatri H. Phosphatidylinositol 3-kinase/AKT-mediated activation of estrogen receptor alpha: a new model for anti-estrogen resistance. J Biol Chem. 2001;276(13):9817–24. Epub 2001/01/15. doi: 10.1074/jbc.M010840200 [DOI] [PubMed] [Google Scholar]
- 41.Mangia A, Chiriatti A, Bellizzi A, Malfettone A, Stea B, Zito FA, et al. Biological role of NHERF1 protein expression in breast cancer. Histopathology. 2009;55(5):600–8. Epub 2009/11/17. doi: 10.1111/j.1365-2559.2009.03424.x [DOI] [PubMed] [Google Scholar]
- 42.Shenolikar S, Voltz JW, Cunningham R, Weinman EJ. Regulation of ion transport by the NHERF family of PDZ proteins. Physiology (Bethesda). 2004;19:362–9. Epub 2004/11/18. [DOI] [PubMed] [Google Scholar]
- 43.Cardone RA, Bellizzi A, Busco G, Weinman EJ, Dell'Aquila ME, Casavola V, et al. The NHERF1 PDZ2 domain regulates PKA-RhoA-p38-mediated NHE1 activation and invasion in breast tumor cells. Mol Biol Cell. 2007;18(5):1768–80. Epub 2007/03/03. doi: 10.1091/mbc.E06-07-0617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Metzen E, Stiehl DP, Doege K, Marxsen JH, Hellwig-Burgel T, Jelkmann W. Regulation of the prolyl hydroxylase domain protein 2 (phd2/egln-1) gene: identification of a functional hypoxia-responsive element. Biochem J. 2005;387(Pt 3):711–7. Epub 2004/11/26. doi: 10.1042/BJ20041736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marxsen JH, Stengel P, Doege K, Heikkinen P, Jokilehto T, Wagner T, et al. Hypoxia-inducible factor-1 (HIF-1) promotes its degradation by induction of HIF-alpha-prolyl-4-hydroxylases. Biochem J. 2004;381(Pt 3):761–7. Epub 2004/04/24. PubMed Central PMCID: PMC1133886. doi: 10.1042/BJ20040620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Couvelard A, Deschamps L, Rebours V, Sauvanet A, Gatter K, Pezzella F, et al. Overexpression of the oxygen sensors PHD-1, PHD-2, PHD-3, and FIH Is associated with tumor aggressiveness in pancreatic endocrine tumors. Clin Cancer Res. 2008;14(20):6634–9. Epub 2008/10/18. doi: 10.1158/1078-0432.CCR-07-5258 [DOI] [PubMed] [Google Scholar]
- 47.Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8(9):705–13. Epub 2009/01/15. doi: 10.1038/nrc2468 [DOI] [PubMed] [Google Scholar]
- 48.Semenza GL, Jiang BH, Leung SW, Passantino R, Concordet JP, Maire P, et al. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271(51):32529–37. Epub 1996/12/20. [DOI] [PubMed] [Google Scholar]
- 49.Zhang D, Tai LK, Wong LL, Chiu LL, Sethi SK, Koay ES. Proteomic study reveals that proteins involved in metabolic and detoxification pathways are highly expressed in HER-2/neu-positive breast cancer. Mol Cell Proteomics. 2005;4(11):1686–96. Epub 2005/07/29. doi: 10.1074/mcp.M400221-MCP200 [DOI] [PubMed] [Google Scholar]
- 50.Sonveaux P, Vegran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest. 2008;118(12):3930–42. Epub 2008/11/27. doi: 10.1172/JCI36843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang ZY, Loo TY, Shen JG, Wang N, Wang DM, Yang DP, et al. LDH-A silencing suppresses breast cancer tumorigenicity through induction of oxidative stress mediated mitochondrial pathway apoptosis. Breast Cancer Res Treat. Epub 2011/04/01. [DOI] [PubMed] [Google Scholar]
- 52.Moritz KM, Lim GB, Wintour EM. Developmental regulation of erythropoietin and erythropoiesis. Am J Physiol. 1997;273(6 Pt 2):R1829–44. Epub 1998/01/22. [DOI] [PubMed] [Google Scholar]
- 53.Maxwell PH, Pugh CW, Ratcliffe PJ. Inducible operation of the erythropoietin 3' enhancer in multiple cell lines: evidence for a widespread oxygen-sensing mechanism. Proc Natl Acad Sci U S A. 1993;90(6):2423–7. Epub 1993/03/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ebert BL, Bunn HF. Regulation of the erythropoietin gene. Blood. 1999;94(6):1864–77. Epub 1999/09/09. [PubMed] [Google Scholar]
- 55.Acs G, Chen M, Xu X, Acs P, Verma A, Koch CJ. Autocrine erythropoietin signaling inhibits hypoxia-induced apoptosis in human breast carcinoma cells. Cancer Lett. 2004;214(2):243–51. Epub 2004/09/15. doi: 10.1016/j.canlet.2004.04.027 [DOI] [PubMed] [Google Scholar]
- 56.Wojchowski DM, Gregory RC, Miller CP, Pandit AK, Pircher TJ. Signal transduction in the erythropoietin receptor system. Exp Cell Res. 1999;253(1):143–56. Epub 1999/12/02. doi: 10.1006/excr.1999.4673 [DOI] [PubMed] [Google Scholar]
- 57.Miura Y, Miura O, Ihle JN, Aoki N. Activation of the mitogen-activated protein kinase pathway by the erythropoietin receptor. J Biol Chem. 1994;269(47):29962–9. Epub 1994/11/25. [PubMed] [Google Scholar]
- 58.Anagnostou A, Liu Z, Steiner M, Chin K, Lee ES, Kessimian N, et al. Erythropoietin receptor mRNA expression in human endothelial cells. Proc Natl Acad Sci U S A. 1994;91(9):3974–8. Epub 1994/04/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang K, Qiu S, Lu Y, Fan Z. Autocrine/paracrine erythropoietin regulates migration and invasion potential and the stemness of human breast cancer cells. Cancer Biol Ther. 2014;15(1):89–98. Epub 2013/10/09. PubMed Central PMCID: PMC3938527. doi: 10.4161/cbt.26717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oikawa M, Abe M, Kurosawa H, Hida W, Shirato K, Sato Y. Hypoxia induces transcription factor ETS-1 via the activity of hypoxia-inducible factor-1. Biochem Biophys Res Commun. 2001;289(1):39–43. Epub 2001/11/16. doi: 10.1006/bbrc.2001.5927 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.