Abstract
MITOMAP (http://www.MITOMAP.org), a database for the human mitochondrial genome, has grown rapidly in data content over the past several years as interest in the role of mitochondrial DNA (mtDNA) variation in human origins, forensics, degenerative diseases, cancer and aging has increased dramatically. To accommodate this information explosion, MITOMAP has implemented a new relational database and an improved search engine, and all programs have been rewritten. System administrative changes have been made to improve security and efficiency, and to make MITOMAP compatible with a new automatic mtDNA sequence analyzer known as Mitomaster.
BACKGROUND
MITOMAP is a comprehensive database of human mitochondrial DNA (mtDNA) variation and its relationship with human evolution and disease. The data content and operational tools of MITOMAP have been expanded over the past five years, as the importance of mtDNA variation to human health has become increasingly apparent.
The mtDNA is a closed circular molecule of 16 569 nt. The numbering system of the ‘Cambridge’ reference sequence permits all the information in MITOMAP to be interrelated through the affected nucleotide (1). The mtDNA codes for 37 genes, including a 12S and 16S rRNA, 22 tRNAs and 13 essential genes for oxidative phosphorylation (OXPHOS) polypeptides. In addition, the mtDNA contains a 1000 nt control region that encompasses transcription and replication regulatory elements (2).
Mitochondria OXPHOS provides most of the cellular energy, generates much of the endogenous reactive oxygen species (ROS) and regulates apoptosis through the mitochondrial permeability transition pore (mtPTP). Each cell contains hundreds to thousands of cytosolic, and thus maternally inherited, mtDNAs. Because of its exposure to ROS, the mtDNA has a very high mutation rate, and new mutations create a mixture of mutant and normal mtDNAs within the cell, called heteroplasmy. As the percentage of mutant mtDNAs increases, mitochondrial energy production declines and ROS increases. Increased ROS acts as a mitogen, but excessive ROS together with reduced energy production can lead to apoptosis (2).
Human mtDNA variation correlates with the geographic origin of the population harboring the mtDNA. This is because mtDNAs harbor ancient polymorphisms that permitted our ancestors to adapt to the increasing cold encountered as they migrated out of Africa and into temperate Eurasia and arctic Siberia [MXB1]. Today, these same variants are known to affect human longevity and predisposition to neurodegenerative diseases such as Alzheimer's disease (AD) and Parkinson's disease (PD) (3).
More recent germline mtDNA mutations have been linked to a wide range of degenerative diseases, preferentially affecting the central nervous system, heart, muscle, renal and endocrine systems. Pathogenic mtDNA mutations include mtDNA rearrangements, base substitution mutations in the coding region and also control-region mutations (2). Somatic mtDNA rearrangement and base substitution mutations have also been associated with aging (2). Indeed, different tissues preferentially accumulate specific control region mutations, suggesting that these mutations may affect important regulatory elements (4,5). This hypothesis has been supported by the recent discovery that somatic mutations in key control-region regulatory elements are elevated in AD brains (6).
Finally, mtDNA coding region and control-region mutations are being found in a wide spectrum of cancers (7). Increasingly, there is an evidence that these mtDNA play an important role in neoplastic transformation (8).
The extraordinary array of clinically relevant mtDNA variants makes it more important than ever that an authoritative database be maintained on human mtDNA variation. Moreover, it is now clear that seemingly unrelated mtDNA population variation, age-related somatic mutations and inherited pathogenic mutations can all contribute to an individual's predisposition to disease. Hence, being able to rapidly make the appropriate associations via the mtDNA sequence of ancient variants and recent mutations is critical for the proper application of mtDNA analysis to forensics, anthropology, and clinical and personalized medicine.
DATA
In MITOMAP, the location of each gene and regulatory-functional element is defined by its beginning and ending nucleotide positions. Descriptions of the gene functions and access to relevant references on the crystal structures of the various respiratory complexes, when available, are provided. A list of all currently known animal mtDNA sequences is also maintained, which permits the alignment of the nucleotide and amino acid sequence of each gene, permitting determination of the evolutionary conservation of population and disease associated amino acid sequence variants. A data file of the diagnostic nucleotide sequence variants that define the various population-specific mtDNA haplogroups is also maintained, as is a complete list of all known sequence polymorphisms.
MITOMAP also maintains a compendium of all known pathogenic mtDNA mutations. These are divided into four classes of mutations, each associated with its respective disease phenotype designation. The pathogenic mutation classes are as follows: polypeptide mutations, protein synthesis (rRNA and tRNA) mutations, rearrangement mutations and control region mutations.
The amount of curated mtDNA data has grown enormously in the past six years since the previous Nucleic Acids Research report. Just in the past two years (September 2002–September 2004), the number of protein-coding gene mutations has increased 44%, the number of protein synthesis mutations has increased 58% and the number of somatic mutations has increased 552% (see Table 1).
Table 1. MITOMAP data.
| 1994 | September 15, 2002 | September 13, 2004 | 2002–2004 # Increase | % Increase | |
|---|---|---|---|---|---|
| References | 629 | 2030 | 2944 | 914 | 45.02 |
| Polymorphisms | 743 | 1062 | 1532 | 470 | 44.26 |
| mRNA mutations | 25 | 59 | 93 | 34 | 57.63 |
| rtRNA mutations | 29 | 87 | 98 | 11 | 12.64 |
| Deletions | 77 | 97 | 106 | 9 | 9.28 |
| Multiple deletions | 64 | 69 | 70 | 1 | 1.45 |
| Insertionsa | 11 | 5 | 6 | 1 | 20.00 |
| Rearrangements | 0 | 7 | 8 | 1 | 14.29 |
| Somatic mutations | 0 | 21 | 137 | 116 | 552.38 |
| Unpublished polymorphisms | 0 | 205 | 648 | 443 | 216.10 |
aInsertions data were divided into Rearrangements and Insertions in 1995.
DATABASE
All of the data for MITOMAP is now managed in PostgreSQL (http://www.postgresql.org/), an open-source database management system (i.e. DBMS) with object-relational modeling capabilities. The Georgia Tech Emory Networked Object Management Environment (i.e. GENOME), used in the previous version of MITOMAP, was an attempt to overcome data-modeling limitations inherent in the DBMS initially available to MITOMAP (9). However, in the intervening years, an assortment of DBMSs with mature data-modeling capabilities have become available which combine aspects of both relational and object-oriented modeling. To accommodate the transition in DBMS, moderate changes were made to the data schema so that it would adhere more strictly to the principles governing relational design. These changes greatly increased the ease of extending MITOMAP to include new types of data and to be integrated with other systems.
Data management for MITOMAP is now performed in two steps. New data extracted by the curator is initially stored, formatted and screened in Excel spreadsheets. Updated versions of the curated data are then periodically uploaded to the server, where Perl programs implementing the ParseExcel and DBI modules are used to parse the values from the spreadsheets and populate the data structures implemented in PostgreSQL. Error screening is performed both by the curator and by the population programs.
The hardware/server foundation of MITOMAP has also been updated. Concurrent, with the relocation of the MITOMAP servers to the University of California, Irvine, MITOMAP has been moved to a dual-processor server running RedHat Linux as its operating system. A second server has also been added to handle the new systems being integrated with MITOMAP.
INTERFACE
MITOMAP implements a dynamic web interface that allows researchers to browse its collection of mtDNA variation and references. Simple searches may be performed on all of the data, while advanced search options are available for searching through the nearly 3000 references of mitochondrial related literature.
The search engine, CGI scripts and all supporting programs of MITOMAP were rewritten in the latest version of the Perl programming language. The previous version of MITOMAP had become a collection of programs in various languages, making adjustments difficult. The new CGI scripts utilize the CGI.pm module (http://stein.cshl.org/WWW/software/CGI/) for ease of development and maintainability (10). The Apache web server was also updated and configured to use modperl, an embedded Perl interpreter, for better security and performance.
EXPERT SYSTEM
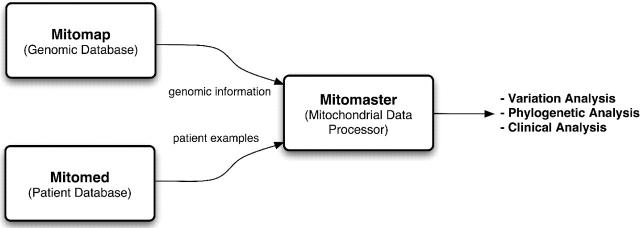
The functionality of MITOMAP is being extended by integration with an analytical system (see Figure 1), called Mitomaster. Mitomaster provides a component for processing and analyzing biological information related to mitochondria, and uses the information stored in MITOMAP as an important part of its knowledgebase. While MITOMAP is a browsable collection of information managed by a curator, Mitomaster is designed to use this information for the analysis of new information submitted by researchers. A clinical database, Mitomed, is also being designed to integrate with these systems.
Figure 1.
Integration of MITOMAP with the analytical system Mitomaster and a clinical database called Mitomed.
CONCLUSION
MITOMAP has fulfilled its original goal of integrating various domains of information related to the mitochondrial genome. However, the proliferation of biological databases and accumulation of vast quantities of data have created new problems of system integration and data analysis. The changes outlined here address these new challenges and will allow MITOMAP to continue to serve as a model system. It is reasonable to expect that as the volume and complexity of information increases in other locus-specific databases, they will benefit from similar efforts at integration and expansion.
Acknowledgments
ACKNOWLEDGEMENTS
This research has been supported by NIH grants NS213L8, HL64017, AG13154 and an Ellison Foundation Senior Investigator Grant (D.C.W.); and NIH Biomedical Informatics Training Grant T15 LM007443, NSF grant EIA-0321390 and a UCI Laurel Wilknening Faculty Innovation Award (P.B.).
REFERENCES
- 1.Anderson S., Bankier,A.T., Barrell,B.G., de Bruijn,M.H., Coulson,A.R., Drouin,J., Eperon,I.C., Nierlich,D.P., Roe,B.A., Sanger,F. et al. (1981) Sequence and organization of the human mitochondrial genome. Nature, 290, 457–465. [DOI] [PubMed] [Google Scholar]
- 2.Wallace D.C. and Lott,M.T. (2002) Mitochondrial genes in degenerative diseases, cancer and aging. In Rimoin,D.L., Connor,J.M., Pyeritz,R.E. and Korf,B.R. (ed.), Emery and Rimoin's Principles and Practice of Medical Genetics. Churchill Livingstone, London, pp. 299–409. [Google Scholar]
- 3.Ruiz-Pesini E., Mishmar,D., Brandon,M., Procaccio,V. and Wallace,D.C. (2004) Effects of purifying and adaptive selection on regional variation in human mtDNA. Science, 303, 223–226. [DOI] [PubMed] [Google Scholar]
- 4.Michikawa Y., Mazzucchelli,F., Bresolin,N., Scarlato,G. and Attardi,G. (1999) Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science, 286, 774–779. [DOI] [PubMed] [Google Scholar]
- 5.Coskun P.E., Ruiz-Pesini,E. and Wallace,D.C. (2003) Control region mtDNA variants: longevity, climatic adaptation, and a forensic conundrum. Proc. Natl Acad. Sci. USA, 100, 2174–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coskun P.E., Beal,M.F. and Wallace,D.C. (2004) Somatic mitochondrial DNA control region mutations are prevalent in Alzheimer Disease brains. Proc. Natl Acad. Sci. USA, 101, 10726–10731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Copeland W.C., Wachsman,J.T., Johnson,F.M. and Penta,J.S. (2002) Mitochondrial DNA alterations in cancer. Cancer Invest., 20, 557–569. [DOI] [PubMed] [Google Scholar]
- 8.Horton T.M., Petros,J.A., Heddi,A., Shoffner,J., Kaufman,A.E., Graham,S.D.,Jr, Gramlich,T. and Wallace,D.C. (1996) Novel mitochondrial DNA deletion found in a renal cell carcinoma. Genes Chromosomes Cancer, 15, 95–101. [DOI] [PubMed] [Google Scholar]
- 9.Kogelnik A.M., Lott,M.T., Brown,M.D., Navathe,S.B. and Wallace,D.C. (1998) MITOMAP: a human mitochondrial genome database—1998 update. Nucleic Acids Res., 26, 112–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stein L. (1998) Official Guide to Programming with CGI.pm. John Wiley & Sons, Inc., NY. [Google Scholar]