Abstract
Molecular markers are required in a broad spectrum of gene screening approaches, ranging from gene-mapping within traditional ‘forward’-genetics approaches through QTL identification studies to genotyping and haplotyping studies. As we enter the post-genomics era, the need for genetic markers does not diminish, even in the species with fully sequenced genomes. PlantMarkers is a genetic marker database that contains a comprehensive pool of predicted molecular markers. We have adopted contemporary techniques to identify putative single nucleotide polymorphism (SNP), simple sequence repeat (SSR) and conserved orthologue set markers. A systematic approach to identify as broad a range of putative markers has been undertaken by screening the available openSputnik unigene consensus sequences from over 50 plant species. A web presence at http://markers.btk.fi provides functionality so that a user may search for species-specific markers on the basis of many specific criteria not limited to non-synonymous SNPs segregating between different varieties or measured polymorphic SSRs. Feedback forms are provided with all sequence entries to enable inclusion of, for example, map location for markers validated by the research community.
INTRODUCTION
The availability of genetic markers is fundamental within plant biology and plant breeding. There are a wide range of uses and applications for molecular markers, but most are associated with the map-based cloning of individual genes, the characterization of quanitative multi-gene traits and the survey and analysis of genetic diversity. With the subsequent association of genes and their related markers they additionally become valuable within the context of both genotyping and haplotyping.
Regardless of the ultimate reason for the application of molecular markers, there is a general need for both high- density and uniform maps that represent whole genomes. While techniques for the classification and typing of alleles have become amenable to high-throughput screening, e.g. microarray-based genotyping, the more important marker discovery steps require significant investments in both time and money. Single nucleotide polymorphism (SNP) markers, for example, have enjoyed massive popularity through their high density within the genome and their ease of characterization. The identification of these markers, however, requires access to reliable DNA sequence from the complete range of plants strains/varieties or ecotypes that will subsequently be used.
Access to a complete genome sequence now has been demonstrated to complement traditional marker-based approaches rather well. In Arabidopsis thaliana, which has a complete genome sequence (1), parallel efforts have since led to the creation of several broad and genome-based resources that will both expedite and facilitate traditional breeding and mapping approaches (2,3).
The widespread application of transcriptome sampling strategies within plant genomics has created an extremely large and clearly redundant sequence collection (4). Although these random sampling approaches are biased and are not truly representative of the whole genome, they do satisfy the core requirements of a broad range of marker discovery approaches. For the plant species with the larger expressed sequence tag (EST) collections, large numbers of genes are represented along with information for several of the more common strains or varieties of the species. This random sequence information along with the underlying redundancy and parental associations has been used in predictive approaches for SNPs (5), simple sequence repeats (SSRs) (6) and conserved orthologue set (COS) markers (7), and the speed and success of marker validation typically exceeds traditional approaches.
The plant EST database at EMBL has recently passed the five million EST sequence level. More than 50 plant species with over 5000 ESTs are represented. These species are of interest to plant breeders and/or to the scientific community. Given the technical abilities to detect in silico molecular markers, there is a truly vast potential collection of molecular markers present within these sequences.
We have created a database resource, PlantMarkers, for the prediction, analysis and display of plant molecular markers. As a sequence substrate, we used the plant EST data from within the EMBL sequence databases (8). After clustering the ESTs to yield a set of species-specific and less redundant unigenes, we have identified SNP, SSR and COS markers from the available sequence collections. Putative markers have been anchored to available protein-coding sequence where possible. Underlying sequence annotations that relate to clone library, strain or variety have been retained, thus allowing for the selection of putative polymorphic sequences that are segregating between different collections.
IMPLEMENTATION AND DATABASE STRUCTURE
The PlantMarkers database has been implemented using the Java programming language and standard JDBC database adapters. No special SQL features or objects are used within the application pipeline to ensure platform and database independence. The PlantMarkers database has been tested with both the MySQL and PostgreSQL relational database management systems; and is running on a Linux platform.
DATABASE CONTENTS
The EST sequences within the PlantMarkers database have been masked, clustered and assembled within the context of the openSputnik database (9,10). EST sequences were clustered using the HPT2 clustering algorithm (Biomax informatics, Martinsried, Germany) and assembled using the CAP3 assembler. The current release of PlantMarkers is based on over four million ESTs from over 50 plant species downloaded from the EMBL sequence database.
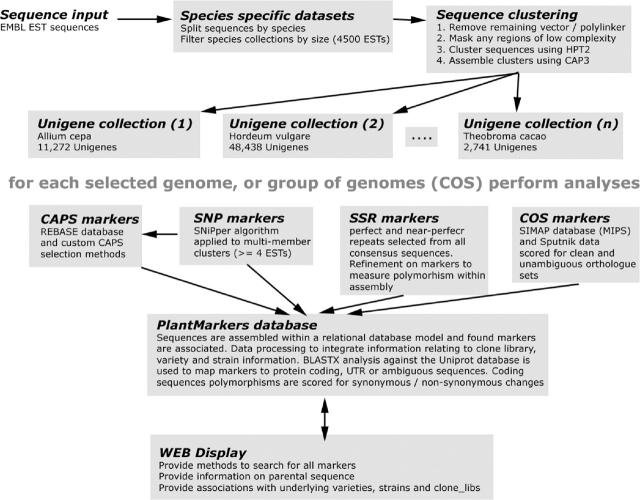
Following CAP3 assembly both the unigene consensus sequence and the underlying assembly (for multi-member sequences) are retained. The sequence assembly and the alignment of the constituent ESTs were used as the source for polymorphism detection while the unigene consensus is a valuable sequence reference. Following the clustering and assembly step, SNP, CAPS and SSR markers are predicted sequentially on each available unigene set. The unigene sets are merged for the selection of candidate COS markers. A generalized flow diagram showing the main steps within the preparation of the PlantMarkers database is shown in Figure 1.
Figure 1.
A schema showing the analysis pipeline used within the PlantMarkers database. The openSputnik sequence analysis pipeline is used to split and size select suitable EST collections from the complete EMBL-EST collections. The sequences are clustered and assembled and each species is placed within its own database. Using each species-specific database sequentially, SNP and SSR markers are predicted. The derived and filtered SNP markers are further refined by performing an in silico restriction digestion to identify candidate CAPS markers. Collections of genomes are aggregated and are placed within reciprocal context using the MIPS SIMAP database. All derived sequences are placed within the relational structure of the PlantMarkers database and markers are further refined. Web display methods interface directly with the PlantMarkers database.
The prediction of markers has been performed using highly permissive parameters. While this will certainly generate a large number of false positive results within the dataset, we prefer to present the user with the largest marker space and allow the user to impose a variety of selected thresholds rather than imposing an arbitrary threshold on the data ourselves.
SSR prediction
SSR markers represent well-established and traditional forms of molecular marker (11). Their detection by computational means is straightforward in that only simple perfect sequence repeats have to be detected. Several mechanistically equivalent methods have been published for the experimental selection of SSR markers from EST sequence data and the validation rate has been high (6,12,13).
The openSputnik unigene collections have been scored for putative microsatellite markers. The search was restricted to di-, tri-, tetra- and penta-nucleotide repeats because these have previously been shown to be the dominant repeat types (6). To maximize the available repeat search space, a permissive requirement for a minimum number of repeat units has been imposed. This is reflected by the requirement for 7, 6, 6 and 6 repeat units for di-, tri-, tetra- and penta-nucleotide repeats, respectively. In addition to scoring perfect repeats, we have also measured near perfect repeats with only slight repeat pattern deviations. This allows for the possibility that there may be a perfect repeat pattern at this locus within different varieties, ecotypes and cultivars. This creates a population of candidate SSR markers. Following the pre-screening for candidate SSR markers a second round of analysis is performed. Individual EST sequences that constitute the unigene consensus sequence are scored and if repeats at the same locus and of different length can be found the candidate SSR is labelled as a probable SSR. In Table 1 of the Supplementary Information, basic statistics for the prevalence of SSR markers, their types and sizes are shown along with the frequency of putative and probable markers for a taxonomically diverse and representative selection of plants.
SNP prediction
Of the molecular marker types currently used SNPs are perhaps the most dominant method (14). While bioinformatics methods have been developed for the selection of SNPs from aligned sequences (15–17), the SNiPper algorithm was developed especially to separate probable SNPs from likely sequencing errors within the context of unigene assemblies (5). Using the SNiPper algorithm SNPs have been selected and validated for both plants and animals.
From within the 50 openSputnik unigene collections we have scored putative SNPs using the SNiPper algorithm. We have imposed a minimum cluster size of four ESTs to score a putative SNP. To maximize the SNP search space, no arbitrary requirements for underlying allele frequencies have been imposed. These thresholds are instead imposed when a user selects a set of putative SNPs. Following the classification of candidate SNPs, the parental unigene sequences have been annotated by performing BLASTX analyses against the UniProt database (18). The results are filtered using the expectation value of 1E−10. This allows us to map SNPs to protein-coding or probable untranslated region sequence in many cases. This further allows us to identify a SNPs position in the codon and to determine if the SNP would result in a synonymous or non-synonymous substitution. In Table 2 of the Supplementary Material, SNP data for several genomes are shown.
The SNP markers have been further processed by identifying the subset that form potential cleaved amplified polymorphic sequence (CAPS) markers. Using the restriction enzyme data for common enzymes presented within the REBASE database (19) in silico sequence digestions were performed using the EMBOSS (20) restrict method on the unigene sequence immediately upstream and downstream of the SNP. Polymorphisms that result in altered restriction patterns have been tagged within the database and CAPS marker frequencies are also shown in Supplementary table 2.
COS marker selection
COS markers were used for comparative mapping between closely related species (7). For a given group of species, a COS is formed by identifying a gene from each species that is orthologous to all other genes in the set. For the purpose of generating markers, paralogues or closely related sequences within one species are a hindrance, hence in contrast to conserved orthologue groups (COGs) we select only sets where there are no close paralogues in any of the species involved.
Bioinformatics-based orthologue detection relies on sequence similarity searches of all genes against all genes of all other species involved. To efficiently manage the computational cost, we utilize the SIMAP (SImilarity MAtrix of Proteins) database (http://mips.gsf.de/proj/simap) (21). This stores the similarity between a given protein sequence and all known sequences, expressed as FASTA scores. The database can be augmented incrementally, and several tools are available to retrieve specialized datasets. For the detection of orthologues, we use best bidirectional hit (BBH) and INPARANOID (22) algorithms to process the data.
Since both algorithms work on pairs of species, we combine the results by selecting a ‘seed’ species, which preferably has a complete genome and full-length protein sequences, and then select the intersection of all pairwise sets between the centre species and each of the other species. The result of this analysis is, for a given group of species, a list of conserved orthologue sets where each set contains a representative protein from each species, and in no species is there a close paralogue. COS markers have been selected within taxonomic clades and using the complete aggregation of all sequences from all species. Not all permutations of EST collections are available within the PlantMarkers database.
QUERY INTERFACE
A query interface to the data has been implemented using the Zope application server software. The PlantMarkers interface has been implemented as a Zope product that directly interfaces with the underlying RDBMS. We have provided analytical interfaces that allow for the simple search of markers on the basis of simple forms. Searches require a few key elements such as unigene collection that will be screened, parameters to define the search space (number of ESTs that form a consensus, minor-allele frequencies and major-allele frequencies for SNPs, repeat type and repeat length for SSRs) and searches can be extended to include or exclude markers that fall within the protein-coding sequences or which could become CAPS markers.
The user interface also provides the possibility that a user can add additional information for any given marker. User submitted information should ideally relate to markers that have been successfully amplified, and in which strains or varieties along with other information such as map position for any mapped markers. Information is added to a plain text field and will be curated by the database administrator to enrich the value of the PlantMarkers database.
DATA AVAILABILITY
All of the data within the database are freely available to the scientific community. In addition to data access through the web interface, markers may be downloaded in an XML format and as an MS Excel compatible tab-delimited files.
FUTURE DIRECTIONS
The PlantMarkers database will remain synchronous with EST and GSS sequence collections that are available through the openSputnik platform. The database will diversify to include other datasets outside of the plant kingdom, and a mammalian sequence release has been planned for the Spring 2005. The database will naturally evolve to suit the needs and requirements of the users. A logical evolution of the resource will involve linking the sequence resource to genomic resources and estimating possible genetic locations on the basis of known syntenic regions within related genomes. A collection of graphical tools will be added to the data display to facilitate visualization of the underlying data and to convert the current predictive resource into a more valuable inter-species marker database. Many of the markers within the database may already have been validated elsewhere. We would be very happy to update the resource with, and credit research groups for, molecular markers that have been either experimentally validated (or not).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
REFERENCES
- 1.The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 2.Jander G., Norris,S.R., Rounsley,S.D., Bush,D.F., Levin,I.M. and Last,R.L. (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol., 129, 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmid K.J., Sorensen,T.R., Stracke,R., Torjek,O., Altmann,T., Mitchell-Olds,T. and Weisshaar,B. (2003) Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Res., 13, 1250–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudd S. (2003) Expressed sequence tags: alternative or complement to whole genome sequences? Trends Plant Sci., 8, 321–329. [DOI] [PubMed] [Google Scholar]
- 5.Kota R., Rudd,S., Facius,A., Kolesov,G., Thiel,T., Zhang,H., Stein,N., Mayer,K. and Graner,A. (2003) Snipping polymorphisms from large EST collections in barley (Hordeum vulgare L.). Mol. Genet. Genomics, 270, 24–33. [DOI] [PubMed] [Google Scholar]
- 6.Thiel T., Michalek,W., Varshney,R.K. and Graner,A. (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet., 106, 411–422. [DOI] [PubMed] [Google Scholar]
- 7.Fulton T.M., Van der Hoeven,R., Eannetta,N.T. and Tanksley,S.D. (2002) Identification, analysis, and utilization of conserved ortholog set markers for comparative genomics in higher plants. Plant Cell, 14, 1457–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kulikova T., Aldebert,P., Althorpe,N., Baker,W., Bates,K., Browne,P., van den Broek,A., Cochrane,G., Duggan,K., Eberhardt,R. et al. (2004) The EMBL Nucleotide Sequence Database. Nucleic Acids Res., 32, D27–D30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudd S., Mewes,H.W. and Mayer,K.F. (2003) Sputnik: a database platform for comparative plant genomics. Nucleic Acids Res., 31, 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rudd S., (2005) Open Sputnik—a database to ESTablish comparitive plant genomics using unsaturated sequence collections. Nucleic Acids Res., 32, D622–D627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tautz D. and Renz,M. (1984) Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res., 12, 4127–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham J., Smith,K., MacKenzie,K., Jorgenson,L., Hackett,C. and Powell,W. (2004) The construction of a genetic linkage map of red raspberry (Rubus idaeus subsp. idaeus) based on AFLPs, genomic-SSR and EST-SSR markers. Theor. Appl. Genet., 109, 740–749. [DOI] [PubMed] [Google Scholar]
- 13.Robinson A.J., Love,C.G., Batley,J., Barker,G. and Edwards,D. (2004) Simple sequence repeat marker loci discovery using SSR primer. Bioinformatics, 20, 1475–1476. [DOI] [PubMed] [Google Scholar]
- 14.Rafalski A. (2002) Applications of single nucleotide polymorphisms in crop genetics. Curr. Opin. Plant Biol., 5, 94–100. [DOI] [PubMed] [Google Scholar]
- 15.Barker G., Batley,J., O'Sullivan,H., Edwards,K.J. and Edwards,D. (2003) Redundancy based detection of sequence polymorphisms in expressed sequence tag data using autoSNP. Bioinformatics, 19, 421–422. [DOI] [PubMed] [Google Scholar]
- 16.Kwok P.Y. and Duan,S. (2003) SNP discovery by direct DNA sequencing. Methods Mol. Biol., 212, 71–84. [DOI] [PubMed] [Google Scholar]
- 17.Weil M.M., Pershad,R., Wang,R. and Zhao,S. (2004) Use of BAC end sequences for SNP discovery. Methods Mol. Biol., 256, 1–6. [DOI] [PubMed] [Google Scholar]
- 18.Apweiler R., Bairoch,A., Wu,C.H., Barker,W.C., Boeckmann,B., Ferro,S., Gasteiger,E., Huang,H., Lopez,R., Magrane,M. et al. (2004) UniProt: the Universal Protein knowledgebase. Nucleic Acids Res., 32, D115–D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts R.J., Vincze,T., Posfai,J. and Macelis,D. (2003) REBASE: restriction enzymes and methyltransferases. Nucleic Acids Res., 31, 418–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rice P., Longden,I. and Bleasby,A. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet., 16, 276–277. [DOI] [PubMed] [Google Scholar]
- 21.Mewes H.W., Amid,C., Arnold,R., Frishman,D., Guldener,U., Mannhaupt,G., Munsterkotter,M., Pagel,P., Strack,N., Stumpflen,V. et al. (2004) MIPS: analysis and annotation of proteins from whole genomes. Nucleic Acids Res., 32, D41–D44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Remm M., Storm,C.E. and Sonnhammer,E.L. (2001) Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol., 314, 1041–1052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data within the database are freely available to the scientific community. In addition to data access through the web interface, markers may be downloaded in an XML format and as an MS Excel compatible tab-delimited files.