Abstract
Within-species sequence variation data are of special interest since they contain information about recent population/species history, and the molecular evolutionary forces currently in action in natural populations. These data, however, are presently dispersed within generalist databases, and are difficult to access. To solve this problem, we have developed Polymorphix, a database dedicated to sequence polymorphism. It contains within-species homologous sequence families built using EMBL/GenBank under suitable similarity and bibliographic criteria. Polymorphix is an ACNUC structured database allowing both simple and complex queries for population genomic studies. Alignments within families as well as phylogenetic trees can be download. When available, outgroups are included in the alignment. Polymorphix contains sequences from the nuclear, mitochondrial and chloroplastic genomes of every eukaryote species represented in EMBL. It can be accessed by a web interface (http://pbil.univ-lyon1.fr/polymorphix/query.php).
INTRODUCTION
Comparative genomics has benefited during the last decade from several specialized databases like HOVERGEN (1) and HOBACGEN (2) to access growing amounts of sequence data. These databases have facilitated the biologists' work by giving access to information on between-species similarity/homology. No such tool, however, exists for within-species polymorphism data, which are currently dispersed within huge generalist databases, making it very difficult to access and re-analyse them.
A polymorphism sequence database would be useful for two main reasons. First, it would help population geneticists to quickly review existing datasets and bibliography in species/taxa of interest. The task of building a complete polymorphism sequence dataset in a taxon of interest (e.g. Drosophila), or for a locus of interest (e.g. cytochrome b) would otherwise typically require several days or weeks. Second, it would make it possible to set up data sets for meta-analyses. Multi-locus analysis is a powerful way to detect adaptation at the population level, and discriminate between selection and demographic effects—selection acts locally in the genome, while genetic drift and migration affect all loci simultaneously (3–6). Large datasets are also required to understand the role of the various forces driving molecular evolution such as mutation, selection, recombination and gene conversion (7–10). Finally, a polymorphism database would allow one to compare amounts and patterns of sequence polymorphism in distinct species, and investigate correlations with species' ecology and life history traits (11,12).
Currently, one can retrieve sequence polymorphism datasets in the PopSet section from the Entrez web interface developed at the NCBI. This database stores alignments submitted by authors, but there is no attempt to cluster sequences in homologous families, and phylogenetic studies are mixed with studies of intra-species polymorphism. Submission uses the Sequin client program. The EBI hosts a similar alignment database, EMBL-Align, accessed by the SRS web interface. Submissions are made by Webin-Align, a web interface. Several databases already exist for genetic polymorphisms. ALFRED (ALlele FREquency Database) stores allelic frequencies, source of the samples and phenotype information, but not sequence alignment (13). It is restricted to human populations and data input relies on authors' submissions. MENDB (14) contains information about sequence variation, primers and PCR conditions for laboratory work. Some basic descriptive statistics are also given, such as intra- and inter-population diversity. Data in MENDB correspond to notes published in Molecular Ecology Notes. Finally, PGDB (Population Genetics Database) is a prototype mitochondrial database (15), but again, relying on authors submissions. None of these databases allow queries that make use of all the information available in EMBL/GenBank.
We now provide Polymorphix, a database dedicated to within-species sequence polymorphism. Polymorphix is an ACNUC database (16) organizing GenBank/EMBL sequences into within-species sequence ‘families’, using both nucleotide similarity and bibliographic criteria. Sequences are aligned within these ‘families’. Outgroups are added, and a phylogenetic tree is built. Polymorphix can be accessed via a client system with a dedicated web interface (http://pbil.univ-lyon1.fr/polymorphix/query.php), or using query_www, an expert web interface allowing complex queries of data on several sequence families databases (17).
METHODS
We used all the eukaryotic genomic DNA sequences in EMBL release 73 after removing the EST, HTG, GSS, SYN, STS and HTC sections. We also removed EMBL entries matching the keywords: ‘tandem repeat’ and ‘satellite repeat’. To build families in a given species, we first extracted all the sequences of length between 100 and 20 000 bp from the species, and performed a similarity search of all sequences against themselves, using the megablast program. Two sequences were clustered into the same family if they shared a >100 bp-long fragment with similarity >80%. Two criteria were used to remove paralogous sequences, and to split paralogous loci into distinct families. First, two sequences were not clustered if there was a mismatch of >50 nt with <80% similarity flanking the megablast match. Such a mismatch is interpreted as evidence that the sequences represent duplicate genes in distinct genomic contexts. Second, a sequence was removed from its family if its bibliographic reference was not shared by at least three other sequences in the family. This aims to focus on data relevant to population genetics by removing sequences coming from, say, genome projects, which may not be allelic.
After clustering, sequences in families were aligned using Mabios (18), a multiple alignment program especially efficient for highly similar sequences. Next, a consensus sequence was built for each alignment, and was used to search for outgroup sequences by performing a blast against the whole EMBL database (current version) minus the EST section. The five highest matches (from distinct species) were selected as outgroups, provided that they shared >78% similarity to the consensus sequence. This 78% threshold is arbitrary. We refrained from including too distant outgroup, which are useless for the goal of orienting polymorphic sites within the ingroup. Outgroup sequences were added without altering the ingroup alignment, by using the profile and quicktree options in Clustalv1.8 (19). A NJ (20) phylogenetic tree (K2P distances) was built for each family.
This treatment was repeated for each species in EMBL—subspecies are considered as the same taxon in Polymorphix. Then the EMBL flat files describing the selected sequences were modified. Several keywords were added to each sequence in Polymorphix, namely the name of the family(ies) it belongs to (e.g. Hsa000001 for family number one in Homo sapiens), and keywords ‘ingroup’ and ‘outgroup’ when appropriate. A sequence can be an ingroup in a family and outgroup in one or several other families. An ACNUC (16) base including the new annotations was finally generated.
An important issue is the problem of updating Polymorphix as new GenBank/EMBL versions are released. Currently, Polymorphix updates are achieved by re-building families de novo for all the species for which new sequences have been added since the previous EMBL version. More elaborate procedures keeping existing families might be required in the future.
POLYMORPHIX CONTENT AND USAGE
Polymorphix database contains 244 million bases, 217 271 sequences and 18 445 bibliographic references. Our clustering procedure has recovered 10 337 families in 5554 eukaryotes species. Polymorphix allows simple or elaborate queries on EMBL/GenBank fields (species names, keywords, etc.), and a blast-assisted selection of families. Alignments (Clustal and MASE formats) and trees can be downloaded. The coding regions of sequences and their ingroup/outgroup status are annotated in the header of MASE (21) alignment files. Trees are less obviously useful than in between-species homology databases, but they help in visualizing potential population structure, distance to outgroup and in checking paralogy. For each family, a short description appears giving the name of the ingroup species and a locus identifier. Users of the elaborate query_www interface have to keep in mind that queries are made on sequences, and not on families. The sequence query result is then projected onto the family space. This means that a family will be selected as soon as at least one sequence of the family satisfies the query. The web interface allows one to select a subset of sequences within a family if required (e.g. ingroup only).
Table 1 shows family distribution among the major lineages of animals, plants and fungi, for the nuclear and organellar genomes. Two tables in Supplementary Material give the detailed family distribution within mammals and angiosperms, respectively. In many animal taxa and mammalian orders, the number of mitochondrial families is higher than the nuclear one, despite the genome size being 2 × 105 times smaller, reflecting the prevalence of mitochondrial DNA as a tool for phylogeographic studies. This is not true for primates and rodentia, because of a greater effort in human and mouse nuclear polymorphism studies, and because of a higher proportion of ‘false’ polymorphic loci in these fully sequenced genomes—e.g. undetected paralogues.
Table 1. Family distribution among living kingdoms and organelles.
| Taxon | Nuclear | Mitochondrion | Chloroplast |
|---|---|---|---|
| Animalia | 2850 | 3784 | |
| Arthropoda | 1052 | 941 | |
| Mollusca | 63 | 225 | |
| Nematoda | 75 | 26 | |
| Annelida | 5 | 10 | |
| Echinodermata | 32 | 59 | |
| Vertebrata | 1519 | 2445 | |
| Chondrichthyes | 17 | 2 | |
| Actinopterygii | 180 | 708 | |
| Amphibia | 22 | 175 | |
| Sauropsida | 90 | 805 | |
| Aves | 63 | 473 | |
| Mammalia | 1210 | 750 | |
| Viridiplantae | 1422 | 14 | 484 |
| Briopsida | 55 | 0 | 42 |
| Moniliformopses (e.g. ferns) | 9 | 0 | 28 |
| Coniferophyta | 39 | 2 | 20 |
| Gnethophyta | 5 | 0 | 1 |
| Gynkgophyta | 1 | 0 | 0 |
| Cycadophyta | 0 | 0 | 0 |
| Angiospermae | 1257 | 12 | 366 |
| Fungi | 1175 | 71 | |
| Ascomycota | 771 | 46 | |
| Basidiomycota | 350 | 24 |
In plants, mitochondrial DNA polymorphism has hardly ever been explored, probably because of technical problems—the plant mitochondrial genome is relatively large, and highly repetitive. Chloroplastic DNA, in contrast, is widely used for polymorphism studies.
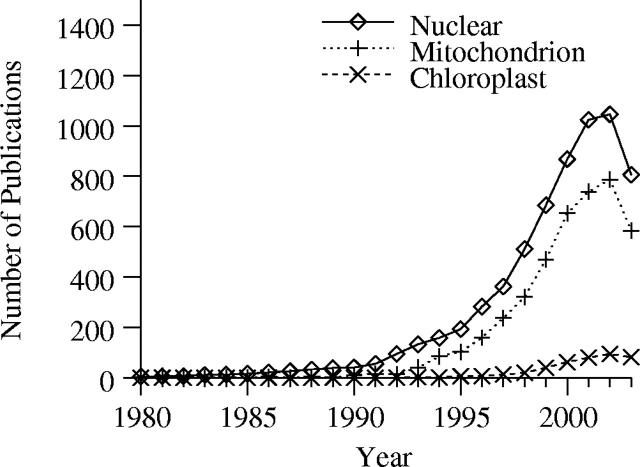
We also plotted the year-wise distribution of sequences publication in Polymorphix (Figure 1) separately for nuclear, mitochondrial and chloroplastic DNA. The number of nuclear and mitochondrial sequence polymorphism publications has been increasing exponentially since 1990. However, in 2003, this number is lower than in 2002, probably because of the delay between sequence submission (usually prior to publication) and update of references by authors in GenBank/EMBL.
Figure 1.
Publication number in Polymorphix as a function of year.
DISCUSSION
The Polymorphix database provides a simple access to sequence polymorphism data for eukaryotic species present in GenBank/EMBL. One important problem when searching for polymorphism data in GenBank/EMBL is to distinguish polymorphic variants from paralogous sequences. In Polymorphix, we used two criteria to try to exclude paralogues: (i) sequence similarity and (ii) bibliographic references. The similarity threshold that we used (>80% identity) is not very stringent, because in some cases, allelic variants can be very divergent (22). But to be included in a family, we require that the bibliographic references of every sequence in the family, is shared with at least three other members of the family. This criteria allows us to exclude most of paralogues and to retain most of allelic variants (except for sequences that are not yet associated with a published article). Note, however, that some paralogous sequences might still be present in Polymorphix families. A manual survey revealed that the proportion of ‘true’ polymorphic loci in Polymorphix is >95% for Drosophila melanogaster nuclear data and mammalian mitochondrial data, but ∼50% for Mus musculus nuclear data. Hence, it is necessary for the user to manually curate the data (sequence annotations, bibliographic references) to exclude paralogues. The web interface provides a number of tools for rapidly checking and manipulating families, including alignments, trees and links to publications.
The main advantage of Polymorphix is that it contains virtually all the polymorphism sequence datasets that were submitted to GenBank/EMBL. Polymorphix will therefore help population geneticists and molecular evolutionary biologists in building exhaustive datasets for meta-analyses. As an illustration, a one-day long manual survey and ‘cleaning’ of Polymorphix was enough to build a dataset of 220 non-coding polymorphic loci in D.melanogaster, a dataset three or four times larger than in published meta-analyses (9,23). Polymorphix can also be useful as a bibliographic tool, e.g. for performing a quick review of the literature of sequence polymorphism in a taxon of interest.
Polymorphix does not include single nucleotide polymorphism (SNP) data, for which species-specific dedicated databases are available (e.g. dbSNP at NCBI). Although typically more voluminous than sequence polymorphism datasets, SNP data are of limited interest for population genetics analyses. This is because (i) they do not provide access to haplotype/allele genealogies, on which many methods are based, (ii) they are available in a small number of taxa and (iii) they do not allow characterization of the molecular evolution of specific genes of interest. However, SNP data are useful for characterizing molecular microevolutionary patterns at the genome level (24,25).
One limitation of Polymorphix is that it does not provide information about the sampling strategy (population subdivision, individual location), or about allele frequencies. These data are crucial for many studies of population genetics, and will have to be obtained from the literature by users prior to data analysis. Some questions, however, can be addressed just with haplotype or allelic type data, such as the use of the McDonald–Kreitman test to detect adaptive evolution (26). It should be noted, however, that the EMBL format supports the submission of this information through the feature key ‘variation’ and the qualifiers ‘/allele=’, ‘/frequency=’ and ‘/country=’. We did not make use of this information because of the low number of entries using these features. We would, however, encourage population geneticists to submit these data, as well as alignments, together with sequences, since this low-cost effort can greatly help in the future re-analysis or meta-analysis of polymorphism sequence data.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Deborah Charlesworth and Sylvain Glemin for critical reading of the manuscript and for their helpful suggestions. We thank Julien Dutheil for providing his Bibliographic manager software for Biologists called B3.
REFERENCES
- 1.Duret L., Mouchiroud,D. and Gouy,M. (1994) HOVERGEN: a database of homologous vertebrate genes. Nucleic Acids Res., 22, 2360–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perriere G., Duret,L. and Gouy,M. (2000) HOBACGEN: database system for comparative genomics in bacteria. Genome Res., 10, 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hudson R., Kreitman,M. and Aguade,M. (1987) A test of neutral molecular evolution based on nucleotide data. Genetics, 116, 153–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galtier N., Depaulis,F. and Barton,N.H. (2000) Detecting bottlenecks and selective sweeps from DNA sequence polymorphism. Genetics, 155, 981–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlötterer C. (2003) Hitchhiking mapping—functional genomics from the population genetics perspective. Trends Genet., 19, 32–38. [DOI] [PubMed] [Google Scholar]
- 6.Mousset S., Brazier,L., Cariou,M.-L., Chartois,F., Depaulis,F. and Veuille,M. (2003) Evidence of a high rate of selective sweeps in African Drosophila melanogaster. Genetics, 163, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akashi H. (1995) Inferring weak selection from patterns of polymorphism and divergence at ‘silent’ sites in Drosophila DNA. Genetics, 139, 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eyre-Walker A. (1999) Evidence of selection on silent site base composition in mammals: potential implications for the evolution of isochores and junk DNA. Genetics, 152, 675–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andolfatto P. and Przeworski,M. (2001) Regions of lower crossing over harbor more rare variants in African populations of Drosophila melanogaster. Genetics, 158, 657–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glinka S., Ometto,L., Mousset,S., Stephan,W. and De Lorenzo,D. (2003) Demography and natural selection have shaped genetic variation in Drosophila melanogaster: a multi-locus approach. Genetics, 165, 1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foltz D.W. (2003) Invertebrate species with nonpelagic larvae have elevated levels of nonsynonymous substitutions and reduced nucleotide diversities. J. Mol. Evol., 57, 607–612. [DOI] [PubMed] [Google Scholar]
- 12.Lynch M. and Conery,J.S. (2003) The origins of genome complexity. Science, 302, 1401–1404. [DOI] [PubMed] [Google Scholar]
- 13.Rajeevan H., Osier,M.V., Cheung,K.H., Deng,H., Druskin,L., Heinzen,R., Kidd,J.R., Stein,S., Pakstis,A.J., Tosches,N.P. et al. (2003) ALFRED: the ALelle FREquency Database. Update. Nucleic Acids Res., 31, 270–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livingstone K. and Rieseberg,L. (2003) MENDB: a database of polymorphic loci from natural populations. Bioinformatics, 19, 663–664. [DOI] [PubMed] [Google Scholar]
- 15.Neigel J.E. and Leberg,P. (2004) A prototype object database for mitochondrial DNA variation. J. Hered., 95, 85–88. [DOI] [PubMed] [Google Scholar]
- 16.Gouy M., Milleret,F., Mugnier,C., Jacobzone,M. and Gautier,C. (1984) ACNUC—a nucleic-acid sequence database and analysis system. Nucleic Acids Res., 12, 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perriere G., Combet,C., Penel,S., Blanchet,C., Thioulouse,J., Geourjon,C., Grassot,J., Charavay,C., Gouy,M., Duret,L. et al. (2003) Integrated databanks access and sequence/structure analysis services at the PBIL. Nucleic Acids Res., 31, 3393–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdeddaim S. (1997) Fast and sound two-step algorithms for multiple alignment of nucleic sequences. Int. J. Artif. Intell. Tools, 6, 179–192. [Google Scholar]
- 19.Higgins D.G., Bleasby,A.J. and Fuchs,R. (1992) CLUSTAL V: improved software for multiple sequence alignment. Comput. Appl. Biosci., 8, 189–191. [DOI] [PubMed] [Google Scholar]
- 20.Saitou N. and Nei,M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol., 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 21.Galtier N., Gouy,M. and Gautier,C. (1996) SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci., 12, 543–548. [DOI] [PubMed] [Google Scholar]
- 22.Kusaba M., Nishio,T., Satta,Y., Hinata,K. and Ockendon,D. (1997) Striking sequence similarity in inter- and intra-specific comparisons of class I SLG alleles from Brassica oleracea and Brassica campestris: implications for the evolution and recognition mechanism. Proc. Natl Acad. Sci. USA, 94, 7673–7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bierne N. and Eyre-Walker,A. (2004) The genomic rate of adaptive amino acid substitution in Drosophila. Mol. Biol. Evol., 21, 1350–1360. [DOI] [PubMed] [Google Scholar]
- 24.Lercher M.J., Smith,N.G.C., Eyre-Walker,A. and Hurst,L.D. (2002) The evolution of isochores: evidence from SNP frequency distributions. Genetics, 162, 1805–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webster M.T. and Smith,N.G.C. (2004) Fixation biases affecting human SNPs. Trends Genet., 20, 122–126. [DOI] [PubMed] [Google Scholar]
- 26.Mcdonald J.H. and Kreitman,M. (1991) Adaptive protein evolution at the Adh locus in Drosophila. Nature, 351, 652–654. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.