Abstract
Copper ions play an important role in ethylene receptor biogenesis and proper function. The copper transporter RESPONSIVE-TO-ANTAGONIST1 (RAN1) is essential for copper ion transport in Arabidopsis thaliana. However it is still unclear how copper ions are delivered to RAN1 and how copper ions affect ethylene receptors. There is not a specific copper chelator which could be used to explore these questions. Here, by chemical genetics, we identified a novel small molecule, triplin, which could cause a triple response phenotype on dark-grown Arabidopsis seedlings through ethylene signaling pathway. ran1-1 and ran1-2 are hypersensitive to triplin. Adding copper ions in growth medium could partially restore the phenotype on plant caused by triplin. Mass spectrometry analysis showed that triplin could bind copper ion. Compared to the known chelators, triplin acts more specifically to copper ion and it suppresses the toxic effects of excess copper ions on plant root growth. We further showed that mutants of ANTIOXIDANT PROTEIN1 (ATX1) are hypersensitive to tiplin, but with less sensitivity comparing with the ones of ran1-1 and ran1-2. Our study provided genetic evidence for the first time that, copper ions necessary for ethylene receptor biogenesis and signaling are transported from ATX1 to RAN1. Considering that triplin could chelate copper ions in Arabidopsis, and copper ions are essential for plant and animal, we believe that, triplin not only could be useful for studying copper ion transport of plants, but also could be useful for copper metabolism study in animal and human.
Author summary
Copper ions are cofactors of protein functions, and their disorder is closely related to many human diseases which drive to develop new copper chelator related drugs. In plants, copper ions are essential for ethylene receptors, but many details are unclear. Researchers need novel specific copper chelators to study these questions. Here, by using plant chemical genetics, we identified a novel chemical triplin, which could activate ethylene signaling pathway by chelating copper ions essential for ethylene receptors. Ethylene resistance mutant etr1-1 and ein2 were resistant to triplin, and copper transporter mutants ran1-1 and ran1-2 were hypersensitive to triplin. Using triplin as a molecular genetics tool, we showed copper chaperone ATX1 acts the upstream of RAN1 which transports copper ions to ethylene receptors. We provide a sample that metabolism could be studied by combining chemical genetics and known signaling pathway. Moreover, triplin could chelate copper ions effectively and specifically in Arabidopsis, and could be a useful tool to study copper ion transport in plant, and valuable for designing copper chelator related drug.
Introduction
The phytohormone ethylene (C2H4) plays important roles in plant growth and development. When exposed to ethylene gas for 3 days, dark-grown Arabidopsis seedling shows a typical triple response phenotype including a short hypocotyl and root, larger diameter hypocotyl, and an exaggerated apical hook [1,2]. ETHYLENE RESPONSE1 (ETR1), ETHYLENE RESPONSE2 (ETR2), ETHYLENE RESPONSE SENSOR1 (ERS1), ETHYLENE RESPONSE SENSOR2 (ERS2) and ETHYLENE INSENSITIVE4 (EIN4) are five ethylene receptors in Arabidopsis which function redundantly and negatively to regulate ethylene signaling and responses. The receptors signal is transferred to a downstream Raf-like protein kinase CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) [3,4]. CTR1 interacts with and phosphorylates an endoplasmic reticulum (ER) membrane-localized Nramp homolog ETHYLENE IN SENSITIVE 2 (EIN2). This prevents EIN2 from activating downstream components of ethylene signaling including ETHYLENE INSENSITIVE3 (EIN3) and EIN3-LIKE1 (EIL1). When ethylene binds to the receptors, the phosphorylation of EIN2 by CTR1 is reduced, leading to accumulation of EIN2 and proteolytic cleavage of the cytosolic C-terminal domain of EIN2, which enters the nucleus to initiate ethylene signaling [5–10].
Copper ions are cofactors that are required for ethylene binding to ETR1. Ethylene insensitive mutant etr1-1 eliminates both ethylene binding and the interaction of copper ion with the receptor [11]. In Arabidopsis, the copper ions required by the ethylene receptors are transported by RAN1, also called HEAVY METAL ATPASE 7 (HMA7), which is a Cu-transporting P-type ATPase. Two weak mutant alleles, ran1-1 and ran1-2, were identified in a mutant screening where they responded to the ethylene receptor antagonist trans-cyclooctene (TCO) with a triple response phenotype which could be partially suppressed by adding copper ion to the plant growth medium. Additionally, ran1-1 and ran1-2 are hypersensitive and display a similar phenotype upon treatment with a low concentration of the copper ion chelator neocuproine. It has been shown that RAN1 is essential for the biogenesis of ethylene receptors in Arabidopsis [12–14].
A similar protein, Ccc2a, has been identified in Saccharomyces cerevisiae where it functions to transport copper ions from the copper chaperone, Atx1, to the secretory pathway. Atx1 is a small metal homeostasis factor that protects cells against reactive oxygen toxicity caused by excess or abnormal distribution of copper ions [15–18]. A homolog of Atx1 is found in humans. This metallochaperone, HUMAN ATX1-LIKE HOMOLOG1 (HAH1), also transports copper ions and interacts with Menkes and the Wilson disease proteins [19–21].
In Arabidopsis, there are two homologs of yeast Atx1, COPPER CHAPERONE (CCH) and ATX1 [22–24]. A T-DNA insertion mutant of ATX1 is specifically hypersensitive to excess copper ion as well as copper deficiency, while the over-expression of ATX1 enhances plant tolerance to excess copper ion and copper deficiency [25]. Previous yeast two-hybrid assays suggested that ATX1 and CCH△ (without C-terminal) interact with RAN1 and HMA5 [24,26]. It has been predicted that copper ions transported by RAN1 that are essential for ethylene receptors may come from ATX1 and CCH, but there is no genetic evidence to support this hypothesis [25,27].
To date, only the chelator neocuproine has been found to cause a plant triple response phenotype, but whether the phenotype could be suppressed by adding copper ions is unknown [14]. Neocuproine should be cautiously used as it may potentiate a cytotoxic effect of endogenous copper on cells [28]. Thus, there is no report about a specific copper ion chelator which causes a triple response phenotype specifically via a reduction in copper ions. We believe that a copper ion chelator with such a property would be useful for investigating the mechanisms of copper ion transport to the ethylene receptors. More ever, chemical genetics approaches are powerful in dealing with the function redundancy of components involved in phytohormone signaling, and have helped scientists to identify ABA receptors and novel signaling components involving in auxin and other phytohormones [29–32].
Here, we have used a chemical genetics approach to uncover a novel synthetic small molecule triplin. We found that triplin could cause a triple response phenotype in dark-grown Arabidopsis seedlings as a copper chelator. By testing the sensitivity of copper ion transport mutants to triplin, we showed that ATX1 acts upstream of RAN1. Our genetic and biochemical results support a model where ATX1 transports copper ions to RAN1 for ethylene receptor biogenesis and signaling. Triplin does not directly act on ethylene binding to receptor. Rather, it perturbs copper ion transport involved in the interaction of RAN1 and ATX1. In addition, triplin suppresses the toxic effects of excess copper ion on plant root growth. Thus, triplin may provide a useful chemical genetics tool to study copper ion transport in plants, and could be a lead structure for drug development for human copper disorders diseases in the future.
Results
Identification and characterization of triplin
To identify novel small molecules that affect ethylene signaling, we used the same plant chemical genetics screening method described previously [32–34]. Arabidopsis wild type Columbia-0 (Col-0) were grown and screened against a synthetic small molecule library with 12,000 compounds in the dark for 3 days. Those compounds that caused a triple response in seedlings were selected and their chemical genetics effects were retested. From this plant chemical screening, we acquired 14 compounds which cause a triple response in dark-grown Arabidopsis seedlings. Among these hits, the 5 strongest hits representing different structures were further assayed. Using the ethylene perception inhibitor, AgNO3, and the ethylene insensitive mutant ein2-5, we found that only a compound we called triplin (1-(1-morpholino-1-(thiophen-2-yl) propan-2-yl)-3-(2-(trifluoromethoxy) phenyl) thiourea) worked through the ethylene signaling pathway (Fig 1 and S1 Fig). We also examined the chemical genetic activities of 38 triplin analogs collected from the library. Our results showed 5 analogs could cause triple response phenotypes at 100 μM, and 11 analogs caused the phenotype at a higher concentration of 200 μM. The structure analysis of these active analogs indicates that the morpholino and thiourea moieties of the molecules could be important for enhancing their chemical genetics activities (S1 Table).
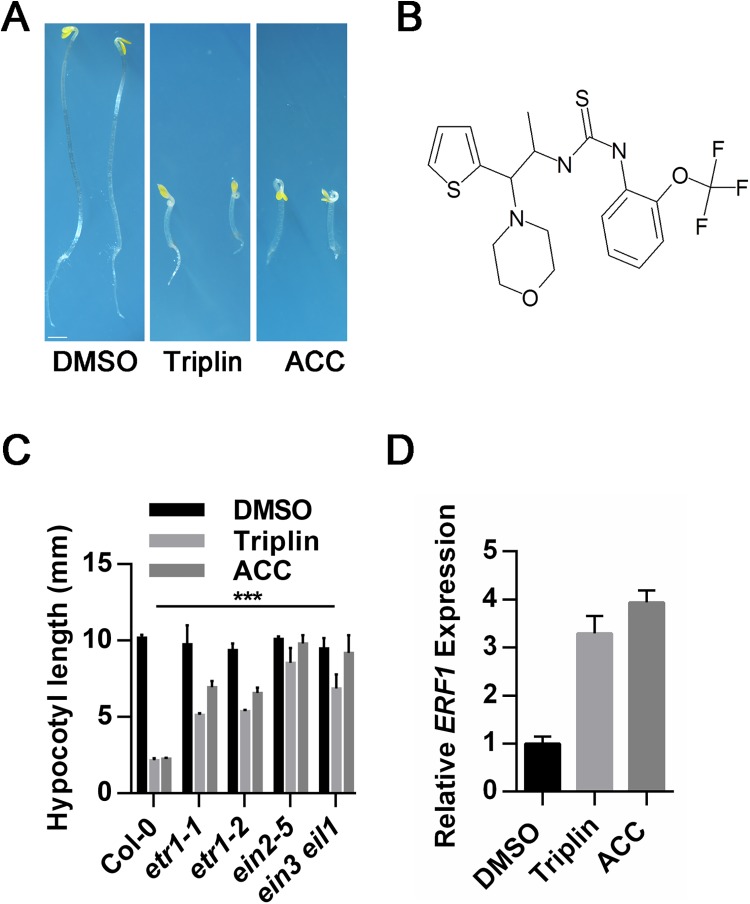
Fig 1. Triplin acts through ethylene signaling pathway to cause a triple response phenotype in Arabidopsis seedlings.
(A) Phenotypes of 3-day-old, dark-grown Col-0 seedlings treated with 100 μM triplin, 50 μM ACC, or 1% (v/v) DMSO as a control. Scale bar represents 1 mm. (B) The chemical structure of triplin. (C) The hypocotyl length of 3-day-old, dark-grown Col-0, etr1-1, etr1-2, ein2-5 and ein3 eil1 seedlings treated with 100 μM triplin, 50 μM ACC, or 1% (v/v) DMSO as a control. Each experiment was repeated three times, more than 30 seedlings were used every time. Error bars represent SEM. ***P < 0.0001 (two-tailed Student’s t-test) indicated a significant difference of the hypocotyl length of the mutants compared to Col-0 treated with 100 μM triplin. (D) qRT-PCR analysis the expression of the ethylene response gene ERF1 treated by 1% (v/v) DMSO, 100 μM triplin, or 50 μM ACC. Each experiment was repeated three times, and error bars represent SEM.
Triplin acts through the ethylene signal transduction pathway
Two strategies were employed to study the mechanism of action of triplin. First, we screened ethyl methanesulfonate (EMS) mutagenized populations of Col-0 for triplin resistant mutants. We acquired nine triplin resistant mutants which also showed resistance to the application of the ethylene biosynthesis precursor, 1-aminocylopropane-1-carboxylic acid (ACC). Among these resistant mutants, two are dominant mutants that have the same mutation as etr1-1 as revealed by DNA sequencing [1]. The other seven mutants are recessive and were genetically determined to be on the EIN2 locus by examining F1s acquired from crossings with ein2 (S2 Fig). We further assayed the effects of triplin treatment on known ethylene signaling mutants. Ethylene resistant mutant etr1-1, etr1-2, ein2-5 and ein3 eil1 showed resistance to triplin (Fig 1C and S3 Fig). Additionally, adding 500 μM AgNO3 blocked triplin’s effects on seedlings (S6C Fig). Consistent with triplin affecting ethylene signaling, the ethylene-responsive gene, ERF1, was up-regulated by triplin treatment (Fig 1D). These results indicated that triplin acts on ethylene signaling. It is possible that triplin also increases ethylene biosynthesis to cause a triple response. To determine if this was occurring, we first examined how alterations in ethylene biosynthesis affect responses to triplin. We found that triplin does not increase ACSs gene expression levels. Neither the ACC synthase (ACS) inhibitor, aminoethoxyvinylglycine (AVG) nor the ethylene biosynthesis mutant cin5 affects triplin responses. Additionally, triplin treatment did not enhance ethylene biosynthesis and actually causes a decrease in ethylene production (S4 Fig). Together, these results indicate that triplin treatments cause triple responses by acting on ethylene signaling but not ethylene biosynthesis.
Triplin genetically acts upstream of ethylene receptors in the ethylene signaling network
To map where triplin acts on the ethylene signaling network, we assayed how triplin treatments affect other ethylene insensitive mutants of ethylene receptors including ers1-1, ers2-1, etr2-1 and ein4. Our results showed they were all resistant to triplin (S3B Fig), indicating that triplin likely acts on or upstream of the ethylene receptors in the ethylene signaling network. Previous research has shown that the copper ion transporter RAN1 is involved in the biogenesis of ethylene receptors and certain ion chelators such as neocuproine can trigger a plant triple response [14]. As we expected, ran1-1 and ran1-2 mutants were hypersensitive to triplin treatments comparable with the published results for neocuproine [14] (S10B and S10C Fig). Low doses of triplin (e.g. less than 10 μM) had no visible effects on wild type but caused a triple response in ran1-1 and ran1-2 (Fig 2A). Further, introducing RAN1 into the mutants rescued a wild type response to triplin (Fig 2B). However, ran1-2 etr1-1 and ran1-2 ein2-5 double mutants were both resistant to triplin treatments similar to the etr1-1 and ein2-5 single mutants (Fig 2C). Together, our chemical genetics results showed that triplin is likely to act upstream of the ethylene receptors and may be involved in altering copper ion transport to the receptors. Our simple postulation is that triplin is a copper ion chelator which causes the triple response phenotype by chelating the copper ions necessary for ethylene receptor biogenesis.
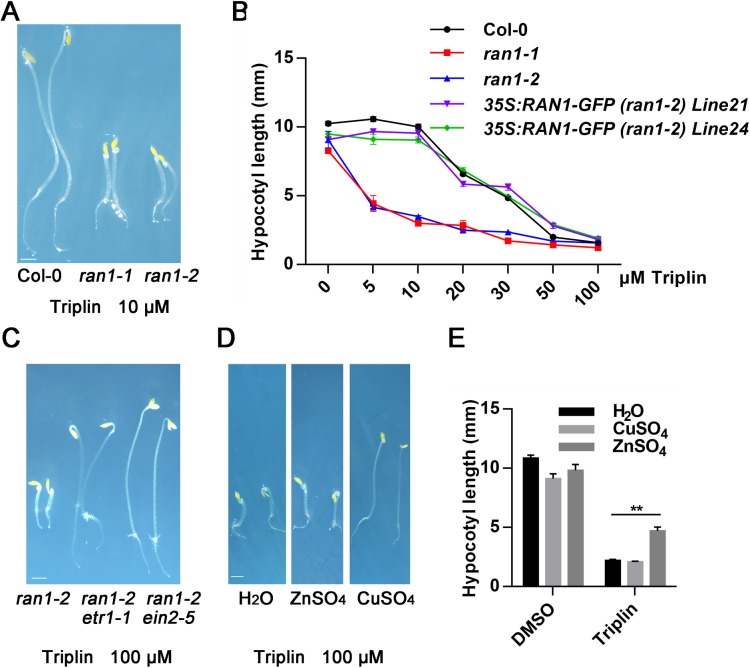
Fig 2. ran1-1 and ran1-2 are hypersensitive to triplin and copper can partially reverse the effects of triplin.
(A) Phenotypes of 3-day-old, dark-grown seedlings of Col-0, ran1-1 and ran1-2 treated with 10 μM triplin. (B) Triplin dose responses of Col-0, ran1-1, ran1-2 and two 35S:RAN1-GFP (ran1-2) transgenic lines. Data is the average hypocotyl length under each condition. Each experiment was repeated three times, more than 30 seedlings were used every time. Error bars represent SEM. (C) Phenotypes of 3-day-old, dark-grown seedlings of ran1-2, ran1-2 etr1-1 and ran1-2 ein2-5 treated with 100 μM triplin. (D) The phenotypes of 3-day-old, dark-grown seedlings of Col-0 treated with 100 μM triplin in the presence of 20 μM ZnSO4, 20 μM CuSO4 or H2O as a control. (E) The hypocotyl length of seedlings as described in (D). Each experiment was repeated three times, more than 30 seedlings were used every time. Error bars represent SEM. **p < 0.01 indicated the difference of the hypocotyl length between the seedlings treated with CuSO4 compared with H2O and ZnSO4 in the presence of 100 μM triplin. Scale bars represent 1 mm.
Triplin chelates copper ions
To determine if triplin acts as a copper ion chelator and its specificity, we examined the effects of copper ions and other ions such as Ag, Zn, Ca, Mg, Mn, Co, Ni, Na, K, Li and Mo on triplin responses in plants. Our results showed that addition of excess Cu2+ (CuSO4) partially reverses the effects of triplin on plants (Fig 2D and 2E). Of the other ions tested, only Ag+ (AgNO3) had a similar effect (S5 Fig and S6C Fig). We also examined the known copper ion chelator neocuproine and found that its effects were reversed by adding Zn2+ (ZnSO4) under our assay conditions (S10A Fig). To further verify that triplin acts by chelating copper ions, we tested whether triplin treatment can alleviate the toxic effects of high levels of copper ions on plant growth. Our results indicated that the growth of Arabidopsis seedlings roots treated with 50 μM CuSO4 was strongly inhibited; addition of 100 μM triplin at the same time partially reversed this inhibition of root growth (Fig 3A and 3B). Higher concentration of CuSO4 causes more severe growth inhibition which can also be partially reversed by adding triplin (S6A and S6B Fig). We next tested if neocuproine has a similar effect as triplin on plant grown with high levels of copper ions. To our surprise, neocuproine aggravated the copper ion toxic effects on plant growth. Another thing is that ZnSO4 could restore the phenotype caused by neocuproine. This indicated that the specificity of neocuproine is questionable (S10A and S10C Fig).
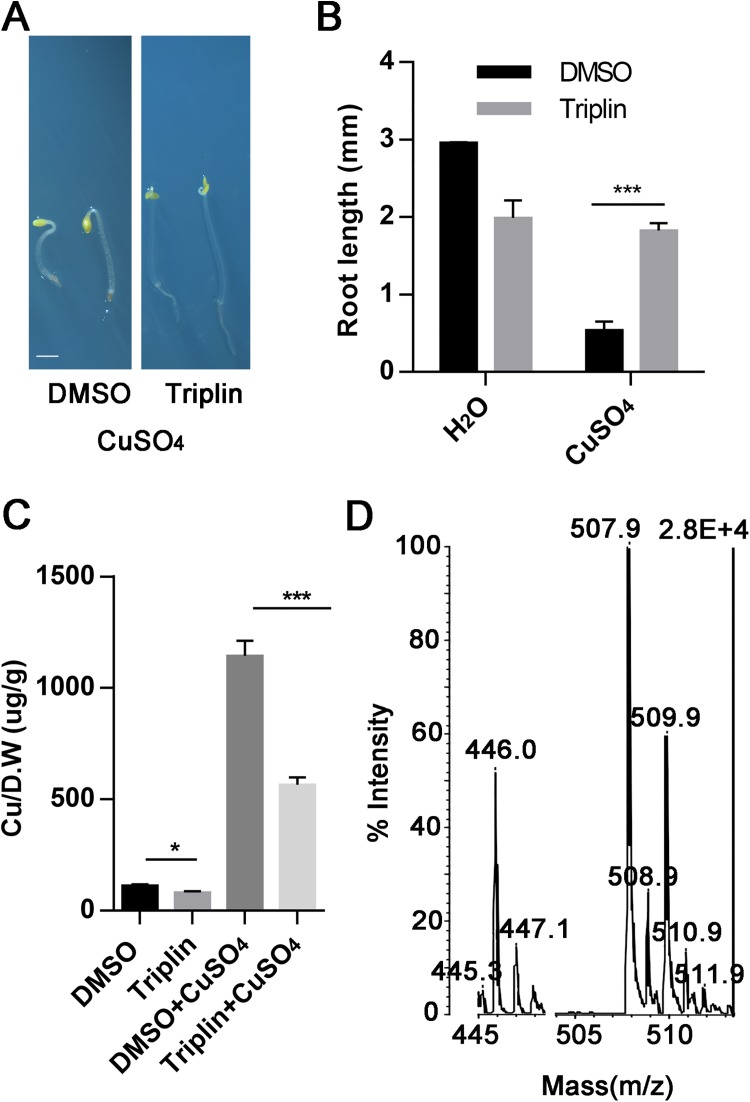
Fig 3. Triplin can chelate copper ions in vitro.
(A) The phenotypes of 3-day-old, dark-grown seedlings of Col-0 treated with 50 μM CuSO4 in the presence of 1% (v/v) DMSO or 100 μM triplin. Scale bar represents 1 mm. (B) The root length of the seedlings as described in (A). Each experiment was repeated three times, more than 30 seedlings were used every time. Error bars represent SEM. (C) The relative copper contents of 3-day-old, dark-grown Col-0 seedlings on 0.5xMS growth medium with or without 100 μM triplin and/or 20 μM CuSO4. D.W represents dry weight of the seedlings. Each experiment was repeated three times (Error bars represent SEM). (D) MALDI-TOF-MS analysis of the mixture of equal volume of 100 μM CuSO4 and 100 μM triplin. Mr of triplin is 446.0g/mol. Mr of triplin+Cu is 510.0g/mol. *P < 0.05 and ***P < 0.0001 (two-tailed Student’s t-test) indicate a significant difference between groups of different treatments.
To better understand the effects of triplin on plants, we measured the relative copper ion content of seedlings grown with copper ions and triplin. When seedlings were grown on a normal 0.5xMS growth medium, adding 100 μM triplin to the medium slightly decreased the copper ion content in the seedlings from 110 μg/g to 80 μg/g. When seedlings were grown on a growth medium containing 20 μM CuSO4, the relative copper ion content in the seedlings increased from 110 μg/g to 1,100 μg/g. Adding 100 μM triplin reduced the ion content from 1,100 μg/g to 560 μg/g (Fig 3C). We tried to mix triplin with different metal ions such as Ag, Cu, Zn and Ca. Unexpectedly, we found that triplin formed a tawny turbidness with CuSO4, and a black precipitate with AgNO3 (S7 Fig). We analyzed the products formed from mixing 100 μM CuSO4 with 100 μM triplin by Matrix-Assisted Laser Desorption/ Ionization Time of Flight Mass Spectrometry (MALDI-TOF-MS) analysis. With this we observed a peak in the m/z of 509.9, which is equal to the sum of molecular weights of triplin (446.0 g/mol) and copper (Fig 3D). Silver ions can also conjugate with triplin, but a higher concentration (e.g. 10 mM) of AgNO3 is needed. Conjugates of triplin with other metal ions including ZnSO4 and FeSO4 were not detected with our mass spectrographic analysis (S8 Fig). Together, our results indicate that triplin is a copper chelator which can also chelate silver to a less degree. This possibly explains why both Cu2+ and Ag+ can block or partially block triplin’s effects on plant growth.
Triplin causes the triple response phenotype by affecting copper ion transport in ethylene signaling
In order to examine how triplin acts as a copper ion chelator to perturb ethylene signaling and cause the triple response phenotype, we first used two copper ion deficient growth medium to grow plants. One included all essential elements for plant growth except copper ions were not added. The other consisted of 0.5xMS growth medium supplemented with 500 μM of the copper ion chelator, bathocuproinedisulfonic acid (BCS). The effects of 20 μM or 50 μM triplin on plant growth in dark were compared on these medium to control medium. The seedlings grown on both copper-deficient growth medium showed more severe triple responses in response to 20 μM triplin than seedlings grown on control medium (Fig 4A and 4B). Under these same growth conditions with different triplin concentration, ein2-5 showed obvious resistance to triplin (S9 Fig). This indicates that the exaggerated triple response in the absence of added copper is also dependent on ethylene signaling.
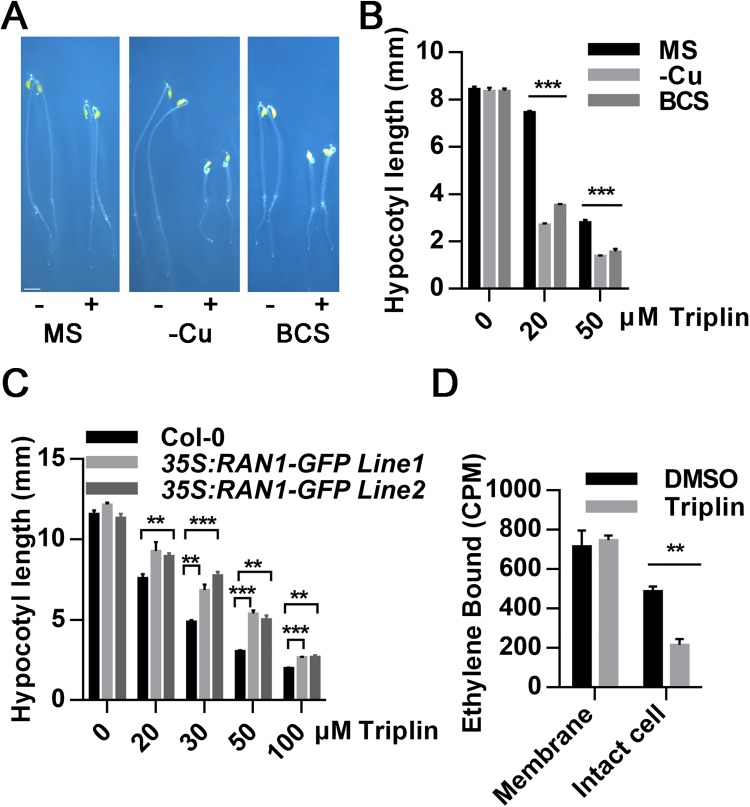
Fig 4. Triplin can affect the transport of copper ions in vivo.
(A) The phenotypes of 3-day-old, dark-grown seedlings of Col-0 on different growth medium with (+) or without (-) 20 μM triplin. -Cu represents the growth medium made of all essential elements needed for plant growth except copper ion. BCS represents the growth medium made of 0.5xMS salt with 500 μM of the copper ion chelator BCS. Scale bar represents 1 mm. (B) The hypocotyl length of 3-day-old seedlings as described in (A). The difference of the hypocotyl lengths represent either the seedlings grown on -Cu or BCS medium compared to the ones grown on 0.5xMS. (C) The hypocotyl length of 3-day-old seedlings of Col-0 and 35S: RAN1-GFP lines grown in dark under different dose of triplin. (D) Triplin’s effects on ethylene-binding to ETR1 expressed in yeast. Saturable ethylene binding to intact yeast cells expressing the ethylene binding domain of ETR1 and membranes isolated from these yeast cells was measured. Ethylene binding is indicated as counts per minute (CPM). Each experiment was repeated three times, and error bars represent SEM. In (B) and (C), experiments were repeated three times, more than 30 seedlings were used every time. Error bars represent SEM. *P < 0.05, **P < 0.01, ***P < 0.0001 (two-tailed Student’s t-test) indicate a significant difference between groups of different treatments.
These results are consistent with our earlier results indicating that triplin can chelate copper ions. However, we also noticed that simply lowering copper levels in the growth medium is not enough to cause a triple response. This led us to speculate that there may be additional actions of triplin on plants to cause the triple response. One possibility we considered was that triplin enters into plant cells to affect copper ion transport. To examine this possibility, we used RAN1 overexpression transgenic lines in the Col-0 background to test their sensitivity to triplin treatment. Our results showed that the overexpression lines are resistant to triplin treatments (Fig 4C and S14 Fig). One possible explanation is that triplin acts as copper ion chelator that competes for copper with the ethylene receptors resulting in less ethylene binding to the receptors. To examine this possibility, we measured ethylene binding to membranes isolated from yeast expressing the ethylene binding domain of ETR1 in the presence and absence of triplin [14]. Triplin had no measurable effect on specific ethylene binding to the receptors (Fig 4D). By contrast, ethylene binding to the receptors was reduced approximately 50% when triplin was added to the growing yeast cells expressing ETR1 (Fig 4D). These results indicate that triplin is not directly affecting ethylene binding but may affect the delivery of copper ions to the receptors by affecting other copper delivery proteins. For example, triplin treatments may reduce the copper ion levels below the optimal levels needed for some copper ion chaperones. Lack of copper ions would result in a lower number of functional receptors leading to the triple response. If this is true, we should find some copper ion transport mutants that are either hypersensitive or resistant to triplin treatment.
ATX1 acts on copper ion transport to ethylene receptors
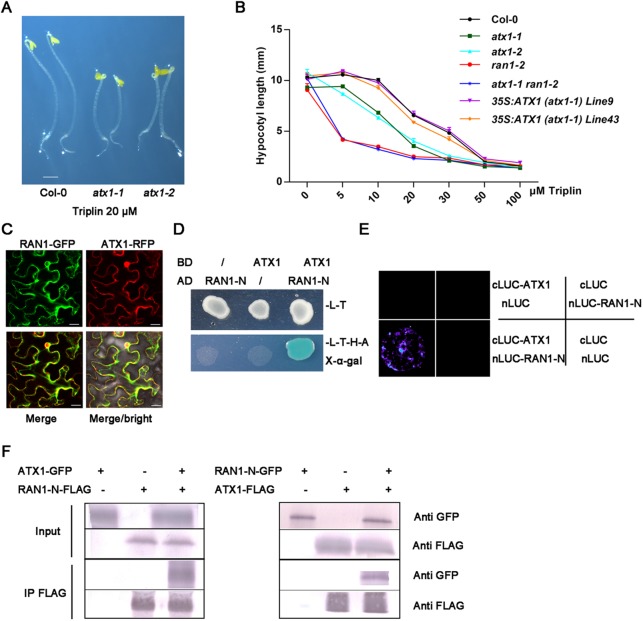
Previous studies showed RAN1 is a key copper ion transporter involved in ethylene receptor biogenesis [12–14]. However the mechanisms for how copper ions are delivered to RAN1 and how RAN1 delivers copper to the ethylene receptors are presently missing. Therefore, we took advantage of triplin to look for the protein(s) that deliver copper to RAN1. We speculated that mutations in such proteins should be hypersensitive to triplin treatment. We therefore tested triplin sensitivity of T-DNA mutants of Arabidopsis that affect copper ion transport, including copt1, copt2, copt4, copt5, hma1, hma5, hma6, ccs, zip2, zip4, cch and atx1. This uncovered that only the copper ion chaperone mutants atx1-1 and atx1-2 are hypersensitive to triplin (Fig 5A and 5B). ATX1 is a copper ion chaperone playing an essential role in copper ion homeostasis that confers plant tolerance to both copper excess and deficiency conditions in Arabidopsis [25]. atx1-1 (SALK_026221) is a knock-out mutant identified previously [25], atx1-2 (SALK_041022) is another knock-out mutant identified in this research (S11 Fig). Both mutants showed hypersensitivity to triplin treatments. Application of 20 μM triplin to atx1-1 and atx1-2 seedlings caused a triple response as seen with the exaggerated apical hook and shorter hypocotyl. By contrast, 20 μM triplin did not cause a triple response phenotype in wild type seedlings. Re-introducing the ATX1 gene into the atx1-1 mutants restored a wild type response to triplin (Fig 5B). The hypersensitivity of atx1-1 to triplin could be partially reduced by adding copper. Furthermore, atx1-2 etr1-1 and atx1-1 ein2-5 double mutants were resistant to triplin treatment comparable with the etr1-1 and ein2-5 single mutants (S11C Fig). This indicates that triplin is acting to result in lower levels of copper delivered to RAN1, which in turn, reduces deliver of copper to the ethylene receptors.
Fig 5. The copper chaperone ATX1 interacts with RAN1.
(A) atx1-1 and atx1-2 are hypersensitive to triplin. The phenotypes of 3-day-old, dark-grown Col-0, atx1-1 and atx1-2 seedlings treated with 20 μM triplin are shown. Scale bar represents 1 mm. (B) Hypocotyl lengths of 3-day-old, dark-grown seedlings of Col-0, atx1-1, atx1-2, ran1-2, atx1-1 ran1-2 and two 35S:ATX1-GFP (atx1-1) transgenic lines treated with different doses of triplin are shown. Each experiment was repeated three times, more than 30 seedlings were used every time. Error bars represent SEM. (C) Subcellular co-localization of ATX1 and RAN1. ATX1-RFP and RAN1-GFP were transiently expressed in N. benthamiana leaves and observed and imaged under a confocal microscope. Scale bars represent 20 μM. (D) ATX1 interacted with RAN1 in yeast two-hybrid assay. ATX1 was fused to a GAL4 DNA-binding domain (BD) and RAN1-N (289 amino-terminal amino acids of RAN1) was ligated to a GAL4 activation domain (AD). The protein interactions were examined on cells grown on synthetic dropout (-Leu/-Trp/-His/-Ade) medium plus X-α-Gal (50mg/L) plates for 3 days. (E) Bimolecular fluorescence complementation assays showed interaction between ATX1 and RAN1 using the split luciferase system. Nicotiana benthamiana leaves were infiltrated with agrobacteria containing different construct combinations harboring both the C- and N-terminal of the luciferase fused to either ATX1 and RAN1-N or just one of them (controls). (F) Co-Immunoprecipitation assays showed interaction between ATX1 and RAN1. Nicotiana benthamiana leaves were infiltrated with agrobacteria containing ATX1-GFP/FLAG and RAN1-N-FLAG/GFP or just one of them (controls). The protein extracts were immunoblotted with anti-FLAG antibody or anti-GFP antibody.
ATX1 physically interacts with RAN1
Previous work showed that the ethylene receptors are localized at ER membrane network [35]. To further examine how ATX1 is involved in RAN1 mediated copper ion transport to ethylene receptors, we compared the subcellular localization of ATX1 and RAN1. First, we used 5-day-old light grown 35S:ATX1-GFP (atx1-1) transgenic lines and observed filar and cloudy GFP signals in the cytoplasm and around the nucleus of the root meristem zone cells. In root elongation zone cells, stronger GFP signal were observed in the cytoplasm and nucleus. Follow up observations using 4', 6-diamidino-2-phenylindole (DAPI) stain confirmed the nuclear signals observed in 35S:ATX1-GFP (atx1-1) transgenic lines [24, 25] (S12A and S12B Fig). For the transgenic lines grown in dark, we observed strong GFP signals in the cytoplasm and nucleus of the hypocotyl cells comparable with what was observed in root elongation zone cells (S12C Fig). We speculate that, in mature cells, the expression and signal of ATX1-GFP is stronger than in younger cells. We then carried out plasmolysis experiments and showed that these GFP signals observed in intact cells are not localized at the cell wall. We next scanned a thin layer of a cell and observed similar filar and cloudy GFP signals, which may be from ATX1-GFP adhered to the cell endomembrane system (S12D Fig). Using a transgenic line expressing both ATX1-GFP and WAK2-mCherry, an ER marker, we observed the ATX1-GFP signals were partially co-localized with the ER markers (S12E Fig).
To confirm this observation, we also used a transient expression system to express ATX1-GFP and WAK2-mCherry in tobacco (Nicotiana benthamiana) leaf epidermal cells to observe the sub-cellular location of ATX1. The observations in these experiments are consistent with our previous observations that ATX1 is localized to the cytoplasm and nucleus and may adhere to ER membranes (S13A Fig). Using this transient expression system, we observed that GmMAN1-mcherry (a Golgi marker) displayed a marked punctate signal and OsREM4.1-mCherry (a plasma membrane marker) showed no filar signals, which are different from the ATX1-GFP pattern. This confirmed that ATX1 did not specifically localize to the plasma membrane and Golgi. Only WAK2-mCherry showed filar signal (S13A Fig). We then examined the subcellular location of RAN1 using the transient expression system to express RAN1-GFP and WAK2-mCherry and we observed that RAN1-GFP was localized to the endomembrane and also was co-localized with the ER marker (S13B Fig). Further we wondered whether ATX1 and RAN1 are co-localized in a cell. To address this question, we used the transient expression system to express RAN1-GFP and ATX1-RFP, and we observed both proteins were co-localized on the ER membrane (Fig 5C). Together, our sub-cellular observations indicate both ATX1 and RAN1 can adhere to ER membranes.
To explore the possibility of ATX1 and RAN1 interacting in planta, we first carried out yeast two-hybrid experiments. ATX1 was fused to a GAL4 DNA-binding domain (BD) and RAN1-N (289 amino-terminal amino acids of RAN1) was ligated to a GAL4 activation domain (AD). Three days after these co-transformed yeast cells were grown on a synthetic dropout (-Leu/-Trp/-His/-Ade) medium with X-α-Gal (50 mg/L), we observed clones became blue, suggesting an interaction between ATX1 and RAN1-N in yeast cells (Fig 5D). This was next confirmed by using bimolecular fluorescence complementation (BiFC) assays in Nicotiana benthamiana leaves. For this, 35S: cLUC-ATX1 and 35S: nLUC-RAN-N constructs were made and co-transformed into the plants, and the results showed that these proteins interact with each other (Fig 5E). The co-immunoprecipitation (Co-IP) assays using ATX1-GFP/FLAG and RAN1-N-FLAG/GFP also showed that ATX1 interacts with RAN1 (Fig 5F). These results indicate that ATX1 physically interacts with RAN1 in planta. Together, the above data supports evidences for the hypothesis that the copper ions required for ethylene receptor biogenesis and signaling are transported through ATX1 to RAN1 and finally to ethylene receptors.
Discussion
The functional characterization of genes using genetic approaches in plants depends on observable phenotypes when the gene being studied is mutated. For ethylene signaling, this mutational approach has long been saturated. It is important and pressing to overcome this problem. Recently developed chemical genetics approaches using specific active small molecules provide reversible, time and strength controllable genetic perturbations for genetic research and gene function characterization [36]. In this research, we applied a high-throughput plant chemical genetics screening and uncovered triplin, a novel small molecule, which cause a triple response in dark-grown Arabidopsis seedlings. Further we demonstrated triplin is a novel copper ion chelator and it alleviates the toxic effects of high copper ion levels on root growth. Additionally, the triple response phenotype resulting from triplin treatment is reduced by adding Cu2+ and Ag+. This is very different from another ion chelator neocuproine. Although neocuproine causes a triple response phenotype, this effect is suppressed by Zn2+ but not Cu2+. There is some white precipitate when we mixed Zn2+ and neocuproine (S7 Fig). While low concentration of copper ions could partially restore the hypersensitivity of ran1-2 to neocuproine (S10B Fig). Another problem with neocuproine is that adding it to plant growth medium exaggerates the toxic effects of high levels of copper ions on plant root growth (S10C Fig). So we speculate that neocuproine could cause triple response phenotype by chelating copper ions like triplin, but its specificity and safety are questionable. Other copper ion chelators such as BCS do not elicit the triple response phenotype. We believe that triplin not only chelates copper ions in the plant growth medium, but also chelates copper ions in plant cells once it enters into plant cells. We predict that this restricts the access of copper ions needed for ethylene receptor biogenesis, and this condition lead to the triple response phenotype. Our study shows that triplin is different from other characterized copper chelators and it has the potential to perturb copper ion transport and to dissect the ethylene signaling network.
We still know little about the chemistry of triplin. One thing to note is triplin does not affect ethylene binding to ETR1 directly (Fig 4D). This may be related to its limited solubility in water and we noticed its precipitation was enhanced by adding CuSO4 or AgNO3 (S11C Fig). One possibility is its low solubility inhibits triplin to affect ethylene binding to ETR1 directly. However triplin could enter growing yeast cells expressing ETR1 when it was added to the yeast growing medium in which triplin decreased the copper level of yeast cells and affected ETR1 biosynthesis. This possibly resulted in producing abnormal ethylene receptors which could not bind ethylene properly (Fig 4D). It is important to characterize triplin in term of its chemistry in future.
Copper ion transport from the copper transporter RAN1 to the ethylene receptors for normal receptor biogenesis and function has been proposed, but many important details in this process are still missing [12–14]. For example, what component provides copper ion to RAN1 and where does this copper ion relay occur? These are some important questions we need to address in copper ion transport coupled ethylene receptor biogenesis and functioning. Our results examining the effects of triplin on known ethylene-related mutants indicates that triplin acts upstream of the receptors but does not affect ethylene biosynthesis. For example, etr1-1 and ein2 are both resistant to triplin. Furthermore, a triplin hypersensitive mutant screening identified both ran1 and atx1 mutants which are posited to function upstream of the receptors to deliver copper for normal receptor biogenesis. The hypersensitivity of both the ran1 and axt1 mutants was abolished by the mutations in downstream ethylene signaling network components such as etr1-1 (Fig 2C and S11C Fig). We have noticed that atx1-1 and atx1-2 are less sensitive to triplin than ran1-1 and ran1-2 (Fig 5B), and the similar results were observed when treated with neocuproine (S10D Fig). These results suggest ATX1 may be positioned in the network prior to RAN1. Our ethylene binding assays indicate that triplin is not directly affecting the ethylene receptors, but appears to be affecting delivery of copper ions to the receptors. This supports the hypothesis that triplin targets copper ion transport upstream of the receptors.
Although RAN1 has been proposed to play a key role in copper ion transport in ethylene receptor biogenesis and signaling, the subcellular location of RAN1 has not been determined [12, 27]. The function of the well-known copper ion chaperone ATX1 is difficult to study by using conventional genetics approaches, since mutants such as atx1-1 do not show a phenotype related to ethylene [25]. By taking advantage of triplin, ethylene signaling mutants and manipulating the concentration of copper ions in the plant growth medium, we provided genetic evidence for the first time of the interaction between RAN1 and ATX1, which contributes to transport copper ions to the ethylene receptors. ATX1 localizes widely in cells, indicating that it likely functions to transport copper ions to other targets in addition to RAN1. This is consistent with the report that ATX1 is essential for copper homeostasis. For example, it may act as a copper-buffer in plant cells [24, 25]. The other copper chaperone in Arabidopsis is CCH, but cch mutants have no ethylene related phenotypes. Additionally, we found that cch mutants had wild type responses to triplin suggesting that CCH is not important for delivery of copper ions to RAN1 and eventually, the ethylene receptors. The subcellular localization of RAN1 was not identified previously, but based on the phenotypes of its mutants it seems likely that RAN1 is active in an endomembrane system compartment, perhaps the ER [12, 27]. Our microscopic observations using ATX1, RAN1, and organelle markers, as well as the results of molecule interaction assays in planta indicate that RAN1 and ATX1 are partially co-localized and interact on the ER. This supports the idea that ATX1 and RAN1 interact as part of copper transport to the ethylene receptors.
Copper ions are important to human health where its disorder can result in various diseases. More and more reports show that copper disorders are closely related to human Alzheimer's disease and cancers. Therefore various copper chelators have been identified and characterized in order to cure these important human diseases [37–40]. Although a few copper ion chelators have been used in therapy, thus far their mechanisms of action are not clear [41]. One problem using these chelators is they are not very specific for copper ions [42], and some chelators, such as neocuproine, might enhance the toxic effects of copper ions on the organism being treated as we show in this study for plant growth. To this end, triplin is a unique copper ion chelator that may provide a new lead structure for copper ion chelators related to developing drug. Thus, the model plant Arabidopsis could be another useful platform to carry out studies to uncover copper ion chelator and explore its action mechanism in vivo.
Materials and methods
Plant chemical genetics screenings
The plant chemical genetics screenings using dark-grown Arabidopsis seedlings against a structure novel and diverse synthetic chemical library of 12,000 small molecules from Life Chemicals Inc was performed as previously reported [32, 33]. For all screenings, surface-sterilized Arabidopsis Col-0 seeds suspended in 0.1% agar were evenly distributed into 96-well plates that contained 0.8% agar, 0.3xMS salts(Sigma-Aldrich), 100 μM individual chemical per well and 1% DMSO (carrier solvent). Seeds were stratified for 3 days in a 4 C refrigerator, transferred to day-light for 1–4 hours then transferred in dark to grow for 3 days at 22 C in a light-tight growth cabinet. Seedlings phenotypes were recorded and imaged using a SZX16 dissecting microscope. Chemicals that caused dark-grown Col-0 seedlings triple response like phenotypes were retested. For each active chemical, its dose effect on the length of the hypocotyls or roots of assayed seedlings were measured using Image J (NIH).
Isolation and characterization of triplin resistant mutants
To acquire triplin resistant mutant, we used a mutant screening strategy that is modified from [30]. Briefly 20,000 M2 seeds from 5,000 ethylmethane sulfonate (EMS)-mutagenized M1 Col-0 plants were surface-sterilized and grown under the same growth conditions and chemical dose as the plant chemical genetics screenings. The phenotypes of seedlings were examined under the microscope, and these with longer hypocotyl or without exaggerative apical hooks were considered as putative triplin resistant mutants, and all these mutants were then retested for their chemical genetics phenotypes in the next generation.
Plant ethylene measurement
Ethylene measurements on plants were performed as previously described [43, 44]. Col-0 seeds were surface sterilized and planted on 0.5xMS solid medium. After stratification at 4 C for 3 days, plates were exposed to light for 1–4 hours and then transferred in dark to grow for 2 days at 22 C. After seed germination, every 22 seedlings per group (n = 3) were transferred to a gas chromatography vial with half volume 0.5xMS growth medium contain 100 μM triplin, 50 μM ACC or 1%DMSO and incubated for 3 days under continual dark. Accumulated ethylene was measured by gas chromatography (Agilent Technologies, 6890N Network GC System) [44]. Seedlings weight is fresh weight.
Plant copper ion content assay
Three-day-old etiolated Col-0 seedlings grown on 0.5xMS solid medium with 20 μM CuSO4 and/or 100 μM triplin were collected. The copper ion contents in plants were then determined by inductively coupled plasma-mass spectrometry as previously described [45].
MALDI-TOF-MS analysis
For these assays, 100 μl of the metal ions (100 μM or 10 mM) and 100 μl triplin (100 μM or10 mM) were mixed for MALDI-TOF-MS analysis according to [46]. A time-of–flight Axima Performance mass spectrometer (Shimadzu, Japan) was used. All mass spectra in this work were acquired at positive ion reflection mode. The data were controlled by a software application of MALDI-MS. The matrix is 5 mg/ml α-Cyano-4-hydroxycinnamic acid (CHCA). Mass spectrum scanning range is 400-1000Da.
Ethylene binding assays
For this, we expressed the first 128 amino acids of ETR1 containing the ethylene binding domain as a fusion protein to GST (glutathione S-transferase) (ETR1 [1–128]-GST) in Pichia pastoris as previously described [47]. Binding assays were then conducted on either intact yeast cells or isolated membranes as previously described using a radioligand binding assay [48, 49]. Briefly, to test the effects of triplin on ethylene binding to its receptors in intact yeast cells, yeast were incubated at 30°C in the presence or absence of 100 μM triplin for 3 days. Intact yeast cells were then harvested and assayed for binding. For binding assays to membranes, cells were disrupted and membranes isolated as previously described [11] and ethylene binding determined in the presence or absence of 100 μM triplin. Saturable ethylene binding is indicated as counts per minute (CPM) and was calculated by subtracting the amount of radioactivity bound in the presence of excess non-radioactive ethylene from what was bound in the absence of unlabeled ethylene. We used western blots with anti-GST antibodies to ensure equal levels of ETR1 [1–128]-GST [11].
Constructs and plant transformation
The constructs and plant transformation were performed as previously described [32]. The coding sequence (CDS) of ATX1 (AT1G66240) and RAN1 (AT5G44790) was amplified from cDNA of Col-0 by PCR using primers listed in S2 Table. Fragments were then cloned into the entry vector pDONR-zeocin by BP reactions by following the instructions of the manufacturer (Life Technology, USA). Then genes were introduced to different pGWB vectors by LR reactions. The vectors were then used to transform Agrobacterium strain GV3101, the transformed agrobacteria were finally used to transform flowering Arabidopsis plants via the floral-dip method [50].
Quantitative real-time RT-PCR (qRT-PCR)
The qRT-PCR was done as previously described [32]. For testing the expression level of ERF1, seeds were surface sterilized and planted on 0.5xMS growth medium contain different chemicals in petri dish plates, after stratification at 4 C for 3 days, plates were exposed to light for 1–4 hours then transferred to dark for 3 days (22 C), and seedlings were collected. For testing the expression level of RAN1 in wild type and transgenic plants, 3-week old plant leaves were collected and total RNAs were extracted and used to synthesize the cDNAs by reverse transcription. Primers for ERF1 were made according to [43]. All primers used were listed in S2 Table. The ACTIN gene was amplified and used as an internal control.
Confocal observation and imaging
pDONR-ATX1 and pDONR-RAN1 were introduced into pGWB vectors as described in [32]. The construct combinations of ATX1:PGWB5/PGWB654 and RAN1:PGWB605 were used to transform Agrobacterium GV3101; then the Agrobacterium were infiltrated into leaves of N. benthamiana with P19 [32]. After 2-day incubation, the transformed plant leaves were observed and imaged under a confocal microscope (Olympus FV1000). For transgenic plants, the 5-day-old light grown roots or 3-day-old dark-grown hypocotyls were imaged under the confocal microscope. However photos in S12C and S12D Fig were imaged by NIKON A1R. ER, Golgi and plasma membrane associated protein markers used is AtWAK2, GmMAN1 and OsREM4.1 respectively [51, 52]. Arabidopsis WAK2-mCherry or GmMAN1-mCherry transgenic seeds are gifts from Chi-Kuang Wen lab of Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences. For DAPI staining, seedlings were fixed in 5% methanol, immersed in 2 μg/ml DAPI in phosphate buffer saline (PBS) and viewed by fluorescence microscopy under UV light.
Yeast two-hybrid assay
Yeast two-hybrid experiments were performed as previously described [32]. pGADT7 (RAN1 N terminal) and pGBKT7 (ATX1) were transformed simultaneously into the yeast strain AH109, the colonies were transferred to a SD (-Leu/-Trp/-His-Ade) solid medium with X-α-gal according to the manufacturer’s protocols (Clontech) for 3 days. The positive clones were identified because they grew well and became blue.
BiFC and Co-IP in N. benthamiana
BiFC and Co-IP assays were performed as previously described [32, 52]. The Split-Luciferase Complementation system was used. The construct combinations of cLuc-ATX1/nLuc-RAN1-N were used to transform Agrobacterium GV3101; then the Agrobacterium were infiltrated into leaves of N. benthamiana with P19 as previously described. After 2-day incubation, 1 mM luciferin (Sigma) was filtrated into the leaves and the pictures were recorded using a CCD imaging system (Berthold, https://www.berthold.com/). For the Co-IP assays, the N. benthamiana leaves were transfected with Agrobacterium containing ATX1-GFP/FLAG and RAN1-N-FLAG/GFP, and incubated for 2 days. Total proteins were extracted with extraction buffer [50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 1% Triton X-100, and 0.1% protease inhibitor cocktail (Promega)] for 20 minutes, and centrifuged at 4°C at 12,000 rpm for 20 min. The supernatant was incubated with pretreated anti-FLAG antibody coupled agarose beads (Abmart) for 2 hours at 4°C. Beads were washed three times with wash buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 1% Triton X-100). The bound proteins were eluted with 2xSDS loading buffer and boiled at 100°C for 5 minutes. The eluted proteins were separated on SDS-PAGE and immunoblotted with anti-FLAG antibody (Abmart) and anti-GFP antibody (Abmart).
Accession numbers
ATX1 (AT1G66240), RAN1 (AT5G44790), CCH (AT3G56240), ETR1 (AT1G66340), EIN2 (AT5G03280), EIN3 (AT3G20770), Triplin (F0655-1171)
Supporting information
The phenotypes of 3-day-old, dark-grown Col-0 and ein2-5 seedlings treated with 100 μM F0617-0170, F0665-0601, F1806-0031 or F2001-0992. For AgNO3 treatment a concentration of 500 μM was used. Scale bars represent 1 mm.
(TIF)
(A) The phenotype of 3-day -old, dark-grown seedlings from the M2 generation of the triplin resistant dominant mutants 45–1 and 45–2 treated with 100 μM triplin. (B) The genome DNA sequencing raw data of mutants 45–1 and 45–2 showing the G194A substitution mutations identical as the mutation in etr1-1. (C) The phenotypes of 3-day-old, dark-grown recessive triplin resistant mutants 39–1, 39–2, 41–1, 41–2, 42, 43 and 44 and their F1s with ein2 treated with 100 μM triplin. For comparison, the phenotypes of Col-0 and ein2 are shown. Scale bars represent 1 mm.
(TIF)
The phenotypes of 3-day-old, dark-grown seedlings without (-) or with (+) 100 μM Triplin. The phenotypes of (A) Col-0, ein2-5 and the ein2-5 complementary lines, com17-2 and com27-7, and (B) ethylene resistant mutants of ethylene receptors, ers1-1, ers2-1, etr2-1 and ein4.Scale bars represent 1 mm.
(TIF)
(A) qRT-PCR analysis of the relative expression levels of ACS5, ACS6 and ACS11. 3-day-old dark-grown Col-0 seedlings were treated with DMSO or 100 μM triplin. Each experiment was repeated three times, and error bars represent SEM. (B) Hypocotyl length of 3-day-old, dark-grown seedlings of Col-0 and cin5-1 treated with DMSO or 100 μM triplin. Each experiments was repeated three times, more than 30 seedlings were used every time. Error bars represent SEM. No significant difference was observed by two-tailed Student’s t-test using 0.05 cut-off. (C) Hypocotyl length of 3-day-old, dark-grown Col-0 seedlings in the presence of triplin without or with 10 μM AVG. The experiments were repeated three times with similar results (n ≥ 30).Values represent means ± SD, and no significant difference was observed by two-tailed Student’s t-test using 0.05 cut-off. (D) Gas chromatography analysis of ethylene production in Col-0 treated by 100 μM triplin, 50 μM ACC, or 1% (v/v) DMSO as a control. n = 3; error bars represent SEM. The scale bars represent 1 mm.
(TIF)
(A) The phenotypes of 3-day-old, dark-grown Col-0 seedlings treated with 100 μM triplin in the presence of the indicated metal salts. The concentrations of metal salts were 200 μM except for NiSO4 where 100 μM was used. The scale bars represent 1 mm. (B) The hypocotyl length of the seedlings in (A). The experiments were repeated three times with similar results (n ≥ 30).Values represent means±SD., and no significant difference was observed using two-tailed Student’s t-test with 0.05 cut-off.
(TIF)
(A) The phenotypes of 3-day-old, dark-grown Col-0 seedlings treated with 100 or 200 μM CuSO4 with or without 100 or 200 μM triplin. (B) The hypocotyl length of the seedlings in (A). Each experiments was repeated three times, more than 30 seedlings were used every time. Error bars represent SEM. *P < 0.05 (two-tailed Student’s t-test) indicated a significant difference between groups of different treatments. (C) The phenotypes of 3-day-old, dark-grown Col-0 seedlings treated with 100 μM triplin in the presence of 0, 100, 200, or 500 μM AgNO3. (D) The hypocotyl length of the seedlings in (C). All experiments were repeated three times with the similar results. Error bars represent SD (n > 30). In (A) and (C), the scale bars represent 1 mm.
(TIF)
The appearance 100 μl of 100 mM metal salts of CaCl2, ZnSO4, CuSO4 or AgNO3 were mixed with 100 μl of 10mM copper ion chelators triplin, neocuproine or BCS in 1.5 ml Eppendof tubes. The scale bar represents 1 mm.
(TIF)
The MALDI-TOF-MS analysis results of the reaction product of 100 μM ZnSO4 and 100 μM triplin (A), the reaction product of 100 μM FeSO4 and 100 μM triplin (B) and the reaction product of 10 mM AgNO3 and 10 mM triplin (C). The metal salts and triplin were mixed as described in S7 Fig.
(TIF)
The phenotypes of 3-day-old, dark-grown ein2-5 seedlings grown on different growth medium with or without 50 μM triplin. -Cu indicates the growth medium was made of plant essential elements except copper ion. BCS indicates the growth medium of 0.5xMS with 500 μM BCS. The scale bar represents 1 mm.
(TIF)
(A) The phenotypes of 3-day-old, dark-grown Col-0 seedlings treated with 100 μM neocuproine in the presence of no metal, 500 μM AgNO3, 100 μM CuSO4 or 100 μM ZnSO4. For comparison, the phenotypes of ein2-5 grown with only 100 μM neocuproine are shown. (B) The phenotypes of 3-day-old, dark-grown ran1-2 seedlings grown on 5 μM neocuproine without or with 10 or 20 μM CuSO4 are shown. (C) The phenotypes of 3-day-old, dark-grown Col-0 seedlings without or with 5, 20, 50 or 100μM neocuproine coupled with 0, 10 or 20 μM CuSO4 are shown. (D) atx1-1 and ran1-2 are hypersensitive to neocuproine. The phenotypes of 3-day-old, dark-grown seedlings of Col-0, atx1-1 and ran1-2 were treated with 5, 10 or 20 μM neocuproine. In all pictures, the scale bars represent 1 mm.
(TIF)
(A) A diagram shows the locations of the ATX1 T-DNA insertion mutants, Salk_026221 (atx1-1) and Salk_041022 (atx1-2). (B) The gene expression of the atx1-1 and atx1-2 were examined by RT-PCR. ACTIN was used as a loading control. (C) The phenotypes of 3-day-old, dark-grown seedlings of atx1-1, atx1-1 etr1-1 and atx1-1 ein2-5 treated with 100 μM triplin. The scale bar represents 1 mm. (D) The phenotypes of 3-day-old, dark-grown seedlings of Col-0 and atx1-1 treated with 20 μM triplin with (+) or without (-) 10μM CuSO4. The scale bars represent 1 mm.
(TIF)
(A) Scanning images of the roots of 35S:ATX1-GFP (atx1-1) seedlings under a confocal microscope (Olympus FV1000). (B) Scanning images of the roots of 35S:ATX1-GFP (atx1-1) seedlings after DAPI staining under a confocal microscope (Olympus FV1000). (C) Scanning images of the epidermal cells of the hypocotyl of 35S:ATX1-GFP (atx1-1) seedling by a confocal microscope (NIKON A1R). (D) Scanning image of the bottom of a cell in the hypocotyl of 35S:ATX1-GFP (atx1-1) seedling by a confocal microscope (NIKON A1R). (E) Scanning images of the hypocotyls of F1 from 35S:ATX1-GFP (atx1-1) and transgenic marker lines with WAK2-mCherry under a confocal microscope (Olympus FV1000). (F) Scanning images of the hypocotyls of F1 from 35S:ATX1-GFP (atx1-1) and transgenic marker lines with GmMAN1-mCherry under a confocal microscope (Olympus FV1000). All scale bars represent 20 μm.
(TIF)
(A) ATX1-GFP and WAK2 / GmMAN1 / OsREM4.1-mCherry were transiently expressed in N. benthamiana leaves and their co-localizations were shown. (B) RAN1-GFP and WAK2 / GmMAN1 / OsREM4.1-mCherry were transiently expressed in N. benthamiana leaves and their co-localizations were shown. All images were made under a confocal microscope (Olympus FV1000).Scale bars represent 20 μM.
(TIF)
(A) The phenotypes of 3-day-old, dark-grown seedlings of Col-0 and two 35S:RAN1-GFP transgenic lines treated with 0, 30, 50 or 100 μM triplin. (B) qRT-PCR analysis of the relative RAN1 expression levels in two 35S:RAN1-GFP lines. Each experiment was repeated three times, and error bars represent SEM.
(TIF)
(DOCX)
(DOCX)
Acknowledgments
We thank Suhong Liu (Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences) for the MALDI-TOF-MS analysis, Hongying Yi (Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences) for plant copper content assay, Xiaoshu Gao (Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences) for help in confocal imaging, Wei Zhang from Wen lab for ethylene measurement, Xuelu Wang (Fudan University) for providing atx1 mutant, and Hong Qiao for providing ein2-5 and its complementary lines. We thank Tsuyoshi Nakagawa (Shimane University) for providing pGWB vectors.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the National Science Foundation of the USA (IOS-1254423, MCB-1517032) to BMB; grants from the National Natural Science Foundation of China (31630014) to LL; and grants from the National Natural Science Foundation of China (31171293 and 31371361) and one hundred talents program of Chinese Academy of Sciences to YZ. The funders have not played any role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bleecker AB, Estelle MA, Somerville C, Kende H. Insensitivity to Ethylene Conferred by a Dominant Mutation in Arabidopsis thaliana. Science. 1988;241(4869):1086–9. doi: 10.1126/science.241.4869.1086 [DOI] [PubMed] [Google Scholar]
- 2.Roman G, Lubarsky B, Kieber JJ, Rothenberg M, Ecker JR. Genetic-Analysis of Ethylene Signal-Transduction in Arabidopsis-Thaliana—5 Novel Mutant Loci Integrated into a Stress-Response Pathway. Genetics. 1995;139(3):1393–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hua J, Meyerowitz EM. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94(2):261–71. [DOI] [PubMed] [Google Scholar]
- 4.Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. Ctr1, a Negative Regulator of the Ethylene Response Pathway in Arabidopsis, Encodes a Member of the Raf Family of Protein-Kinases. Cell. 1993;72(3):427–41. [DOI] [PubMed] [Google Scholar]
- 5.Ju CL, Yoon GM, Shemansky JM, Lin DY, Ying ZI, Chang JH, et al. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. P Natl Acad Sci USA. 2012;109(47):19486–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiao H, Shen Z, Huang SS, Schmitz RJ, Urich MA, Briggs SP, et al. Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science. 2012;338(6105):390–3. doi: 10.1126/science.1225974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell. 1997;89(7):1133–44. [DOI] [PubMed] [Google Scholar]
- 8.Guo HW, Ecker JR. Plant responses to ethylene gas are mediated by SCF (EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell. 2003;115(6):667–77. [DOI] [PubMed] [Google Scholar]
- 9.Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, et al. EIN3-dependent regulation of plant ethylene hormone signaling by two arabidopsis F box proteins: EBF1 and EBF2. Cell. 2003;115(6):679–89. [DOI] [PubMed] [Google Scholar]
- 10.Gagne JM, Smalle J, Gingerich DJ, Walker JM, Yoo SD, Yanagisawa S, et al. Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. P Natl Acad Sci USA. 2004;101(17):6803–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez FI, Esch JJ, Hall AE, Binder BM, Schaller GE, Bleecker AB. A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science. 1999;283(5404):996–8. [DOI] [PubMed] [Google Scholar]
- 12.Hirayama T, Kieber JJ, Hirayama N, Kogan M, Guzman P, Nourizadeh S, et al. Responsive-to-antagonist1, a Menkes/Wilson disease-related copper transporter, is required for ethylene signaling in Arabidopsis. Cell. 1999;97(3):383–93. [DOI] [PubMed] [Google Scholar]
- 13.Woeste KE, Kieber JJ. A strong loss-of-function mutation in RAN1 results in constitutive activation of the ethylene response pathway as well as a rosette-lethal phenotype. Plant Cell. 2000;12(3):443–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Binder BM, Rodriguez FI, Bleecker AB. The Copper Transporter RAN1 Is Essential for Biogenesis of Ethylene Receptors in Arabidopsis. J Biol Chem. 2010;285(48):37263–70. doi: 10.1074/jbc.M110.170027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin SJ, Culotta VC. The ATX1 gene of Saccharomyces cerevisiae encodes a small metal homeostasis factor that protects cells against reactive oxygen toxicity. Proc Natl Acad Sci U S A. 1995;92(9):3784–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huffman DL, O'Halloran TV. Energetics of copper trafficking between the Atx1 metallochaperone and the intracellular copper transporter, Ccc2. J Biol Chem. 2000;275(25):18611–4. doi: 10.1074/jbc.C000172200 [DOI] [PubMed] [Google Scholar]
- 17.Fu DD, Beeler TJ, Dunn TM. Sequence, Mapping and Disruption of Ccc2, a Gene That Cross-Complements the Ca2+-Sensitive Phenotype of Csg1 Mutants and Encodes a P-Type Atpase Belonging to the Cu2+-Atpase Subfamily. Yeast. 1995;11(3):283–92. doi: 10.1002/yea.320110310 [DOI] [PubMed] [Google Scholar]
- 18.Banci L, Bertini I, Cantini F, Felli IC, Gonnelli L, Hadjiliadis N, et al. The Atx1-Ccc2 complex is a metal-mediated protein-protein interaction. Nature Chemical Biology. 2006;2(7):367–8. doi: 10.1038/nchembio797 [DOI] [PubMed] [Google Scholar]
- 19.Hamza I, Schaefer M, Klomp LWJ, Gitlin JD. Interaction of the copper chaperone HAH1 with the Wilson disease protein is essential for copper homeostasis. P Natl Acad Sci USA. 1999;96(23):13363–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anastassopoulou I, Banci L, Bertini I, Cantini F, Katsari E, Rosato A. Solution structure of the Apo and copper(I)-loaded human metallochaperone HAH1. Biochemistry-Us. 2004;43(41):13046–53. [DOI] [PubMed] [Google Scholar]
- 21.Setty SRG, Tenza D, Sviderskaya EV, Bennett DC, Raposo G, Marks MS. Cell-specific ATP7A transport sustains copper-dependent tyrosinase activity in melanosomes. Faseb J. 2009;23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Himelblau E, Mira H, Lin SJ, Culotta VC, Penarrubia L, Amasino RM. Identification of a functional homolog of the yeast copper homeostasis gene ATX1 from Arabidopsis. Plant Physiol. 1998;117(4):1227–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mira H, Martinez-Garcia F, Penarrubia L. Evidence for the plant-specific intercellular transport of the Arabidopsis copper chaperone CCH. Plant J. 2001;25(5):521–8. [DOI] [PubMed] [Google Scholar]
- 24.Puig S, Mira H, Dorcey E, Sancenon V, Andres-Colas N, Garcia-Molina A, et al. Higher plants possess two different types of ATX1-like copper chaperones. Biochem Biophys Res Commun. 2007;354(2):385–90. doi: 10.1016/j.bbrc.2006.12.215 [DOI] [PubMed] [Google Scholar]
- 25.Shin LJ, Lo JC, Yeh KC. Copper chaperone antioxidant protein1 is essential for copper homeostasis. Plant Physiol. 2012;159(3):1099–110. doi: 10.1104/pp.112.195974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andres-Colas N, Sancenon V, Rodriguez-Navarro S, Mayo S, Thiele DJ, Ecker JR, et al. The Arabidopsis heavy metal P-type ATPase HMA5 interacts with metallochaperones and functions in copper detoxification of roots. Plant J. 2006;45(2):225–36. doi: 10.1111/j.1365-313X.2005.02601.x [DOI] [PubMed] [Google Scholar]
- 27.Burkhead JL, Reynolds KAG, Abdel-Ghany SE, Cohu CM, Pilon M. Copper homeostasis. New Phytol. 2009;182(4):799–816. doi: 10.1111/j.1469-8137.2009.02846.x [DOI] [PubMed] [Google Scholar]
- 28.Chen SH, Lin JK, Liu SH, Liang YC, Lin-Shiau SY. Apoptosis of cultured astrocytes induced by the copper and neocuproine complex through oxidative stress and JNK activation. Toxicol Sci. 2008;102(1):138–49. doi: 10.1093/toxsci/kfm292 [DOI] [PubMed] [Google Scholar]
- 29.Zhang XY, Chen YT, Lin X, Hong XY, Zhu Y, Li WY, et al. Adenine Phosphoribosyl Transferase 1 is a Key Enzyme Catalyzing Cytokinin Conversion from Nucleobases to Nucleotides in Arabidopsis. Mol Plant. 2013;6(5):1661–72. doi: 10.1093/mp/sst071 [DOI] [PubMed] [Google Scholar]
- 30.Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, et al. Abscisic Acid Inhibits Type 2C Protein Phosphatases via the PYR/PYL Family of START Proteins. Science. 2009;324(5930):1068–71. doi: 10.1126/science.1173041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savaldi-Goldstein S, Baiga TJ, Pojer F, Dabi T, Butterfield C, Parry G, et al. New auxin analogs with growth-promoting effects in intact plants reveal a chemical strategy to improve hormone delivery. Proc Natl Acad Sci U S A. 2008;105(39):15190–5. doi: 10.1073/pnas.0806324105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye YJ, Gong ZY, Lu X, Miao DY, Shi JM, Lu J, et al. Germostatin resistance locus 1 encodes a PHD finger protein involved in auxin-mediated seed dormancy and germination. Plant Journal. 2016;85(1):3–15. doi: 10.1111/tpj.13086 [DOI] [PubMed] [Google Scholar]
- 33.Zhao Y, Chow TF, Puckrin RS, Alfred SE, Korir AK, Larive CK, et al. Chemical genetic interrogation of natural variation uncovers a molecule that is glycoactivated. Nature Chemical Biology. 2007;3(11):716–21. doi: 10.1038/nchembio.2007.32 [DOI] [PubMed] [Google Scholar]
- 34.Zhao Y. A chemical genetics method to uncover small molecules for dissecting the mechanism of ABA responses in Arabidopsis seed germination. Methods Mol Biol. 2012;876:107–16. doi: 10.1007/978-1-61779-809-2_8 [DOI] [PubMed] [Google Scholar]
- 35.Dong CH, Rivarola M, Resnick JS, Maggin BD, Chang C. Subcellular co-localization of Arabidopsis RTE1 and ETR1 supports a regulatory role for RTE1 in ETR1 ethylene signaling. Plant Journal. 2008;53(2):275–86. doi: 10.1111/j.1365-313X.2007.03339.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toth R, van der Hoorn RA. Emerging principles in plant chemical genetics. Trends Plant Sci. 2010;15(2):81–8. doi: 10.1016/j.tplants.2009.11.005 [DOI] [PubMed] [Google Scholar]
- 37.Arnal N, de Alaniz MJT, Marra CA. Carnosine and neocuproine as neutralizing agents for copper overload-induced damages in cultured human cells. Chem-Biol Interact. 2011;192(3):257–63. doi: 10.1016/j.cbi.2011.03.017 [DOI] [PubMed] [Google Scholar]
- 38.Kenche VB, Barnham KJ. Alzheimer's disease & metals: therapeutic opportunities. Br J Pharmacol. 2011;163(2):211–9. doi: 10.1111/j.1476-5381.2011.01221.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cherny RA, Atwood CS, Xilinas ME, Gray DN, Jones WD, McLean CA, et al. Treatment with a copper-zinc chelator markedly and rapidly inhibits beta-amyloid accumulation in Alzheimer's disease transgenic mice. Neuron. 2001;30(3):665–76. [DOI] [PubMed] [Google Scholar]
- 40.Franz KJ. Application of inorganic chemistry for non-cancer therapeutics. Dalton T. 2012;41(21):6333–4. [DOI] [PubMed] [Google Scholar]
- 41.Pushie MJ, Nienaber KH, Summers KL, Cotelesage JJH, Ponomarenko O, Nichol HK, et al. The solution structure of the copper clioquinol complex. J Inorg Biochem. 2014;133:50–6. doi: 10.1016/j.jinorgbio.2014.01.003 [DOI] [PubMed] [Google Scholar]
- 42.Cherny RA, Atwood CS, Xilinas ME, Gray DN, Jones WD, McLean CA, et al. Treatment with a copper-zinc chelator markedly and rapidly inhibits beta-amyloid accumulation in Alzheimer's disease transgenic mice. Neuron. 2001;30(3):665–76. [DOI] [PubMed] [Google Scholar]
- 43.Tan ST, Xue HW. Casein kinase 1 regulates ethylene synthesis by phosphorylating and promoting the turnover of ACS5. Cell Rep. 2014;9(5):1692–702. doi: 10.1016/j.celrep.2014.10.047 [DOI] [PubMed] [Google Scholar]
- 44.Zhang W, Wen CK. Preparation of ethylene gas and comparison of ethylene responses induced by ethylene, ACC, and ethephon. Plant Physiol Bioch. 2010;48(1):45–53. [DOI] [PubMed] [Google Scholar]
- 45.Li JY, Fu YL, Pike SM, Bao J, Tian W, Zhang Y, et al. The Arabidopsis Nitrate Transporter NRT1.8 Functions in Nitrate Removal from the Xylem Sap and Mediates Cadmium Tolerance. Plant Cell. 2010;22(5):1633–46. doi: 10.1105/tpc.110.075242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu L, Zhang J, Guo YL. Enhanced detection and desalting free protocol for phosphopeptides eluted from immobilized Fe (III) affinity chromatography in direct MALDI TOF analysis. J Proteomics. 2014;96:360–5. doi: 10.1016/j.jprot.2013.12.001 [DOI] [PubMed] [Google Scholar]
- 47.McDaniel BK, Binder BM. Ethylene Receptor 1 (ETR1) Is Sufficient and Has the Predominant Role in Mediating Inhibition of Ethylene Responses by Silver in Arabidopsis thaliana. J Biol Chem. 2012;287(31):26094–103. doi: 10.1074/jbc.M112.383034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaller GE, Bleecker AB. Ethylene-Binding Sites Generated in Yeast Expressing the Arabidopsis Etr1 Gene. Science. 1995;270(5243):1809–11. [DOI] [PubMed] [Google Scholar]
- 49.Sisler EC. Measurement of Ethylene Binding in Plant-Tissue. Plant Physiology. 1979;64(4):538–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal. 1998;16(6):735–43. [DOI] [PubMed] [Google Scholar]
- 51.Nelson BK, Cai X, Nebenfuhr A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51(6):1126–36. doi: 10.1111/j.1365-313X.2007.03212.x [DOI] [PubMed] [Google Scholar]
- 52.Gui J, Zheng S, Liu C, Shen J, Li J, Li L. OsREM4.1 Interacts with OsSERK1 to Coordinate the Interlinking between Abscisic Acid and Brassinosteroid Signaling in Rice. Dev Cell. 2016;38(2):201–13. doi: 10.1016/j.devcel.2016.06.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The phenotypes of 3-day-old, dark-grown Col-0 and ein2-5 seedlings treated with 100 μM F0617-0170, F0665-0601, F1806-0031 or F2001-0992. For AgNO3 treatment a concentration of 500 μM was used. Scale bars represent 1 mm.
(TIF)
(A) The phenotype of 3-day -old, dark-grown seedlings from the M2 generation of the triplin resistant dominant mutants 45–1 and 45–2 treated with 100 μM triplin. (B) The genome DNA sequencing raw data of mutants 45–1 and 45–2 showing the G194A substitution mutations identical as the mutation in etr1-1. (C) The phenotypes of 3-day-old, dark-grown recessive triplin resistant mutants 39–1, 39–2, 41–1, 41–2, 42, 43 and 44 and their F1s with ein2 treated with 100 μM triplin. For comparison, the phenotypes of Col-0 and ein2 are shown. Scale bars represent 1 mm.
(TIF)
The phenotypes of 3-day-old, dark-grown seedlings without (-) or with (+) 100 μM Triplin. The phenotypes of (A) Col-0, ein2-5 and the ein2-5 complementary lines, com17-2 and com27-7, and (B) ethylene resistant mutants of ethylene receptors, ers1-1, ers2-1, etr2-1 and ein4.Scale bars represent 1 mm.
(TIF)
(A) qRT-PCR analysis of the relative expression levels of ACS5, ACS6 and ACS11. 3-day-old dark-grown Col-0 seedlings were treated with DMSO or 100 μM triplin. Each experiment was repeated three times, and error bars represent SEM. (B) Hypocotyl length of 3-day-old, dark-grown seedlings of Col-0 and cin5-1 treated with DMSO or 100 μM triplin. Each experiments was repeated three times, more than 30 seedlings were used every time. Error bars represent SEM. No significant difference was observed by two-tailed Student’s t-test using 0.05 cut-off. (C) Hypocotyl length of 3-day-old, dark-grown Col-0 seedlings in the presence of triplin without or with 10 μM AVG. The experiments were repeated three times with similar results (n ≥ 30).Values represent means ± SD, and no significant difference was observed by two-tailed Student’s t-test using 0.05 cut-off. (D) Gas chromatography analysis of ethylene production in Col-0 treated by 100 μM triplin, 50 μM ACC, or 1% (v/v) DMSO as a control. n = 3; error bars represent SEM. The scale bars represent 1 mm.
(TIF)
(A) The phenotypes of 3-day-old, dark-grown Col-0 seedlings treated with 100 μM triplin in the presence of the indicated metal salts. The concentrations of metal salts were 200 μM except for NiSO4 where 100 μM was used. The scale bars represent 1 mm. (B) The hypocotyl length of the seedlings in (A). The experiments were repeated three times with similar results (n ≥ 30).Values represent means±SD., and no significant difference was observed using two-tailed Student’s t-test with 0.05 cut-off.
(TIF)
(A) The phenotypes of 3-day-old, dark-grown Col-0 seedlings treated with 100 or 200 μM CuSO4 with or without 100 or 200 μM triplin. (B) The hypocotyl length of the seedlings in (A). Each experiments was repeated three times, more than 30 seedlings were used every time. Error bars represent SEM. *P < 0.05 (two-tailed Student’s t-test) indicated a significant difference between groups of different treatments. (C) The phenotypes of 3-day-old, dark-grown Col-0 seedlings treated with 100 μM triplin in the presence of 0, 100, 200, or 500 μM AgNO3. (D) The hypocotyl length of the seedlings in (C). All experiments were repeated three times with the similar results. Error bars represent SD (n > 30). In (A) and (C), the scale bars represent 1 mm.
(TIF)
The appearance 100 μl of 100 mM metal salts of CaCl2, ZnSO4, CuSO4 or AgNO3 were mixed with 100 μl of 10mM copper ion chelators triplin, neocuproine or BCS in 1.5 ml Eppendof tubes. The scale bar represents 1 mm.
(TIF)
The MALDI-TOF-MS analysis results of the reaction product of 100 μM ZnSO4 and 100 μM triplin (A), the reaction product of 100 μM FeSO4 and 100 μM triplin (B) and the reaction product of 10 mM AgNO3 and 10 mM triplin (C). The metal salts and triplin were mixed as described in S7 Fig.
(TIF)
The phenotypes of 3-day-old, dark-grown ein2-5 seedlings grown on different growth medium with or without 50 μM triplin. -Cu indicates the growth medium was made of plant essential elements except copper ion. BCS indicates the growth medium of 0.5xMS with 500 μM BCS. The scale bar represents 1 mm.
(TIF)
(A) The phenotypes of 3-day-old, dark-grown Col-0 seedlings treated with 100 μM neocuproine in the presence of no metal, 500 μM AgNO3, 100 μM CuSO4 or 100 μM ZnSO4. For comparison, the phenotypes of ein2-5 grown with only 100 μM neocuproine are shown. (B) The phenotypes of 3-day-old, dark-grown ran1-2 seedlings grown on 5 μM neocuproine without or with 10 or 20 μM CuSO4 are shown. (C) The phenotypes of 3-day-old, dark-grown Col-0 seedlings without or with 5, 20, 50 or 100μM neocuproine coupled with 0, 10 or 20 μM CuSO4 are shown. (D) atx1-1 and ran1-2 are hypersensitive to neocuproine. The phenotypes of 3-day-old, dark-grown seedlings of Col-0, atx1-1 and ran1-2 were treated with 5, 10 or 20 μM neocuproine. In all pictures, the scale bars represent 1 mm.
(TIF)
(A) A diagram shows the locations of the ATX1 T-DNA insertion mutants, Salk_026221 (atx1-1) and Salk_041022 (atx1-2). (B) The gene expression of the atx1-1 and atx1-2 were examined by RT-PCR. ACTIN was used as a loading control. (C) The phenotypes of 3-day-old, dark-grown seedlings of atx1-1, atx1-1 etr1-1 and atx1-1 ein2-5 treated with 100 μM triplin. The scale bar represents 1 mm. (D) The phenotypes of 3-day-old, dark-grown seedlings of Col-0 and atx1-1 treated with 20 μM triplin with (+) or without (-) 10μM CuSO4. The scale bars represent 1 mm.
(TIF)
(A) Scanning images of the roots of 35S:ATX1-GFP (atx1-1) seedlings under a confocal microscope (Olympus FV1000). (B) Scanning images of the roots of 35S:ATX1-GFP (atx1-1) seedlings after DAPI staining under a confocal microscope (Olympus FV1000). (C) Scanning images of the epidermal cells of the hypocotyl of 35S:ATX1-GFP (atx1-1) seedling by a confocal microscope (NIKON A1R). (D) Scanning image of the bottom of a cell in the hypocotyl of 35S:ATX1-GFP (atx1-1) seedling by a confocal microscope (NIKON A1R). (E) Scanning images of the hypocotyls of F1 from 35S:ATX1-GFP (atx1-1) and transgenic marker lines with WAK2-mCherry under a confocal microscope (Olympus FV1000). (F) Scanning images of the hypocotyls of F1 from 35S:ATX1-GFP (atx1-1) and transgenic marker lines with GmMAN1-mCherry under a confocal microscope (Olympus FV1000). All scale bars represent 20 μm.
(TIF)
(A) ATX1-GFP and WAK2 / GmMAN1 / OsREM4.1-mCherry were transiently expressed in N. benthamiana leaves and their co-localizations were shown. (B) RAN1-GFP and WAK2 / GmMAN1 / OsREM4.1-mCherry were transiently expressed in N. benthamiana leaves and their co-localizations were shown. All images were made under a confocal microscope (Olympus FV1000).Scale bars represent 20 μM.
(TIF)
(A) The phenotypes of 3-day-old, dark-grown seedlings of Col-0 and two 35S:RAN1-GFP transgenic lines treated with 0, 30, 50 or 100 μM triplin. (B) qRT-PCR analysis of the relative RAN1 expression levels in two 35S:RAN1-GFP lines. Each experiment was repeated three times, and error bars represent SEM.
(TIF)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.