Abstract
Fruits are the defining feature of angiosperms, likely have contributed to angiosperm successes by protecting and dispersing seeds, and provide foods to humans and other animals, with many morphological types and important ecological and agricultural implications. Rosaceae is a family with ∼3000 species and an extraordinary spectrum of distinct fruits, including fleshy peach, apple, and strawberry prized by their consumers, as well as dry achenetum and follicetum with features facilitating seed dispersal, excellent for studying fruit evolution. To address Rosaceae fruit evolution and other questions, we generated 125 new transcriptomic and genomic datasets and identified hundreds of nuclear genes to reconstruct a well-resolved Rosaceae phylogeny with highly supported monophyly of all subfamilies and tribes. Molecular clock analysis revealed an estimated age of ∼101.6 Ma for crown Rosaceae and divergence times of tribes and genera, providing a geological and climate context for fruit evolution. Phylogenomic analysis yielded strong evidence for numerous whole genome duplications (WGDs), supporting the hypothesis that the apple tribe had a WGD and revealing another one shared by fleshy fruit-bearing members of this tribe, with moderate support for WGDs in the peach tribe and other groups. Ancestral character reconstruction for fruit types supports independent origins of fleshy fruits from dry-fruit ancestors, including the evolution of drupes (e.g., peach) and pomes (e.g., apple) from follicetum, and drupetum (raspberry and blackberry) from achenetum. We propose that WGDs and environmental factors, including animals, contributed to the evolution of the many fruits in Rosaceae, which provide a foundation for understanding fruit evolution.
Keywords: coalescence, fruit evolution, molecular clock, nuclear phylogeny, Rosaceae, genome duplication
Introduction
Angiosperms (flowering plants) differ from all other plants by producing seeds within the fruits, which serve to protect developing seeds and also facilitate seed dispersal, by animals, wind, or water (Ridley 1930; Seymour et al. 2013). The fruit wall insulates the tender young seeds from harsh environments, protecting the seeds against damage from pathogens, water loss, and other stresses. Fruits of many plants are highly nutritious and are consumed by animals, long before there were humans; the fruits collected or incompletely eaten by animals are often transported away from the plants that produce them (Fleming and Kress 2011). Sometimes, undigested seeds are excreted by the animals and allowed to germinate subsequently (Ridley 1930). Some fruits develop appendages that attach to passing animals, and are carried elsewhere. Many plants produce fruits with extensions such as wings (as in maple) or pappus (as in Asteraceae members) that allow easy dispersal by wind. Plants like coconut produce fruits that can travel along currents to distant coasts. The protection provided by the fruits and the increased seed dispersal due to fruit characteristics have likely contributed to the unparalleled successes of angiosperms, totaling over 300,000 extant species (Fuentes and Vivian-Smith 2009; Judd et al. 2015; Spooner 2016). In addition, humans have benefited greatly from fruits of both wild and cultivated species, including many fleshy fruits, such as apple, peach, orange, melon, grapes, and date palm, but also dry fruits such as grains, beans, and chestnuts. Therefore, investigating fruit evolution is important for the understanding of the evolution of angiosperms, and can impact other fields such as ecology and agriculture.
In some angiosperm families, such as the Brassicaceae (cabbage and relatives), Fabaceae (legumes), Poaceae (grasses), and Vitaceae (grape), different species of the same family produce relatively similar fruits belonging to the same types with variation in size and shape, whereas other families contain species producing a few fruit types. In contrast, Rosaceae species produce several highly distinctive types of fruits (Potter et al. 2007a; Phipps 2014) (fig. 1), including fleshy pomes (with a relatively soft core and multiple seeds: apple, pear), drupes (with a hard central shell and a single seed: peach, plum, cherry), and dry achene (with a thin wall and a single seed). Furthermore, some species produce aggregate fruits such as drupetum (a group of tiny drupelets loosely attached to a central structure, as in raspberry), achenetum (multiple achenes from a single flower), sometimes with a fleshy enlarged receptacle (strawberry) or an enveloping hypanthium (fused lower portions of the sepals, petals and stamens, as in rose), and follicetum (several pod-like structures each with one or more seeds, from a single flower). In particular, Rosaceae members produce many economically important fruits (apple, pear, peach, plum, cherry, strawberry, and raspberry), which have increased in size and sweetness due to human domestication efforts. Therefore, Rosaceae provide an excellent system to conduct comparative and evolutionary studies of fruits.
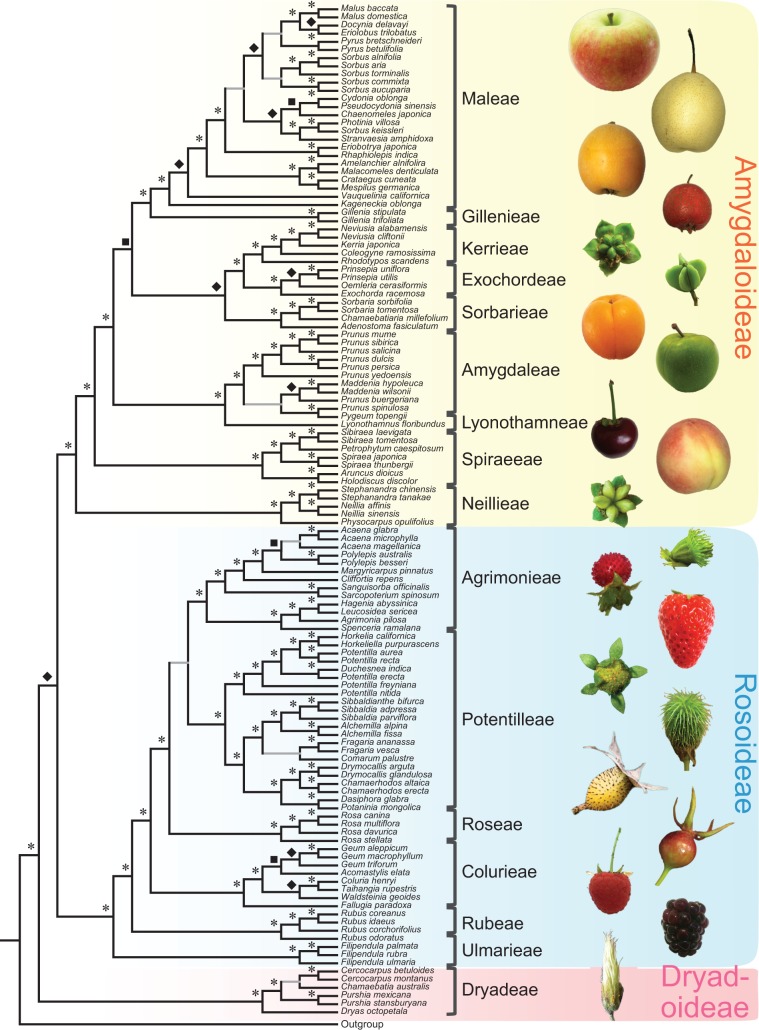
Fig. 1.
A summary of Rosaceae phylogeny and Rosaceae fruit morphologies. On the left is a summary tree with results from five coalescence analyses of 882, 571, 444, 256, and 113 gene sets, respectively, and a concatenation analysis using the 113-gene supermatrix. Topologies consistent in all six trees are drawn in black lines. Grey lines show uncertain relationships, with some trees support the topology. For further information see figure 3 and supplementary figure S14, Supplementary Material online. Asterisks (*) indicate 100% supports in all six trees. Diamonds indicate more than 90% supports in at least five trees and more than 85% supports in all six trees. Squares indicate more than 80% supports in at least three trees and more than 40% supports in all six trees. Plant photographs on the right show the diversity of Rosaceae fruits. The left row (from the top) includes Malus pumila (apple), Eriobotrya japonica (loquat), Kerria japonica, Prunus armeniaca (almond), Prunus sp. (cherry), Spiraea thunbergii, Duchesnea indica, Potentilla supina, Rosa laevigata, Rubus sp. (raspberry), and Dryas octopetala. The right row (from the top) includes Pyrus bretschneideri (pear), Crataegus pinnatifida, Exochorda racemosa, Prunus salicina (plum), Prunus persica (peach), Agrimonia pilosa, Fragaria × ananassa (strawberry), Geum aleppicum, Rosa sp., and Rubus fruticosus (blackberry).
Rosaceae is a moderately large angiosperm family in the order Rosales, with about 3000 species, 3 subfamilies, 16 tribes, and 88–100 genera (Hummer and Janick 2009; Phipps 2014). The family has a worldwide distribution, with particular diverse presence in Northern Hemisphere temperate forests, in which woody Rosaceae members are important forest trees, providing habitats and foods for birds, mammals, and other animals (Hummer and Janick 2009). In addition to important commercial fruit species, Rosaceae also include many ornamental flowers (roses, meadowsweets, hawthorns, crabapples, and rowans). Furthermore, five Rosaceae genomes have been published, including woodland strawberry (Fragaria vesca), domesticated apple (Malus × domestica), pear (Pyrus bretschneideri), peach (Prunus persica), and Mei (Prunus mume, related to apricot) (Velasco et al. 2010; Shulaev et al. 2011; Zhang et al. 2012b; Verde et al. 2013; Wu et al. 2013), providing important resources for comparative analyses.
Rosaceae species are classified into three subfamilies (Potter et al. 2007a), with two large ones, Rosoideae and Amygdaloideae, having ∼2000 and ∼1000 species, respectively, and a small one, Dryadoideae, with fewer than 30 species. Previously, largely according to fruit and other morphological characteristics, Rosaceae were divided into four subfamilies, Rosoideae (s. l., with aggregate fruits), Maloideae (= tribe Maleae/Pyreae with over 30 genera and at least 500 species including apple/pear and relatives with pomes; Maleae hereafter for simplicity), Amygdaloideae (s. s. = tribe Amygdaleae, with drupes), and Spiraeaoideae (Schulze-Menz 1964). Recent molecular analyses support the separation of the former Rosoideae (s. l.) into Rosoideae (s. s.) and Dryadoideae, and the combination of the previous Maloideae, Amygdaloideae (s. s.), and Spiraeaoideae into the current Amygdaloideae (s. l.) (Morgan et al. 1994; Potter et al. 2007a; Chin et al. 2014; Phipps 2014). To avoid confusion, we will use Rosoideae, Amygdaloideae and Dryadoideae in the current sense, and use the tribal names Maleae and Amygdaleae to refer to the respective groups, rather than using the older subfamily names. This chloroplast-gene based system of three subfamilies supports complex evolution of Rosaceae fruits, with multiple fruit types in each of several clades (Potter et al. 2007a); however, the unresolved relationships among tribes and genera have hindered the understanding of evolutionary history of fruit types in Rosaceae.
Importantly, the species richness of Rosaceae could be partly related to polyploidization and species radiation in the family history, with evidence for polyploidy events in the two larger subfamilies (Talent and Dickinson 2005; Vamosi and Dickinson 2006; Dickinson et al. 2007; Rousseau-Gueutin et al. 2009; Lo et al. 2010; Schmidt-Lebuhn et al. 2010; Considine et al. 2012; Burgess et al. 2014; Fougere-Danezan et al. 2015). In particular, the ancestor of Maleae was proposed to be a hybrid of the ancestors of the Spiraeoideae and the Amygdaleae, in part because all Maleae members have a base chromosome number of 17, with an exception of an early branching genus Vauquelinia having a base chromosome number of 15, whereas the putative parents have chromosome number of 9 and 8 for Spiraeoideae and Amygdaleae, respectively (Robertson et al. 1991). More recently, an alternative hypothesis was proposed that Maleae originated with a whole-genome duplication (WGD) event in a relative of the ancestor of Gillenia (x = 9) (Evans et al. 2000; Evans and Campbell 2002; Velasco et al. 2010; Verde et al. 2013). However, the possible occurrence and timing of other WGDs are unclear. Therefore, investigation of the polyploidy/WGD events will contribute to the understanding of Rosaceae evolution.
Both the study of fruit evolution and analysis of WGDs depend on a well-resolved phylogeny of Rosaceae. Specifically, the subfamily Amygdaloideae alone has multiple tribes, with different fruit types, such as pome, drupe, and follicetum; however, the phylogenetic relationships among the tribes are unclear (Potter et al. 2007a). In the first molecular phylogenetic study of Rosaceae based on rbcL sequences, Maleae (e.g., apples and pears), Amygdaleae (e.g., peaches and plums), and Rosoideae (e.g., roses, strawberries, raspberries, and others) are monophyletic clades but the previous Spiraeoideae were not monophyletic, with several distinct clades (Morgan et al. 1994). Additional phylogenetic studies have provided valuable information for subgroups in Rosaceae, including the tribes Maleae, Amygdaleae, Spiraeeae, and Potentilleae, and the genera Geum, Potentilla, Rosa, Rubus, Neillia, and Prunus (Alice and Campbell 1999; Lee and Wen 2001; Smedmark and Eriksson 2002; Aldasoro et al. 2005; Campbell et al. 2007; Potter et al. 2007b; Gehrke et al. 2008; Lundberg et al. 2009; Dobes and Paule 2010; Lo and Donoghue 2012; Oh 2013; Chin et al. 2014; Zhu et al. 2015). However, relationships among Rosaceae tribes and genera remain unclear, in part because of the polyploidy events and rapid separation/diversification among some clades.
In recent years, phylogenetics using dozens or more nuclear genes have been successful for reconstructing phylogeny of difficult angiosperm groups with rapid radiations, such as those of five major angiosperm lineages, of Brassicaceae, and of Caryophyllales (Zhang et al. 2012a; Zimmer and Wen 2013; Zeng et al. 2014; Yang et al. 2015; Huang et al. 2016). The rapid advances of high-throughput sequencing technologies have greatly facilitated transcriptome sequencing, allowing numerous phylogenetic markers to be identified (Wen et al. 2015; Zimmer and Wen 2015). Furthermore, nuclear genes provide information from bi-parental inherence and allow the detection of WGDs and other events. To reconstruct the Rosaceae phylogeny, we sequenced 123 transcriptomes (115 Rosaceae species and 8 other Rosales species), representing all Rosaceae tribes and nearly all multi-species genera. Multiple datasets of low-copy putative orthologous genes were identified and then used for phylogenetic reconstruction, resulting in a highly supported Rosaceae phylogeny, which is consistent with well resolved relationships obtained previously and presents newly resolved relationships. The nuclear gene datasets were also used for molecular clock estimates of the origin of the crown Rosaceae at the boundary of Early and Late Cretaceous (∼100 Ma), and those of the subfamilies and major tribes (between Late Cretaceous to Eiocene), providing possible geological timeframe and climate conditions for the diversification of tribes and genera. In addition, ancestral character reconstructions of fruit types and other morphologies were performed, allowing the proposal of the origins and histories of multiple fleshy fruit types from dry fruit types. Specifically, drupes evolved from follicetum at least twice, once for Prunus (peach, plum and their relatives) and the other for Prinsepia; pomes (apple, pear, and other fleshy-fruited members of Maleae) evolved from coccetum, which was in turn derived from follicetum; multiple drupelets from the same flower also evolved at least twice, from follicetum (nuculanium, for Rhodotypos, with a few drupelets) and from achenetum (drupetum, for Rubus, with many drupelets). The newly resolved Rosaceae phylogeny also allows the placement of WGD events, including two near the origin of Maleae, presenting a comprehensive understanding of Rosaceae fruit evolution in the context of dramatic genome changes and geological times with changing environments.
Results and Discussion
Taxon Sampling and Transcriptomes for Gene Marker Identification
In this study, a total of 124 Rosaceae species were included (supplementary table S1, Supplementary Material online), covering all three subfamilies (6 Dryadoideae, 54 Rosoideae, 64 Amygdaloideae) and 16 tribes in Rosaceae, as well as nearly all multi-species genera, which represented more than 98% of Rosaceae species. For genera that were reported to be nonmonophyletic (Potentilla and Sorbus), species representing more than one clade were sampled. For tribes (Roseae, Rubeae, and Ulmarieae) with only one genus and uncertain relationships with other tribes, more than one species were sampled. In addition, 24 other angiosperm species were used as outgroups (supplementary table S1, Supplementary Material online), including 8 for other families in Rosales and 7 in Fagales, an order relatively close to Rosales within Rosids, as well as others from four orders (Saxifragales, Caryophyllales, Ranunculales, and Laurales) with varying evolutionary distances from Rosaceae (The Angiosperm Phylogeny Group 2016).
Among the 124 Rosaceae datasets, 115 were new transcriptome datasets generated for this study (6 Dryadoideae, 50 Rosoideae, and 59 Amygdaloideae) (supplementary table S2, Supplementary Material online); for outgroups, transcriptomes for 8 species in other Rosales families were also generated (supplementary table S2, Supplementary Material online). For these transcriptomes, young leaves and/or floral buds and/or fruits were sampled and used for total RNA isolation. Transcriptomes with 11,057,450–102,770,184 reads were obtained using the Illumina technologies and the reads for each species were assembled into 20,532–67,177 contigs of cDNA sequences (supplementary table S2, Supplementary Material online). The contigs have median lengths of 726–1632 nucleotides, with N50 values ranging from 510 to 1095 nucleotides. In addition to the new transcriptomes, five genome sequence datasets and two Sequence Read Archive transcriptome datasets were retrieved for Rosaceae species from public databases. Moreover, newly generated shotgun genomic sequences for two Rosoideae species (Cliffortia repens and Hagenia abyssinica) were also included.
Selection of Low-Copy Candidate Orthologous Nuclear Genes
Low-copy candidate orthologous genes were identified for use as phylogenetic markers (for details see “Materials and Methods” section). To avoid possible biases of specific gene sets, candidate marker genes were identified using two approaches. The first approach began with a set of 931 candidate orthologous genes previously identified for a study of an angiosperm deep phylogeny (Zeng et al. 2014) (see “Materials and Methods” section), with low copy nuclear genes shared by nine angiosperm species (Arabidopsis thaliana, Populus trichocarpa, Glycine max, Medicago truncatula, Vitis vinifera, Solanum lycopersicum, Oryza sativa, Sorghum bicolor, and Zea mays), among which Arabidopsis thaliana, Populus trichocarpa, Glycine max, and Medicago truncatula represent taxa in Rosids. These 931 genes were then filtered using (1) a minimal length of 1000 base pairs (bps) in four sequenced genomes in Rosaceae and an outgroup species and (2) the presence in six or more of 10 selected taxa (eight Rosaceae members plus two outgroup species), yielding 546 orthologous groups (OGs) (see “Materials and Methods” section).
It was reported that Rosaceae members had undergone polyploidization events (Vamosi and Dickinson 2006; Zhao et al. 2016), resulting in many duplicate genes. If the duplicate genes subsequently returned to single-copy status due to independent losses of distinct paralogues in different lineages, the remaining genes are referred to as hidden paralogues. To avoid such hidden paralogues, we constructed gene phylogenies of the above-mentioned 546 OGs using sequences from eight representative Rosaceae species with well-supported relationships and two outgroup species (see “Materials and Methods” section). The OGs with single gene trees whose well-supported topologies were not contradictory to the known species relationships were retained, yielding 407 candidate orthologous genes.
Our second approach started from a set of 3863 single copy genes shared by three Rosaceae species with sequenced genomes (Fragaria vesca, Prunus persica, and Prunus mume) and Cucumis sativus in Cucurbitaceae, a family closely related to Rosaceae. To avoid selecting the genes from the first approach again, those in the previous set were not considered further in this approach. After filtering with minimal length of 1000 bps in genes from Cucumis sativus, 2124 genes were retained. Considering that Maleae has undergone at least one recent lineage-specific WGD, marker genes with only one or two copies in Malus × domestica and Pyrus bretschneideri were retained from among the 2124 genes, ultimately yielding 475 genes. The two approaches yielded a total of 882 genes (supplementary table S3, Supplementary Material online); when these genes were used to reconstruct the Rosaceae phylogeny using the coalescence method (see below), all subfamilies and tribes were monophyletic.
Because much of the molecular phylogenetic analysis has been performed using concatenation of multiple genes (supermatrices) and such datasets are needed for molecular clock estimates, we also wanted to use this approach. It has been argued that the use of supermatrices of hundreds of genes with methods such as maximum likelihood (ML) could produce highly supported but wrong topologies (Seo 2008). Furthermore, the use of relatively small numbers of genes can save computational time and facilitate the inclusion of additional taxa. Thus, selection of subsets of genes were made by examining gene tree topologies for hidden paralogues using memberships in the same order (including outgroups), Rosaceae subfamily, and tribe, successively trimming to subsets of 571, 444 and 256 genes, respectively (see “Materials and Methods” section). For even a smaller subset, further selections were made using topological information of four groups (see “Materials and Methods” section) within Maleae to avoid possible hidden paralogues, resulting in 113 genes, which are less prone to systematic errors when using maximum likelihood analyses with supermatrices. In addition, among the 475 genes from the second approach, 163 genes were selected after eliminating hidden paralogues using gene trees from 31 species, including 25 in Maleae and 6 other Rosaceae members, for relationships within Maleae (see “Materials and Methods” section).
A Well Resolved Rosaceae Phylogeny Supported by Multiple Analyses
To reconstruct the Rosaceae phylogeny, we used multiple datasets to minimize possible biases of specific genes. First, we used the combined 882-gene set with the coalescence method implemented in ASTRAL v4.4.4 (Mirarab et al. 2014), yielding a tree with strong support for most nodes (fig. 1; supplementary fig. S1, Supplementary Material online). Largely consistent results were also obtained using the coalescence method with the 571, 444, 256, and 113 gene sets (fig. 1; supplementary figs. S2–S5, Supplementary Material online). A maximum likelihood (ML) analysis was also performed with the 113-gene supermatrix (fig. 2), because the concatenation method with relatively small number of genes has less risk of systemic errors than those using much larger numbers of genes, such as several hundreds. The relationships for the vast majority of taxa in the trees from the six analyses are consistent, as summarized in figure 1, although there are some differences between the trees for a few taxa (see below for more details).
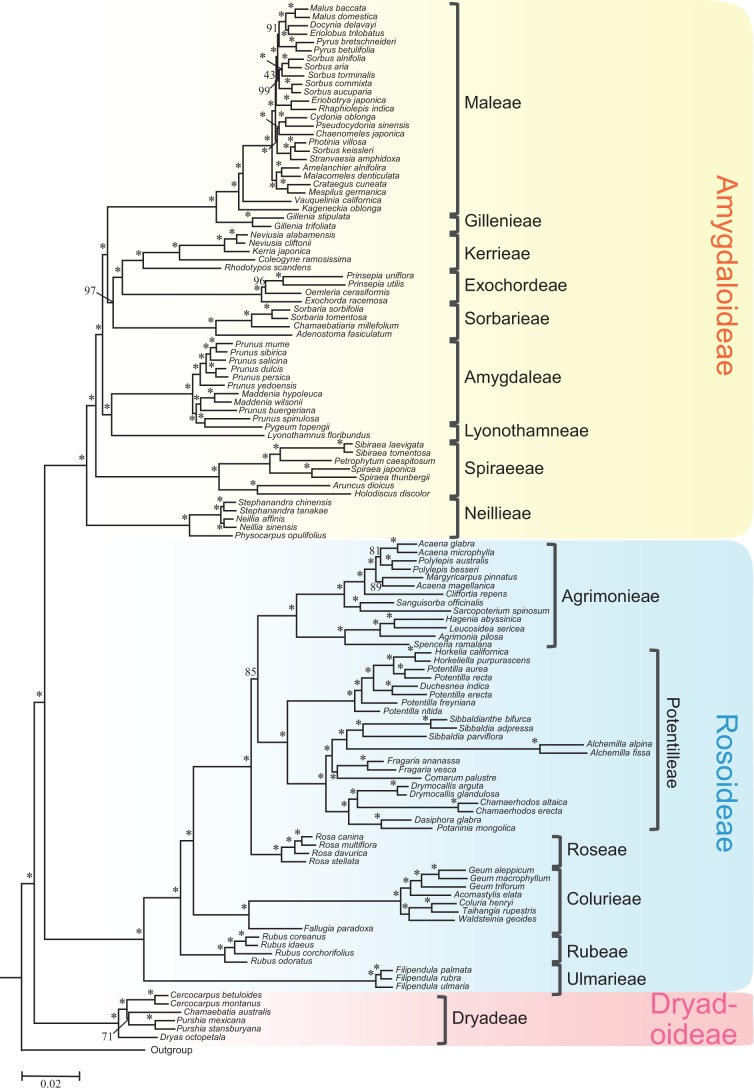
Fig. 2.
A phylogeny from ML analysis using a dataset of concatenated 113 gene sequences. Numbers associated with nodes indicate bootstrap values obtained by Maximum Likelihood (ML) analyses. Asterisks (*) indicate 100% support.
We also obtained a 797-gene dataset after removing some genes that placed outgroup species within a clade of Rosaceae species and used these genes to produce a phylogeny with the coalescence method (supplementary fig. S6, Supplementary Material online). In addition, we reconstructed phylogenies using the 407 (supplementary fig. S7, Supplementary Material online) and 475 (supplementary fig. S8, Supplementary Material online) genes obtained from the first and the second approaches, respectively. To test the effectiveness of smaller gene sets, we have divided the 407 (into 241 and 166 genes) and 475 (into 312 and 163 genes) genes into smaller sets and used them for additional phylogenies (supplementary figs. S9–S12, Supplementary Material online). These phylogenies are largely consistent with that shown in figure 1, but do not always support those relationships that are still somewhat uncertain (fig. 3; see below for more details and discussion).
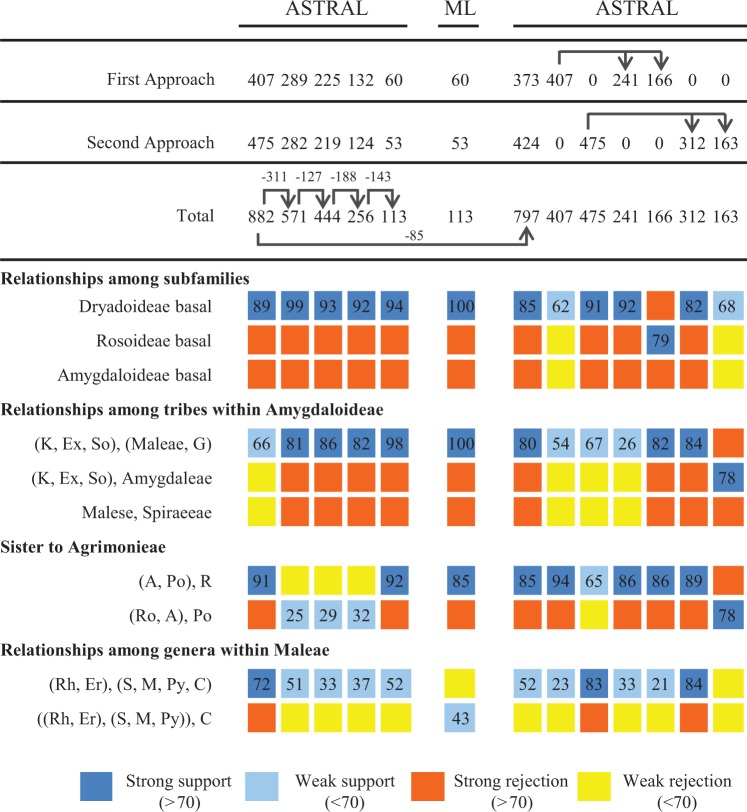
Fig. 3.
A summary of alternative topologies in results from 12 coalescence analyses and one concatenation analysis. At the top are phylogenetic methods and numbers of genes in various sets and relationships between gene sets (see “Results” and “Materials and Methods” sections, and supplementary fig. S24, Supplementary Material online for additional information). The column on the left indicates possible topologies. Designations: Dryadoideae basal: Dryadoideae at the basal position, as sister to the combined clade of Amygdaloideae and Rosoideae, others similarly. K: Kerrieae. Ex: Exochordeae. So: Sorbarieae. G: Gillenieae. A: Agrimonieae. Po: Potentilleae. Ro: Roseae. Rh: Rhaphiolepis. Er: Eriobotrya. S: a combined clade of Sorbus alnifolia, Sorbus aria, Sorbus torminalis, Sorbus commixta, and Sorbus aucuparia. M: a clade including Malus baccata and its three nearest relatives (as shown in fig. 1). Py: Pyrus. C: Cydonia and its five nearest relatives (as in fig. 1). Number in each square refers to values in support of a particular topology indicated in the left column. Strong support refers to support values of at least 70%. Weak support refers to values less than 70%. If there is a strong support for a topology in a particular node, other topologies at this node are strongly rejected. If there is a weak support for a topology in a particular node, other topologies at this node are weakly rejected.
In agreement with the previous molecular phylogenetic studies, all three subfamilies and 16 tribes form monophyletic clades with 100% support in each of the thirteen topologies (fig. 2; supplementary figs. S1–S12, Supplementary Material online). Importantly, our results strongly support the hypothesis that Dryadoideae is the basal clade of Rosaceae with more than 90% support in five topologies, more than 85% support in all six topologies for figure 1; however, Rosoideae was inferred in a previous study to be the sister lineage to the combined clade of the other two subfamilies (Potter et al. 2007a). To test whether the hypothesis of Dryadoideae being basal is supported by various subsets of marker genes, we examined all 13 topologies (fig. 2, supplementary figs. S1–S12, Supplementary Material online) and summarize the results in figure 3. Among the 13 trees, 10 strongly supported and two weakly supported Dryadoideae as the sister to the remainder of the family, only one, with 166 genes, supports Rosoideae being basal (fig. 3). One possibility was that different gene sets contained genes with different topologies. Detailed examination of single-gene trees revealed that, in the 241, 312, and 163 gene sets, the numbers of genes supporting (with more than 50 bootstrap value) Dryadoideae as sister of the combined clade of Rosoideae and Amygdaloideae are 74, 91, and 47, respectively; these numbers are larger than that of genes supporting Rosoideae (55, 85, and 38) or Amygdaloideae (48, 66, and 38) as the basal clade (supplementary fig. S13, Supplementary Material online). On the other hand, in the 166-gene set, 46 genes support Rosoideae as the basal clade, only slightly more than 39 and 42 genes, respectively, for Dryadoideae or Amygdaloideae being basal. Therefore, there were greater numbers of single-gene trees supporting Dryadoideae being first lineage to separate, and variation in the number of genes with specific topologies could at least provide partially explanation for the difference among the results from different gene sets.
Within Dryadoideae our sampling included four genera, with strong support for Dryas being the basal genus, in agreement with previous results (Potter et al. 2007a). Also, Purshia is sister to the combined clade of Cercocarpus and Chamaebatia, with strong supports from most of the analyses (supplementary fig. S14, Supplementary Material online). Within Rosoideae, the tribes Ulmarieae, Rubeae, and Colurieae occupy the basalmost and the next two basal positions, respectively; these relationships are largely consistent with previous results that they are the three successive basalmost lineages, although the relatively positions of Rubeae and Colurieae were uncertain previously (Potter et al. 2007a). The remaining three tribes Roseae, Potentilleae, and Agrimonieae form a strongly supported clade (fig. 1), and support values for Roseae being the basal lineage in this clade were strong in eight of the 13 analyses (fig. 3). The relationships between genera in Rosoideae were also very well resolved (fig. 1). To test whether the process of filtering orthologous genes from 882 to 113 has removed paralogous genes supporting a particular topology, we have examined the position of Roseae, Potentilleae, and Agrimonieae in each single gene tree with more than 70% bootstrap value in each gene set (supplementary fig. S13, Supplementary Material online). In the 882-gene coalescence tree (fig. 3, supplementary fig. S1, Supplementary Material online), Roseae is highly supported as the basal clade; this is consistent with the number of single gene trees supporting Roseae being basal (RB) vs. Potentilleae being basal (PB) (183 vs. 173). After filtering from 882 to 571, the numbers of single-gene trees supporting RB was reduced by 63, greater than 55, the number reduced for PB. Among the remaining 571 genes, the numbers of gene trees supported RB (120) and PB (118) were nearly identical, in agreement with the weak support for PB in the 571 coalescence tree (fig. 3, supplementary fig. S2, Supplementary Material online). After other filtering processes, in the 113 gene set, the single-gene trees supporting RB and PB were 36 and 24, respectively, with a ratio of 3 to 2 (supplementary fig. S13, Supplementary Material online), consistent with the 113-gene coalescence tree strongly supporting RB (fig. 3, supplementary fig. S5, Supplementary Material online). In short, the changes in number of genes supporting specific topologies might be one reason for the differences between the coalescence trees generated using different gene sets.
In the subfamily Amygdaloideae, there are nine tribes and the relationships for several of them were uncertain previously (Potter et al. 2007a). From our results, the tribe Neillieae with follicetum fruit type is maximally supported in all 13 topologies as the basalmost lineage of Amygdaloideae, followed by the tribe Spiraeeae with follicle fruit (100 support value in 12 trees, and 97 in one) (fig. 2, supplementary figs. S1–S12, Supplementary Material online). Lyonothamneae (with a single species), which was previously placed as the basal lineage of the subfamily, is now strongly supported (100 support value in 11 trees, with 99 and 94, respectively, in the other two) as sister to Amygdaleae (peach, plum, and cherry); they together occupy the third basal position in the subfamily. Next along the backbone is a clade with three tribes, with Sorbarieae being sister to the combined clade of Exochordeae and Kerrieae. The close relationship of Exochordeae and Kerrieae was also supported previously (Potter et al. 2007a). Finally, the fleshy-fruited clade Maleae and dry-fruited Gillenieae are sisters with 100% support in all 13 topologies for figure 2 and other phylogenies (supplementary figs. S1–S12, Supplementary Material online). In addition, the sister relationship of the [Sorbarieae, (Exochordeae, Kerrieae)] and (Maleae, Gillenieae) clades is strongly supported in eight of the thirteen topologies, and weakly supported in four others (fig. 3). Within Maleae, Kageneckia and Vauquelinia occupy the successive basal positions, with a highly supported clade of four genera (Amelanchier, Malacomeles, Crataegus, and Mespilus) being the next basal group (fig. 1). These four genera were also found in a well-supported clade previously, but the relationships of this clade with other genera of Maleae were not clear (Potter et al. 2007a; Lo and Donoghue 2012). The remaining 12 genera sampled here for Maleae form a large and very well supported clade, including several smaller clades that are each well-supported (fig. 1), with support for the clade of Eriobotrya and Rhaphiolepis being sister to the clade with the remaining 10 genera (fig. 3). As presented below, near the origin of Maleae there were two successive events of whole genome duplication, making the complex history of this tribe difficult to resolve, with some relationships requiring further analyses (supplementary fig. S14, Supplementary Material online).
Because several relationships are supported by some, but not all, of the analyses here, we then performed approximately unbiased (AU) test to examine the robustness of the relevant results, in two phases (Shimodaira and Hasegawa 2001). The first was to verify the relationships among three subfamilies and the tribes in Amygdaloideae (supplementary table S4, Supplementary Material online), with the topology using the 113-gene supermatrix (supplementary fig. S6, Supplementary Material online) and 20 alternative topologies. All 20 alternative topologies were rejected (P ≤ 0.05), and only the topology of 113-gene supermatrix analysis was not rejected, with a notably high P value of 0.998, supporting Dryadoideae as the basal clade of Rosaceae and the sister relationship of the Kerrieae–Exochordeae–Sorbarieae clade and the Maleae–Gillenieae clade. In contrast, the topologies with Rosoideae as the basal lineage had P values of 0.001 or less. The alternative topology of (Kerrieae–Exochordeae–Sorbarieae, Amygdaleae) and Maleae–Gillenieae being sisters had a P value of only 0.005. In the second phase, with the relationships among subfamilies and tribes fixed, the hypothesis from the 113-gene supermatrix (supplementary fig. S6, Supplementary Material online) and 47 alternative topologies for various relationships within individual tribes were analyzed (supplementary table S5, Supplementary Material online). Again the topology of 113-genes concatenation analysis has the highest P value of 0.820, and only four other topologies could not be rejected (P ≥ 0.05; AU test), including sister relationship of any two genera among Cercocarpus, Chamaebatia, and Purshia in Dryadoideae and Roseae, instead of Potentilleae, being sister to Agrimonieae. Hybridizations, incomplete lineage sorting and other possible factors might contribute to these uncertainties of current topology (Johnson and Soltis 1998; Wendel and Doyle 1998; Philippe et al. 2005; Jeffroy et al. 2006; Avise and Robinson 2008); future analysis with more taxon sampling in these groups might be needed to resolve their relationships.
Rosaceae Likely Originated near the Boundary of Early to Late Cretaceous
To estimate ages of Rosaceae lineages, we used sequences from all 124 of the Rosaceae species in this study and 24 additional outgroups for a total of 148 species with 19 fossil calibrations, including 13 belongs to Rosaceae. We used the tree topology of figure 1 and the sequence matrix of the 113 genes for the estimation inferred by penalized likelihood method implemented using r8s (Sanderson 2003). Assignments of the fossil constraints are provided in the “Materials and Methods” section and supplementary table S6, Supplementary Material online.
The age of crown Rosaceae in our estimation was about 101.6 Ma with the separation of Dryadoideae, followed by an immediate divergence of the two largest subfamilies at 100.7 Ma (fig. 4). Töpel et al. (2012) also performed divergence time estimation using two chloroplast sequences along with eight fossil constraints and one secondary calibration. They used 10 plastid and two nuclear genes and seven fossil calibrations and set a uniform prior with a maximum age of 115 Ma for all calibration points, and then calibrated stem Rosales to 104–115 Ma according to Wang et al. (2009), resulting in an age of crown Rosaceae being slightly older than 100 Ma, very close to our result. Our age predates the result of a previous study (Chin et al. 2014), which used one secondary calibration as the fixed age of crown Rosales as well as six fossil calibrations according to a phylogeny using four chloroplast genes and the ITS sequence. They found the age of crown Rosaceae to be 90.8–85.7 Ma using the minimum (90 Ma) and maximum age (96 Ma) of crown Rosales reported by Wang et al. (2009) as two sets of fixed age constraints, which limited the age estimates. Nevertheless, all the dating results indicate that Rosaceae originated around the boundary between Early and Late Cretaceous and that the three subfamilies diverged within a short period.
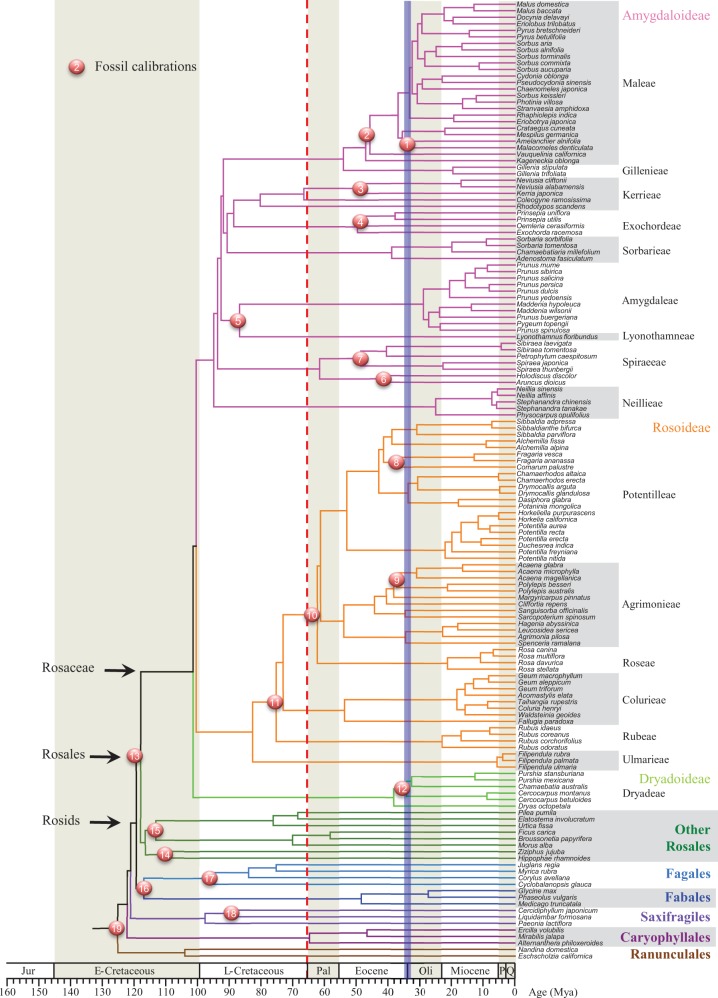
Fig. 4.
Chronogram of Rosaceae using 148 species phylogeny with 19 fossil constraints. Positions of the fossil calibrations are depicted with numbered circles. Crown nodes of Rosids, Rosales and Rosaceae are indicated with arrows. The red dashed line and blue strip highlight the Cretaceous-Paleogene and Eocene-Oligocene boundaries, respectively. Ages are presented as millions of years (Myr). There are two possible topologies within Dryadoideae (relationships among Purshia, Cercocarpus and Chamaebatia) having similar support from our analyses of various gene sets (see supplementary fig. S14, Supplementary Material online); here we only present the result from the analysis with the 113-gene concatenation. Jur: Jurassic. E- and L-Cretaceous mean Early and Late Cretaceous. Pal: Paleocene. Oli: Oligocene. P: Pliocene. Q: Quaternary.
After the split of the three subfamilies, there was a rapid series of nested branching of seven tribes of Amygdaloideae (∼95–90 Ma; fig. 4) during the early portion of Late-Cretaceous; on the contrary, the two most derived tribes, Maleae and Gillenieae, separated at ∼54 Ma just after the Paleocene–Eocene boundary, exhibiting a lag of ∼38 Ma from the split at ∼92 Ma from the clade of Kerrieae–Exochordeae–Sorbarieae. On the other hand, divergences of Rosoideae tribes were gradual, spanning from the middle of Late-Cretaceous (∼82 Ma) to Mid-Paleocene (∼62 Ma). In addition, there was a very long delay of ∼63 Ma before the separation of extent Dryadoideae lineages at ∼38 Ma in late Eocene, possibly due to extinction of early branches of this subfamily. Among the three largest tribes having over 500 species, Rubeae diverged earlier at 75 Ma in Late Cretaceous; the other two tribes originate under relatively stressful conditions than current environment: Potentilleae during Paleocene shortly after a global catastrophe at the boundary of Cretaceous and Paleocene (66 Ma), and Maleae during the hottest and most humid period in the Cenozoic Era.
Evidence for Multiple Whole-Genome Duplication Events across Rosaceae with Two Close Events Shared by Members of Maleae
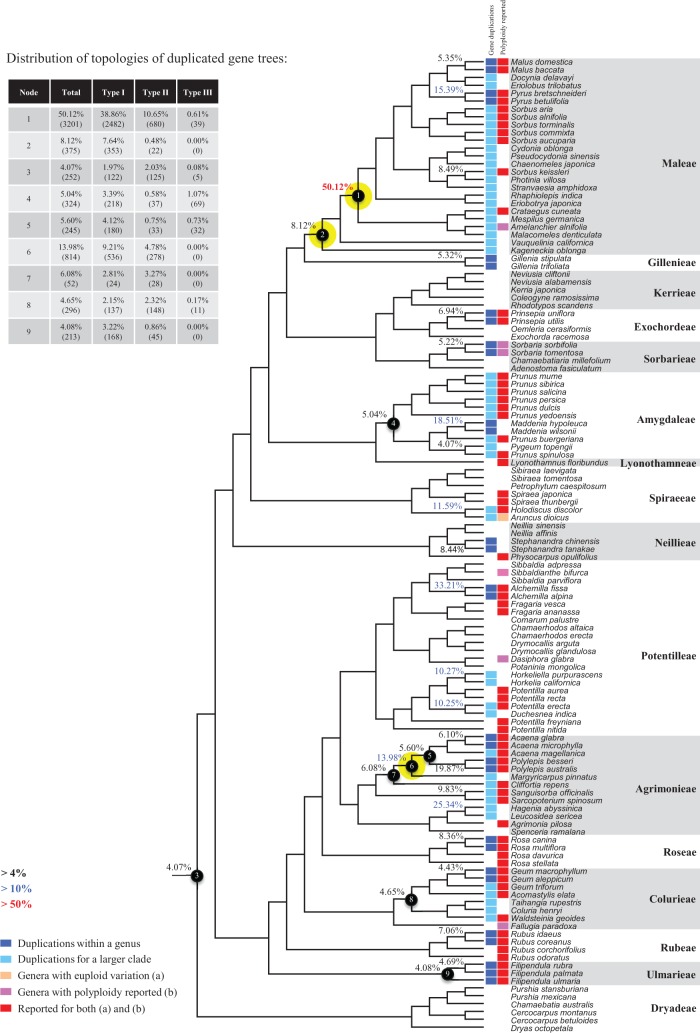
Genome sequences of Rosaceae members suggested that this family might have experienced one or more whole-genome duplications (Dickinson et al. 2007; Velasco et al. 2010; Wu et al. 2013; Chin et al. 2014; Zhao et al. 2016), but the timing of such event was not clear. WGD was also proposed for Maleae based on the chromosome number (Vamosi and Dickinson 2006; Dickinson et al. 2007), but there has not been support from phylogenomic analyses. To address these questions, we used a phylogenomic analysis to detect potential WGD events, as was used in several recent studies (Jiao et al. 2011, 2012; Cannon et al. 2015; Li et al. 2015; Yang et al. 2015) and recognized as an effective method (Kellogg 2016). The basic rationale is to construct gene trees on a transcriptome-wide scale and then to compare gene tree topologies with that of the organismal tree, thereby mapping duplications in each gene family tree onto the organismal tree; the detection of large numbers of gene duplications at specific nodes on the organismal tree is then seen as evidence for WGD events. We also examine the topologies of the gene tree at duplication event to evaluate the strength of support. For a node in the organismal tree with three or more species, we classified the observed topologies into three types regarding the gene retention in each of the duplicated subclades following the node of interest: type I retains both gene copies in both the large and small subclades, whereas type II and III lack duplicates for the entire small or large subclades, respectively. Among them, the type I topology provide stronger evidence than the other two types due to the presence of more genes to infer a correct phylogeny.
Our results identified evidence for a duplication event (fig. 5, node 2) shared by all Maleae members with 8.12% (375 pairs) gene families showing duplications here and 7.64% (353 pairs) with strong support (type I). Much stronger signal was detected for a WGD event (node 1) shared by most of the members of Maleae except Vauquelinia and Kageneckia, supported by 50.12% (3201 pairs) gene families duplicated at this node with 38.86% having type I topology. Duplication events associated with members of Maleae have been reported previously. Based on analysis with Malus × domestica genome, Velasco et al. (2010) reported a WGD dated 30–45 Ma when the age for the split of grape and rosids were fixed at 115 Ma. Wu et al. (2013) reporting the genome sequence of a pear (Pyrus bretschneideri) also observed a WGD shared by pears and apple but not strawberry. Our findings support two possible WGD events in succession near the origin of Maleae, with the large number of resulting new gene copies as materials for functional divergences and innovations. In addition, evidence for polyploids was also found previously within Maleae for members of genera Sorbus, Crataegus, and Amelanchier (red boxes, fig. 5), all included in the clade derived from node 1. In addition, a recent WGD might have happened within Pyrus, with 15.39% (585 pairs) gene families duplicated before separation of P. bretschneideri and P. betulifolia. There was also a likely WGD event at crown node of Amygdaleae, which include members of Maddenia and Pygeum, with 5.04% (324 pairs) gene duplications and 4.12% (180 pairs) in type I gene trees. In the subfamily Rosoideae, we also detected possible WGDs within Agrimonieae, shared by Acaena, Polylepis, and Margyricarpus (nodes 5 and 6). Although 6.08% of gene families were duplicated at node 7, they account for only 52 paralogue pairs and do not provide strong support for WGD. Further, WGD events might have occurred for the tribes Colurieae, with support of 4.65% (296 pairs) of gene trees, and Ulmarieae with 4.08% (213 pairs) of gene trees, consistent with previous reports of polyploidy of these groups (Smedmark and Eriksson 2002; Vamosi and Dickinson 2006). In addition, 4.07% (252 pairs) of gene trees supported a possible WGD shared by all Rosaceae members, in agreement with previous proposal of paleopolyploidy of Rosaceae (Vamosi and Dickinson 2006; Dickinson et al. 2007).
Fig. 5.
Detecting and positioning large-scale gene duplications by comparing gene family trees to the species tree. Percentage of duplicated gene families (among those containing genes from both species lineages from the node of interest) are presented adjacent to each node when there were higher than 4% gene families duplicated at the corresponding node. Numbers higher than 10% and 50% are highlighted with blue and red, respectively. Distributions of three possible topologies for each of the duplicated gene trees are shown in the upper left table, with both the percentage and the actual numbers of gene families recorded. By the diagnosis of tree topologies, nodes with strong evidence for WGD are marked with yellow. Colored boxes indicate species potentially having gene duplications within a genus (dark blue) or shared across genera (light blue); genera having euploid variations summarized by Dickinson et al. (2007) (orange) or polyploid species observed by Vamosi and Dickinson (2006) (pink) or both (red) are also highlighted.
Additional putative WGDs with more than 10% supporting gene trees were detected in more recent lineages of one or two genera, including the clades of Holodiscus and Aruncus, of Horkeliella and Horkelia, of Hagenia and Leucosidea, of Potentilla erecta and Duchesnea indica, and also the clades of genera Maddenia and Alchemilla. Among these potential events, Holodiscus, Aruncus, and Alchemilla were reported to have polyploids (Mishima et al. 2002; Gu et al. 2003; Vamosi and Dickinson 2006; Dickinson et al. 2007; Phipps 2014), supporting our hypotheses. More nodes with 10% to 4% duplicated gene families can also be observed, including those of genera Malus, Gillenia, Prinsepia, Sorbaria, Maddenia, Stephanandra, Acaena, Polylepis, Rosa, Geum, and Rubus. Although these percentages are not high, polyploids have been found in many of these genera, except Gillenia and Stephanandra (Gu et al. 2003; Vamosi and Dickinson 2006; Dickinson et al. 2007; Phipps 2014).
Ancestral State Reconstruction for Fruit Types and Other Morphological Characters
The well-resolved Rosaceae phylogeny provided an excellent opportunity to reconstruct ancestral states of fruit types and other morphological characters. Rosaceae members produce dry fruits, fleshy fruits, or aggregate fruits (Potter et al. 2007a) (fig. 6, supplementary fig. S15, Supplementary Material online). Dry fruits include achene, which has a thin fruit wall with a single seed; some species produce several to many achenes from a single flower, or achenetum (aggregate achenes). Another dry fruit is follicle, which produce one or more seeds within a thin fruit wall that split open at maturity to release the seeds. Again, multiple separate follicle-like fruits within a single flower are referred to as follicetum. Fleshy fruits include drupe, with a thick fleshy fruit wall enveloping a hard shell-like inner fruit wall that surrounds a single seed, and pome, which has thick fleshy accessory tissue (largely derived from the hypanthium, a cup-shaped fusion of lower portions of sepals, petals, and stamens) outside a relatively soft fruit wall containing several seeds. In Rosaceae pomes, the central core is often divided into five small chambers, each with one or a few seeds.
Fig. 6.
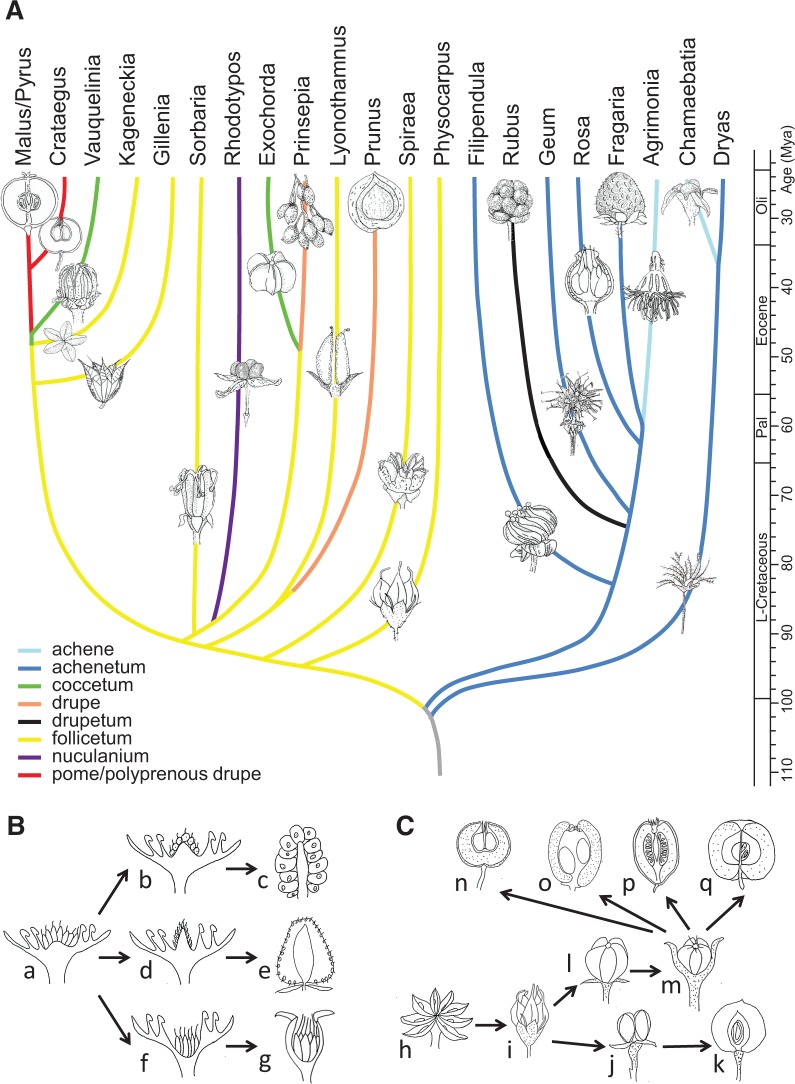
Evolutionary histories of fruit types in Rosaceae. (A) The evolutionary history of different fruit types in Rosaceae in the context of an abbreviated phylogeny from than shown in figure 1. Grey line represents ancestral fruit type of Rosaceae. Other line colors represent different fruit types of extant taxa. The positions of nodes and branches correspond with geological time scale on the right redrawn from the molecular clock analysis shown in figure 4. The positions of fruit drawings do not represent the history or age of the fruit types. (B) Proposed histories of three fruit types in Rosoideae. a: a hypothetical ancestral fruit of Rosoideae (achenetum); b: a hypothetical ancestral fruit of Rubus (drupetum, with less flesh); c: Rubus (drupetum, with more flesh); d: a hypothetical ancestral fruit of Fragaria (achenetum, without enlargement of receptacle); e: Fragaria (achenetum, with fleshy enlargement of receptacle); f: a hypothetical ancestral fruit of Rosa (achenetum, with partial enclosure of fruits by the hypanthium); g: Rosa (achenetum, with full enclosure by the hypanthium, and increased fruit size). (C) Proposed histories of several fruit types in Amygdaloideae. h, i: Hypothetical ancestral fruits of Amygdaloideae, evolving from a follicetum with many (indefinite) carpels (h) to that with five or fewer carpels (i). j: A hypothetical ancestor of Amygdaleae (nuculanium, with a few carpels). k: Prunus (drupe, with a single carpel). l, m: hypothetical ancestral fruits of Pyrinae. l evolved into m via the partial “sinking” of the ovary into the hypanthium and fusing with it. n: Crataegus (with the endocarp still near the top). o: Eriobotrya (with relatively thin flesh). p: Pseudocydonia (with many ovules for a carpel, a likely ancestral character). q: Malus (with centrally located endocarp along the vertical axis, thick flesh, and one or two seeds per carpel, all likely derived features).
In Dryadoideae, achenes or achenetum (such as in Dryas) are the main fruit types. Ancestral character reconstruction supports that achenetum with an enlarged and hemispheric hypanthium, as well as with a very large number of separate carpels in a flower, is the ancestral fruit type of this subfamily (fig. 6A, supplementary fig. S15, Supplementary Material online), consistent with the fruit character reconstruction in Potter et al. (2007a) and Dryas being the basal lineage of the subfamily. Rosoideae also include species with dry fruits such as some achenetum (Ulmarieae, Colurieae, Roseae, and Potentilleae) and achenes (Potaninia and Agrimonieae), as well as fleshy drupetum, or aggregate small drupes (Rubeae). Achenetum with a flat hypanthium such as that in Filipendula is the ancestral character, and it seemed to have evolved into three different complex fruit types (fig. 6B). In one type, the receptacle becomes somewhat enlarged and hemispherical (such as Potentilleae and Colurieae), and then larger and fleshy with many achenes, such as strawberry (Fragaria) and Duchesnea. In the second type, the hypanthium extends upward to become urceolate (urn-like) with multiple achenes enclosed within, such as that in Rosa. In the third type, the wall of each fruit changes from dry and thin to fleshy and thickened, so that the aggregate fruit has evolved into a drupetum (of multiple small drupelets), such as in raspberry and blackberry (Rubus).
In Amygdaloideae, members of several tribes produce the fruit type follicetum (Lyonothamneae, Neillieae, Sorbarieae, Gillenieae, and Kageneckia in Maleae), whereas others have one of several distinct types: achenetum (Neviusia and Holodiscus), coccetum (Exochorda and Vauquelinia), pome (many genera in Maleae), nuculanium (most of genera in Kerrieae), and drupe (Amygdaleae) (fig. 6A, supplementary fig. S15, Supplementary Material online). The ancestral fruits of the subfamily were likely pentamerous aggregate follicles (follicetum), which evolved from an earlier type with an indefinite and larger number of carpels; subsequently, there have been independent further reduction of carpel number for some tribes (such as Amygdaleae and Lyonothamneae). Enlarged and fleshy fruits likely evolved via two distinct ways (fig. 6C). In one, the endocarp (inner fruit wall) became hard, forming nuculanium; in addition, the previously dry pericarp (middle fruit wall) became fleshy and the number of carpels decreased to one or two, finally forming the drupe (Prunus: peach, plum, cherry, and apricot; Prinsepia). Alternatively, after connation of five carpels as coccetum (such as Vauquelinia), the hypanthium become urceolate and fleshy and further closed up with the carpels, evolving into partially inferior (such as Crataegus) or fully inferior (such as apple, Malus) ovaries. It is worth noting that the newly revolved phylogeny of the subfamily places Amygdaleae with single-carpel fruits as sister to Lyonothamnus, which produces follicetum fruits with two separate carpels, suggesting that the reduction of carpel number might be a gradual process.
We have also examined the ancestral states of other characters (supplementary figs. S16–S23, Supplementary Material online). The habit of Rosaceae species most probably evolved from shrubs (supplementary fig. S16, Supplementary Material online). Most Rosoideae species tend to diminish their sizes to perennial or annual herbs with small compound leaves (supplementary fig. S17, Supplementary Material online) and a mass of small dry seeds. On the contrary, trees originated independently in Maleae and Amygdaleae in the subfamily Amygdaloideae, becoming much larger in size with numerous photosynthetic leaves that produce nutrients to support the production of many fleshy fruits. These trees are important members of forests, providing habitats of birds and other animals, which gather fruits and seeds, sometimes for future use in locations away from the plants, thereby facilitating the spread of seeds. In addition to changes in size and number of leaves, compound leaves also evolved a few times independently within Amydaloideae (supplementary fig. S17, Supplementary Material online).
As for flowers, most Rosaceae species share some common ancestral characters (supplementary figs. S18–S23, Supplementary Material online), such as bearing a hypanthium, which is a connation of the receptacle and the basal part of perianth and stamens, and having pentamerous sepals and petals (wind-spread species tend to have no petals). However, pistils are distinctive in different clades. An apocarpous pistil with superior ovary and numerous carpels is the ancestral character of Rosaceae. Most Rosoideae species have maintained this character, while carpel number tends to diminish to five (such as most species in Amygdaloideae) and even to one (such as species in Amygdaleae and some species in Dryadoideae), allowing greater supply of nutrients to each fruit. Furthermore, Maleae species produce inferior pistil, probably ensuring better protection for ovules.
Possible Effects of WGD and Climate Changes on Fruit Evolution
The nuclear phylogenetic analyses here produced a robust Rosaceae phylogeny, which has served as a framework for further analyses, including molecular clock estimates of ages of Rosaceae lineages and genome-scale analyses of gene family evolution with strong evidence for multiple WGDs. Furthermore, ancestral character reconstruction for fruit types support independent origins of several fleshy fruit types from ancestral dry fruits, including different forms of pomes in many genera of Maleae, drupes in Prunus species (peach, plum, cherry, and apricot), nuculanium with 1–5 small fruits in many species in Kerrieae, and drupetum with multiple drupelets in Rubus (such as raspberry and blackberry). In addition, strawberry has evolved a complex fruit structure with a fleshy hypanthium with many achenes attached to its surface. The evolution of fleshiness includes the enlargements of pericarp (drupe, drupetum, or nuculanium), hypanthium/receptacle (strawberry-like), or both (pome).
The fleshy fruits in the subfamily Amygdaloideae are concentrated in the tribes Maleae and Amygdaleae, both of which also have evolved trees, whereas the aggregate fruits with fleshy tissues in Rosoideae are from tribes with herbaceous (Potentilleae; Fragaria) or bushy (Rubeae; Rubus) habits. The Maleae and Amygdaleae lineages with fleshy fruits exhibit strong evidence of having had lineage-specific WGDs because of the divergence times of these tribes from others. In particular, the large and diverse tribe Maleae probably had two WGDs, one shared by all genera examined here and was likely in the most recent common ancestor (MRCA) of the tribe, and the other was shared by all fleshy-fruit bearing members included here, and likely occurred in the MRCA of the subtribe Pyrinae. The successive WGDs might have provided many more genes to allow for functional innovations after the duplications via dosage effects, neofunctionalization, or subfunctionalization (Blanc and Wolfe 2004; Pontes et al. 2004; Madlung et al. 2005; Cusack and Wolfe 2007; Freeling 2009; Bekaert et al. 2011; Hudson et al. 2011). This is corroborated by the evolution of multiple sub-types of pomes in Maleae, with an evolutionary trajectory from superior to inferior ovaries, from five distinct carpels to fused carpels, from thin and nonfleshy hypanthium/pericarp to fleshy tissues, and possibly from multiple ovules to few ovules attached to each carpel (fig. 6).
With the support of highly resolved relationships between genera (figs. 1 and 6), we propose the following scenario for fruit evolution in Maleae: (1) the ancestor produced follicetum fruits with five carpels, as also seen in the basal lineage Kageneckia; (2) in the next basal clade, Crataegus and related genera share a form that has a semi-inferior ovary separated into five parts; this might be an intermediate form of fleshy fruit during evolution; (3) in the next clade, Eriobotrya has a fruit with a fleshy tissue being thinner at the top, suggesting that the enclosure of the ovary by the cup-shaped hypanthium is incomplete; (4) in the clade with Cydonia, Chaenomeles, and Pseudocydonia, these three genera produce fruits with several ovules for each carpel, suggesting some degree of primitiveness; and (5) the fruits of Sorbus, Pyrus, and Malus all have inferior ovaries, suggesting more derived forms.
In the tribe Maleae, the timing of fruit character transitions is closely correlated with those of WGDs and climate events, suggesting possible impacts of WGD and climate factors on fruit evolution. Molecular clock estimates suggest that the tribe Maleae split from Gillenieae at around 54 Ma just after the Paleocene–Eocene boundary, with further divergences within Maleae beginning shortly afterwards. We found evidence for two closely spaced WGDs near the origin of Maleae. The earlier WGD shared by all Maleae members occurred at early Eocene, which was the hottest period in the Cenozoic Era, including both the Paleocene–Eocene Thermal Maximum (PETM) and Early Eocene Climate Optimum (EECO) (Zachos et al. 2008). Within Maleae, after the separation of Kageneckia (with fruit type of follicetum), the ancestor of Vauquelinia and other genera likely produced the fruit type coccetum, with a short lag period from the early WGD event. The second WGD was shared by the fleshy-fruited genera of Maleae (fig. 5) and occurred in late Eocene (fig. 4), when the Earth experienced continuous decrease in temperature and humidity, closely followed by a short glaciation period with extinctions of many species in Europe (Zachos et al. 2001; Hooker et al. 2004). The unusually high percentage (50.12%) of retained gene pairs of the WGD and the rapid taxon separation/diversification after the WGD suggest that the duplicate genes from the WGD contributed to the diversification of the decent Maleae genera. Thus it is possible that the new gene copies from two Maleae WGDs allowed the descendant Maleae members to evolved new fruit types under the selective forces of both the dramatic climate changes from early to late Eocene and the interactions with animals that lived in the forests of woody Maleae members and ate their fleshy fruits.
The potential for new gene functions offered by the WGDs might also have contributed to the evolution of relatively large trees in Maleae and Amygdaleae, as compared with herbs and bushes in other Rosaceae tribes. The tree habit that can reach greater heights for light exposure and that have more leaves to harvest the light energy could also give these tribes greater advantages, by allowing them to produce many more fruits per plant, and more fleshy fruits to attract animals. In contrast, the fleshy fruits of Rubus and Fragaria seems to have evolved via different paths, and are not associated with obvious lineage-specific WGDs or tree habit. Human efforts have resulted in the domestication of several fleshy-fruited Rosaceae species, increasing the sizes and sweetness of the fruits. Still other Rosaceae tribes/genera have retained the earlier dry fruit types of achene and follicle (or related achenetum and follicetum, respectively), yet some of them have developed appendages to facilitate attachment to animals (such as hooks in Geum) or spread by wind (Dryas). The multiple independent origins of distinct fleshy fruits discussed in this study suggest a general trend of fleshy fruit evolution, including the decrease of carpel number, the extension of hypanthium/receptacle to enclose and, sometimes, fuse with the ovary, as well as the enlargement and softening of ovary wall, hypanthium, or receptacle. Such scenario for the evolution of fleshy fruits might also exist in other families, especially those with both dry and fleshy fruits, such as citrus, melons, and tomatoes (Gu et al. 2003). This study provides a foundation combining molecular phylogeny with molecular clock estimates and evidence for WGDs, placing the evolution of Rosaceae fruit types in the context of geological ages and related climate changes, as well as genome-scale changes that allow potential functional innovation. The wide range of fruit types, available genomic resources of Rosaceae species and model system for functional analyses, and the phylogenetic framework presented here all contribute to making Rosaceae an excellent system for studying fruit evolution, an important problem for understanding angiosperm evolution and improving fruit crops for horticulture.
Materials and methods
Tissue Collection, RNA Isolation, Transcriptome Sequencing, and Sequence Retrieval
Young leaves, buds or fruits were collected and frozen at −80 °C. RNA isolation, cDNA synthesis, high-throughput sequencing, sequence assembly were carried out as previously described (Huang et al. 2016). Genomes and Sequence Read Archive data sets were retrieved from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html; last accessed November 12, 2016), Mei Genome Project website (http://prunusmumegenome.bjfu.edu.cn/; last accessed June 13, 2016), Pear Genome Project website (http://peargenome.njau.edu.cn/; last accessed November 12, 2016), and GenBank (http://www.ncbi.nlm.nih.gov/genbank/; last accessed November 12, 2016).
Identification of Low Copy Candidate Orthologous Genes
In the following investigations, sequence retrieval from public or in-house datasets was performed using HaMStR v13.2.3 (Ebersberger et al. 2009) with E value of less than e−20. Maximum likelihood (ML) phylogenetic analysis for single-gene families was performed using RAxML v7.0.4 (Stamatakis 2006) with 100 bootstrap replicates under GTRGAMMAI model as recommended by jModelTest v2.1 (Darriba et al. 2012) (see next section for more on phylogenetic analysis). The scripts for the bioinformatics analyses here are available if requested.
For use as phylogenetic markers, low copy nuclear genes were identified from two groups of genes (supplementary fig. S24A, Supplementary Material online). The first group contains 931 candidate orthologous genes (Zeng et al. 2014) in an overlapping set of orthogroups between two datasets (using the Arabidopsis thaliana gene ID numbers present in both sets): one includes 4180 orthologous genes shared by nine angiosperm genomes (Arabidopsis thaliana, Populus trichocarpa, Glycine max, Medicago truncatula, Vitis vinifera, Solanum lycopersicum, Oryza sativa, Sorghum bicolor, and Zea mays) retrieved from the Deep Metazoan Phylogeny (http://www.deep-phylogeny.org/hamstr/; last accessed November 12, 2016) website, the other includes 1989 orthologous genes obtained from seven genomes (Arabidopsis thaliana, Populus trichocarpa, Prunus persica, Vitis vinifera, Mimulus guttatus, Oryza sativa, and Sorghum bicolor) using OrthoMCL (Li et al. 2003). Using the 931 seeds generated in Zeng et al. (2014) and HaMStR with E value of less than e−20, we obtained homologs of these 931 genes from eight representative Rosaceae species (Fragaria vesca, Rubus coreanus, Crataegus pinnatifida, Malus × domestica, Spiraea japonica, Prunus dulcis, Prunus persica, and Prunus mume) and two outgroup species (Medicago truncatula and Cucumis sativus), which have well-established relationships. Among these, 546 genes were retained that had length of 1000 base pairs or more in each of four Rosaceae genomes (Fragaria vesca, Malus × domestica, Prunus persica, and Prunus mume) and Medicago truncatula, as an outgroup, and coverage of more than 60% of the above mentioned 10 species (8 Rosaceae and 2 outgroups) using in-house scripts. To avoid hidden paralogs, single-gene trees of the 546 OGs from the ten species were reconstructed using RAxML then inspected manually and 407 genes whose tree topologies did not contradict with the organismal tree (supplementary fig. S24B, Supplementary Material online) of the 10 species were retained.
In addition, 3863 single copy nuclear genes that were shared by four species (Fragaria vesca, Prunus Persica, Prunus mume, and Cucumis sativus) were identified by using MCScan v0.8 (Wang et al. 2012). To avoid selecting the same genes, the 3863 orthologs were identified in Arabidopsis thaliana, and by comparing gene IDs, those overlapping with the above-mentioned 4180 gene set were excluded. Then 2124 genes with sequence length of 1000 base pairs or more in Cucumis sativus were selected using in-house scripts. Among the 2124 genes, those with only one or two copies in Malus × domestica and Pyrus bretschneideri were retained, resulting in 475 genes.
The combined set (407 + 475) of 882 genes was retrieved from 124 Rosaceae species and 24 outgroup species using 407 seeds from Zeng et al. and 475 seeds generated in this study respectively by HaMStR, and was used to construct single-gene family trees by RAxML. The tree topologies were examined manually for inconsistencies with well-established known relationships; 311 genes that grouped members of different orders together were removed (resulting in 571 genes), then 127 genes failing to group members of Rosaceae subfamily were removed (yielding 444 genes), then 188 genes with conflict for monophyly of tribes were eliminated (with 256 remaining). Among the relationships in Maleae, four clades of two or three species received maximum support from all of the 882, 571, 444, and 256 gene sets: (Malus × domestica, Malus baccata), (Eriobotrya japonica, Rhaphiolepis indica), (Crataegus cuneata, Mespilus germanica), and (Photinia villosa, Sorbus keissleri, Stranvaesia amphidoxa). Among the 256 single-gene trees, 113 genes yielded single-gene trees with these four clades and were remained for further phylogenetic and other molecular evolutionary analyses.
To test whether the recent WGD events in Maleae affected the identification of orthologous genes in Maleae species, a gene set contains 484 genes with only one copy in Pyrus bretschneideri were selected from 2124 genes using in-house scripts. Then 484 gene family trees were constructed using RAxML v7.0.4 with homologous sequences retrieved using HaMStR from 31 species including 25 species in Maleae (supplementary table S1, Supplementary Material online) and, as outgroups, Gillenia stipulata, Gillenia trifoliata, Prunus persica, Prunus mume, Neillia sinensis, and Physocarpus opulifolius. These gene trees were examined manually and treated as follows: (1) if most of the species have only one copy in the single gene tree, even if some species have more than one copy from very recent lineage-specific duplication, this kind of genes were retained and (2) if most of the species have more than one copy, the clade with more species was retained. Finally, 163 genes remained with at most one copy per species.
Alignment and Phylogenetic Analysis
Phylogenetic reconstruction was performed with all gene sets using coalescence method implemented in ASTRAL v4.4.4 (Mirarab et al. 2014). A maximum likelihood (ML) analysis was also performed with the 113 gene sequence supermatrix using RAxML v7.0.4 (Stamatakis 2006) under GTRGAMMAI model as recommended by jModelTest v2.1 (Darriba et al. 2012). 882 orthologous genes from 148 species were identified using HaMStR. Nucleotide sequences of each orthologous genes were aligned using MUSCLE v3.8.31 (Edgar 2004) with default parameters. Then the regions poorly aligned were further trimmed using trimAl v1.2 (Capella-Gutiérrez et al. 2009). Single-gene trees were reconstructed using RAxML v7.0.4 (Stamatakis 2006) under GTRCAT model. For each gene groups, 100 bootstrap replicates were generated respectively for the coalescent analysis (Mirarab et al. 2014). The nucleotide sequence alignments of 882 orthologous genes in 148 species are accessible in TreeBASE website (http://treebase.org/treebase-web/home.html; last accessed November 12, 2016), with the submission number of 19726.
AU Test
CONSEL v0.1j (Shimodaira and Hasegawa 2001) was used to test the alternative topologies in two phases as previously described in Huang et al. (2016).
Molecular Clock Estimation of Geological Ages
Divergence times were estimated using the 113-gene ML tree and 19 fossil constraints (information for all fossils and their assignments can be found in supplementary table S3, Supplementary Material online), with the penalized likelihood (PL) method implemented in r8s (Sanderson 2003) and the optimum smoothing value of 0.01 according to the build-in cross validation procedure to correct rate heterogeneity among lineages. The r8s method was used instead of BEAST because the former has been shown to be effective in recent studies (Carbonell-Caballero et al. 2015; de Casas et al. 2016; Feng et al. 2016; Plazzi et al. 2016) and the latter would require much more computational time with this large datasets here. The fossils used here were from a survey of literature and online resources on Paleobiology Database website (https://www.paleobiodb.org/; last accessed November 12, 2016) for the oldest fossil of the corresponding MRCA nodes. Among the 19 fossil constraints, 13 were for Rosaceae, described as follows (numbers correspond to those in fig. 4): (1) Fossils of Amelanchier peritula and Amelanchier scudderi were discovered in the Florissant Formation, Colorado, USA, and have been dated to Chadronian in Late Eocene (37.2–33.9 Ma) (Cockerell 1911; MacGinitie 1953). We assigned the fossils to stem Amelanchier with a minimum age of 33.9 Ma. (2) Fossil Vauquelinia comptonifolia found in the Green River Formation in Wyoming, USA, has been dated to 46.2–40.4 Ma in Uintan, Middle Eocene (MacGinitie 1969). Thus, we constrained stem Vauquelinia with a minimum age of 40.4 Ma. (3 and 7) Fossils of Neviusia and Spiraea sp. found in Republic, Washington, USA have been dated to 49–50 Ma in Early Eocene (Mathews 1964; Wehr and Hopkins 1994). Both of them were assigned to the stem node correspondingly with minimum age of 48.6 Ma corresponding to the upper boundary of Early Eocene. (4) Fossils of Oemleria janhartfordae were uncovered in the Klondike Mountain Formation, Washington, USA and were dated to 49.42 ± 0.54 Ma in Early Eocene (Wolfe et al. 2003; Greenwood et al. 2005; DeVore and Pigg 2007; Benedict et al. 2011). The minimum age of stem Oemleria was constrained to 48.6 Ma corresponding to the upper boundary of Early Eocene. (5) Fossil Prunus wutuensis found in the Wutu Formation, Shandong Province, China, has been dated to 55 Ma in Early Eocene (Li et al. 2011). We used this age (55 Ma) to constrain stem Prunus. (6) Holodiscus lisii found in Florissant, Colorado, USA, has been dated to 34.1 Ma in Late Eocene (Schorn 1998; McIntosh and Chapin 2004). We set the minimum age of stem Holodiscus to 34.1 Ma for this fossil calibration. (8) Macrofossils of Fragaria were discovered in the Beaufort formation, Prince Patrick Island in the Canadian Arctic, and the age was considered to be about 2.96 Ma in Late Pliocene, corresponding to the age of Lost Chicken tephra in Alaska (Matthews and Ovenden 1990; Matthews et al. 2003). We used this fossil to calibrate the minimum age of stem Fragaria to 2.96 Ma. (9) Microfossils of Acaena sp. were found in Cullen Formation in Tierra del Fuego, Argentina, which were dated to 48.6–37.2 Ma in Middle Eocene (Zetter et al. 1999). This fossil was used to calibrate stem Acaena with 37.2 Ma as a minimum age constraint. (10) Mesofossil of Rosa germerensis belongs to the Challis Volcanics Formation in Custer County, Idaho, which has been dated to 55.8–48.6 Ma in Early Eocene (Edelman 1975). We calibrated this fossil to stem Rosa with a minimum age of 48.6 Ma. (11) Rubus acutiformis was discovered in Dorset, United Kingdom and dated to 47.8–41.3 Ma in Lutetian, Middle Eocene (Chandler 1963). Thus, we constrain the minimum age of stem Rubus to 41.3 Ma. (12) Fossil Cercocarpus myricaefolius found in the Florissant Formation, Colorado, USA has been dated by a single-crystal 40Ar/39Ar analysis of sanidine from pumice in sandstone and debris flow deposits, resulted in a mean age of 34.07 Ma in Late Eocene (MacGinitie 1953; Evanoff et al. 2001). This mean age was used here to calibrate the minimum age of stem Cercocarpus. (13) The oldest fossil of Rosaceae is a macrofossil found in the Aspen Shale Formation, Wyoming, USA and been dated to 113.0–100.5 Ma in Albian, Early Cretaceous (Peppe et al. 2008). We used this fossil to calibrate the minimum age of stem Rosaceae to 100.5 Ma in our analysis.
Detecting Whole Genome Duplication by Paralog Gene Trees
We performed all-against-all BLASTP with e-value cutoff of 10−5 among gene sequences of 124 Rosaceae species and 24 outgroups (supplementary table S1, Supplementary Material online) to search for homologous genes, and removed genes showing sequence identity < 50%. These genes were then divided into orthologous groups using OrthoMCL v1.4 (Li et al. 2003) with inflation value of 2.0. Ortho groups (OGs), in addition, taxon coverage of 85% or greater was used to balance between the needs to maximize gene number and minimize missing data, totaling 9482 gene families, were retained for subsequent analyses. Amino acid sequences predicted from genes in each of the OGs were aligned by MUSCLE v3.8.31 (Edgar 2004) with default parameters. These alignments were used for phylogenetic reconstructions using RAxML under the GTRCAT model, and the resulting gene family trees were compared with organismal tree in this study (fig. 1) to map the positions of gene duplications. When a node gaining BP support > 50% and having the same two species in each of its duplicated subclades, a gene duplication was mapped and counted to the corresponding position of the organismal tree. We also required the last common ancestor (LCA) of the subclades to share the same or close depth with the differences smaller than one step, while the steps were determined as the stages traveling from a node to the root in the species tree. The above criteria were implemented literately for all gene family trees, and the numbers of duplicated trees were summarized for each node.
Ancestral Character Reconstruction
The morphological information of the characters (supplementary table S7, Supplementary Material online) was obtained from the book Flora of North America (floranorthamerica.org), Flora Republicae Popularis Sinicae (http://frps.eflora.cn/; last accessed November 12, 2016), and Global Plants JSTOR (http://plants.jstor.org/; last accessed November 12, 2016). A monophyletic genus was identified as a unit to construct a morphological character matrix. For a genus that is not monophyletic, more than one unit was used to present characters of different clades. The ancestral characters were analyzed using Mesquite v3.04 (Maddison and Maddison 2004) based on the topology in figure 1.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Note Added in Proof
The whole-genome duplication (WGD) analyses performed here used a method developed by Dr Ji Qi at Fudan. During the reviewing process of this paper, a paper describing the WGD analysis was published and should be the reference for the method: Huang C.-H., Zhang, C., Liu, M., Hu, Y., Gao, T., Qi, J.*, Ma, H.* 2016. Multiple paleopolyploidization events across Asteraceae with two nested events in the early history revealed by nuclear phylogenomics. Mol Biol Evol. 33(11):2820–2835. (* co-corresponding authors)
Supplementary Material
Acknowledgments
We thank the Arnold Arboretum at Harvard University, the Beijing Botanical Garden, the Wuhan Botanical Garden, the Shanghai Botanical Garden, the UC Berkeley Botanical Garden, the University of Bonn Botanical Garden, the United States Botanical Garden and the United States National Arboretum, and Drs Holly Forbes, Kun Sun, Xun Gong, Stefan Lura, William McLaufflin, Zheng Meng, Weimin Ni, Yi Ren, Kyle Wallick, Fusheng Yang, Jinqing Wu, Shoujun Zhang, Yuanping Fang, Yingchun Wang, Zhiping Song, Wenju Zhang, Yaqiong Wang, Fenqin Zhang, Liye Zhang, Ning Zhang, Liqing Zhao, and Ms Liming Cai, for help with plant materials, and Drs Ji Qi, Maoteng Li, Longjiang Yu, Fu Xiang, Chenjiang You, Caifei Zhang, Ren Ren, Xinping Qi, and Mian Liu for technical assistance and discussion. This work was supported by funds from grants from the National Natural Science Foundation of China (91531301 and 31670209), from State Key Laboratory of Genetic Engineering at Fudan University, and the National Top Talent Undergraduate Training Program for outstanding undergraduates and the Wangdao Program for undergraduate research at Fudan.
References
- Aldasoro JJ, Aedo C, Navarro C.. 2005. Phylogenetic and phytogeographical relationships in Maloideae (Rosaceae) based on morphological and anatomical characters. Blumea 50:3–32. [Google Scholar]
- Alice LA, Campbell CS.. 1999. Phylogeny of Rubus (Rosaceae) based on nuclear ribosomal DNA internal transcribed spacer region sequences. Am J Bot. 86:81–97. [PubMed] [Google Scholar]
- Avise JC, Robinson TJ.. 2008. Hemiplasy: a new term in the lexicon of phylogenetics. Syst Biol. 57:503–507. [DOI] [PubMed] [Google Scholar]
- Bekaert M, Edger PP, Pires JC, Conant GC.. 2011. Two-phase resolution of polyploidy in the Arabidopsis metabolic network gives rise to relative and absolute dosage constraints. Plant Cell 23:1719–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict JC, DeVore ML, Pigg KB.. 2011. Prunus and Oemleria (Rosaceae) flowers from the late early Eocene Republic flora of northeastern Washington State, U.S.A. Int J Plant Sci. 172:948–958. [Google Scholar]
- Blanc G, Wolfe KH.. 2004. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16:1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess MB, Cushman KR, Doucette ET, Talent N, Frye CT, Campbell CS.. 2014. Effects of apomixis and polyploidy on diversification and geographic distribution in Amelanchier (Rosaceae). Am J Bot. 101:1375–1387. [DOI] [PubMed] [Google Scholar]
- Campbell CS, Evans RC, Morgan DR, Dickinson TA, Arsenault MP.. 2007. Phylogeny of subtribe Pyrinae (formerly the Maloideae, Rosaceae): limited resolution of a complex evolutionary history. Plant Syst Evol. 266:119–145. [Google Scholar]
- Cannon SB, McKain MR, Harkess A, Nelson MN, Dash S, Deyholos MK, Peng Y, Joyce B, Stewart CN, Rolf M, et al. 2015. Multiple polyploidy events in the early radiation of nodulating and nonnodulating legumes. Mol Biol Evol. 32:193–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T, 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell-Caballero J, Alonso R, Ibañez V, Terol J, Talon M, Dopazo J.. 2015. A phylogenetic analysis of 34 chloroplast genomes elucidates the relationships between wild and domestic species within the genus Citrus. Mol Biol Evol. 32:2015–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler MEJ. 1963. The lower Tertiary floras of southern England III. London: British Museum (Natural History; ). [Google Scholar]
- Chin SW, Shaw J, Haberle R, Wen J, Potter D.. 2014. Diversification of almonds, peaches, plums and cherries—molecular systematics and biogeographic history of Prunus (Rosaceae). Mol Phylogen Evol. 76:34–48. [DOI] [PubMed] [Google Scholar]
- Cockerell TDA. 1911. Fossil insects from Florissant, Colorado. Bull Am Mus Nat Hist. 30:71–82. [Google Scholar]
- Considine MJ, Wan Y, D'Antuono MF, Zhou Q, Han M, Gao H, Wang M.. 2012. Molecular genetic features of polyploidization and aneuploidization reveal unique patterns for genome duplication in diploid Malus. PLoS One 7:e29449.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack BP, Wolfe KH.. 2007. When gene marriages don't work out: divorce by subfunctionalization. Trends Genet. 23:270–272. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Casas R, Mort M, Soltis D.. 2016. The influence of habitat on the evolution of plants: a case study across Saxifragales. Ann Bot. doi: 10.1093/aob/mcw160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVore LM, Pigg BK.. 2007. A brief review of the fossil history of the family Rosaceae with a focus on the Eocene Okanogan Highlands of eastern Washington State, USA, and British Columbia, Canada. Plant Syst Evol. 266:45–57. [Google Scholar]
- Dickinson TA, Lo E, Talent N.. 2007. Polyploidy, reproductive biology, and Rosaceae: understanding evolution and making classifications. Plant Syst Evol. 266:59–78. [Google Scholar]
- Dobes C, Paule J.. 2010. A comprehensive chloroplast DNA-based phylogeny of the genus Potentilla (Rosaceae): implications for its geographic origin, phylogeography and generic circumscription. Mol Phylogen Evol. 56:156–175. [DOI] [PubMed] [Google Scholar]
- Ebersberger I, Strauss S, von Haeseler A.. 2009. HaMStR: profile hidden markov model based search for orthologs in ESTs. BMC Evol Biol. 9:157.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman DW. 1975. The Eocene Germer Basin Flora of South-Central Idaho. [M.S. Thesis]. Moscow: University of Idaho.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanoff E, McIntosh WC, Murphey PC.. 2001. Fossil flora and stratigraphy of the Florissant Formation, Colorado. Proc Denver Mus Nat Sci. 4:1–16. [Google Scholar]
- Evans RC, Alice LA, Campbell CS, Kellogg EA, Dickinson TA.. 2000. The granule-bound starch synthase (GBSSI) gene in the Rosaceae: multiple loci and phylogenetic utility. Mol Phylogen Evol. 17:388–400. [DOI] [PubMed] [Google Scholar]
- Evans RC, Campbell CS.. 2002. The origin of the apple subfamily (Rosaceae: Maloideae) is clarified by DNA sequence data from duplicated GBSSI genes. Am J Bot. 89:1478–1484. [DOI] [PubMed] [Google Scholar]
- Feng Y-L, Wicke S, Li J-W, Han Y, Lin C-S, Li D-Z, Zhou T-T, Huang W-C, Huang L-Q, Jin X-H.. 2016. Lineage-specific reductions of plastid genomes in an orchid tribe with partially and fully mycoheterotrophic species. Genome Biol Evol. 8:2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming TH, Kress WJ.. 2011. A brief history of fruits and frugivores. Acta Oecol. 37:521–530. [Google Scholar]
- Fougere-Danezan M, Joly S, Bruneau A, Gao XF, Zhang LB.. 2015. Phylogeny and biogeography of wild roses with specific attention to polyploids. Ann Bot. 115:275–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. 2009. Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol. 60:433–453. [DOI] [PubMed] [Google Scholar]
- Fuentes S, Vivian-Smith A.. 2009. Fertilisation and fruit initiation In: Østergaard L, editor. Annual Plant Reviews Volume 38: Fruit Development and Seed Dispersal. Oxford, UK: A John Wiley & Sons, Ltd; p. 107–171. [Google Scholar]
- Gehrke B, Brauchler C, Romoleroux K, Lundberg M, Heubl G, Eriksson T.. 2008. Molecular phylogenetics of Alchemilla, Aphanes and Lachemilla (Rosaceae) inferred from plastid and nuclear intron and spacer DNA sequences, with comments on generic classification. Mol. Phylogen Evol. 47:1030–1044. [DOI] [PubMed] [Google Scholar]
- Greenwood DR, Archibald SB, Mathewes RW, Moss PT.. 2005. Fossil biotas from the Okanagan Highlands, southern British Columbia and northeastern Washington State: climates and ecosystems across an Eocene landscape. Can J Earth Sci. 42:167–185. [Google Scholar]
- Gu C, Li C, Lu L, Jiang S, Alexander C, Bartholomew B, Brach AR, Boufford DE, Ikeda H, Ohba H, et al. 2003. Rosaceae In: Wu Z, Raven P, Hong D, editors. Flora of China. Beijing: Science Press; p. 46–434. [Google Scholar]
- Hooker JJ, Collinson ME, Sille NP.. 2004. Eocene-Oligocene mammalian faunal turnover in the Hampshire Basin, UK: calibration to the global time scale and the major cooling event. J Geol Soc Lond. 161:161–172. [Google Scholar]
- Huang CH, Sun R, Hu Y, Zeng L, Zhang N, Cai L, Zhang Q, Koch MA, Al-Shehbaz I, Edger PP, et al. 2016. Resolution of Brassicaceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolution. Mol Biol Evol. 33:394–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson CM, Puckett EE, Bekaert M, Pires JC, Conant GC.. 2011. Selection for higher gene copy number after different types of plant gene duplications. Genome Biol Evol. 3:1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummer KE, Janick J.. 2009. Rosaceae: taxonomy, economic importance, genomics In: Folta KM, Gardiner SE, editors. Genetics and Genomics of Rosaceae. New York, USA: Springer; p. 1–17. [Google Scholar]
- Jeffroy O, Brinkmann H, Delsuc F, Philippe H.. 2006. Phylogenomics: the beginning of incongruence? Trends Genet. 22:225–231. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Leebens-Mack J, Ayyampalayam S, Bowers JE, McKain MR, McNeal J, Rolf M, Ruzicka DR, Wafula E, Wickett NJ, et al. 2012. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 13:R3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473:97–100. [DOI] [PubMed] [Google Scholar]
- Johnson LA, Soltis DE.. 1998. Assessing congruence: empirical examples from molecular data In: Soltis DE, Soltis PS, Doyle JJ, editors. Molecular systematics of plants II: DNA sequencing. Boston: Kluwer; p. 297–348. [Google Scholar]
- Judd W, Campbell C, Kellogg E, Stevens P, Donoghue M.. 2015. Plant systematics: a phylogenetic approach, 4th edn. Sunderland: Sinauer Associates. [Google Scholar]
- Kellogg EA. 2016. Has the connection between polyploidy and diversification actually been tested?. Curr Opin Plant Biol. 30:25–32. [DOI] [PubMed] [Google Scholar]
- Lee S, Wen J.. 2001. A phylogenetic analysis of Prunus and the Amygdaloideae (Rosaceae) using ITS sequences of nuclear ribosomal DNA. Am J Bot. 88:150–160. [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS.. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Smith T, Liu C-J, Awasthi N, Yang J, Wang Y-F, Li C-S.. 2011. Endocarps of Prunus (Rosaceae: Prunoideae) from the early Eocene of Wutu, Shandong Province, China. Taxon 60:555–564. [Google Scholar]
- Li Z, Baniaga AE, Sessa EB, Scascitelli M, Graham SW, Rieseberg LH, Barker MS.. 2015. Early genome duplications in conifers and other seed plants. Sci Adv. 1:e1501084.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo EY, Donoghue MJ.. 2012. Expanded phylogenetic and dating analyses of the apples and their relatives (Pyreae, Rosaceae). Mol Phylogen Evol. 63:230–243. [DOI] [PubMed] [Google Scholar]
- Lo EY, Stefanovic S, Dickinson TA.. 2010. Reconstructing reticulation history in a phylogenetic framework and the potential of allopatric speciation driven by polyploidy in an agamic complex in Crataegus (Rosaceae). Evolution 64:3593–3608. [DOI] [PubMed] [Google Scholar]
- Lundberg M, Topel M, Eriksen B, Nylander JA, Eriksson T.. 2009. Allopolyploidy in Fragariinae (Rosaceae): comparing four DNA sequence regions, with comments on classification. Mol Phylogen Evol. 51:269–280. [DOI] [PubMed] [Google Scholar]
- MacGinitie HD. 1969. The Eocene Green River Flora of northwestern Colorado and northeastern Utah. Univ Cal Publ Geol Sci. 83:1–140. [Google Scholar]
- MacGinitie HD. 1953. Fossil plants of the Florissant beds Colorado. Carn Inst Wash, Contribut Paleon. 599:1–198. [Google Scholar]
- Maddison WP, Maddison DR.. 2004. Mesquite: a modular system for evolutionary analysis. Available from: http://mesquiteproject.org
- Madlung A, Tyagi AP, Watson B, Jiang H, Kagochi T, Doerge RW, Martienssen R, Comai L.. 2005. Genomic changes in synthetic Arabidopsis polyploids. Plant J. 41:221–230. [DOI] [PubMed] [Google Scholar]
- Mathews WH. 1964. Potassium-Argon age determination of Cenozoic volcanic rocks from British Columbia. Geol Soc Am Bull. 75:465–468. [Google Scholar]
- Matthews JV, Ovenden LE.. 1990. Late Tertiary plant macrofossils from localities in arctic/subarctic North America: a review of the data. Arctic 43:364–392. [Google Scholar]
- Matthews JV, Westgate JA, Ovenden L, Carter LD, Fouch T.. 2003. Stratigraphy, fossils, and age of sediments at the upper pit of the Lost Chicken gold mine: new information on the late Pliocene environment of east central Alaska. Quat Res. 60:9–18. [Google Scholar]
- McIntosh WC, Chapin CE.. 2004. Geochronology of the central Colorado volcanic field. New Mexico Bureau Geol Min Res Bull. 160:205–237. [Google Scholar]
- Mirarab S, Reaz R, Bayzid MS, Zimmermann T, Swenson MS, Warnow T.. 2014. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics 30:i541–i548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima M, Ohmido N, Fukui K, Yahara T.. 2002. Trends in site-number change of rDNA loci during polyploid evolution in Sanguisorba (Rosaceae). Chromosoma 110:550–558. [DOI] [PubMed] [Google Scholar]
- Morgan DR, Soltis DE, Robertson KR.. 1994. Systematic and evolutionary implications of rbcL sequence variation in Rosaceae. Am J Bot. 81:890–903. [Google Scholar]
- Oh SH. 2013. Phylogenetic analysis of PISTILLATA sequences in Neillia (Rosaceae). J Plant Biol. 56:145–151. [Google Scholar]
- Peppe DJ, Hickey LJ, Miller IM, Green WA.. 2008. A morphotype catalogue, floristic analysis and stratigraphic description of the Aspen Shale flora (Cretaceous-Albian) of Southwestern Wyoming. Bull Peabody Mus Nat History 49:181–208. [Google Scholar]
- Philippe H, Delsuc F, Brinkmann H, Lartillot N.. 2005. Phylogenomics. Annu Rev Ecol Evol Syst. 36:541–562. [Google Scholar]
- Phipps JB. 2014. Flora of North America North of Mexico, Vol. 9, Magnoliophyta: Picramniaceae to Rosaceae. New York and Oxford: Oxford University Press. [Google Scholar]
- Plazzi F, Puccio G, Passamonti M.. 2016. Comparative large-scale mitogenomics evidences clade-specific evolutionary trends in mitochondrial DNAs of Bivalvia. Genome Biol Evol. 8:2544–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Neves N, Silva M, Lewis MS, Madlung A, Comai L, Viegas W, Pikaard CS.. 2004. Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc Natl Acad Sci U S A. 101:18240–18245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter D, Eriksson T, Evans RC, Oh SH, Smedmark JEE, Morgan DR, Kerr M, Robertson KR, Arsenault MP, Dickinson TA, et al. 2007a. Phylogeny and classification of Rosaceae. Plant Syst Evol. 266:5–43. [Google Scholar]
- Potter D, Still SM, Grebenc T, Ballian D, Božič G, Franjiæ J, Kraigher H.. 2007b. Phylogenetic relationships in tribe Spiraeeae (Rosaceae) inferred from nucleotide sequence data. Plant Syst Evol. 266:105–118. [Google Scholar]
- Ridley HN. 1930. The dispersal of plants throughout the world. Ashford, UK: L. Reeve & Co., Ltd. [Google Scholar]
- Robertson KR, Phipps JB, Rohrer JR, Smith PG.. 1991. A synopsis of genera in Maloideae (Rosaceae). Syst Bot. 2:376–394. [Google Scholar]
- Rousseau-Gueutin M, Gaston A, Ainouche A, Ainouche ML, Olbricht K, Staudt G, Richard L, Denoyes-Rothan B.. 2009. Tracking the evolutionary history of polyploidy in Fragaria L. (strawberry): new insights from phylogenetic analyses of low-copy nuclear genes. Mol Phylogen Evol. 51:515–530. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ. 2003. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19:301–302. [DOI] [PubMed] [Google Scholar]
- Schmidt-Lebuhn AN, Fuchs J, Hertel D, Hirsch H, Toivonen J, Kessler M.. 2010. An Andean radiation: polyploidy in the tree genus Polylepis (Rosaceae, Sanguisorbeae). Plant Biol. 12:917–926. [DOI] [PubMed] [Google Scholar]
- Schorn HE. 1998. Holodiscus lisii (Rosaceae): a new species of ocean spray from the late Eocene Florissant Formation, Colorado, USA. PaleoBios 18:21–24. [Google Scholar]
- Schulze-Menz GK. 1964. Rosaceae In: Melchior H, editor. Engler's Syllabus der Pflanzenfamilien. Berlin: Gebrüder Borntraeger; p. 209–218. [Google Scholar]
- Seo TK. 2008. Calculating bootstrap probabilities of phylogeny using multilocus sequence data. Mol Biol Evol. 25:960–971. [DOI] [PubMed] [Google Scholar]
- Seymour GB, Ostergaard L, Chapman NH, Knapp S, Martin C.. 2013. Fruit development and ripening. Annu Rev Plant Biol. 64:219–241. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M.. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17:1246–1247. [DOI] [PubMed] [Google Scholar]
- Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP, et al. 2011. The genome of woodland strawberry (Fragaria vesca). Nat Genet. 43:109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedmark JEE, Eriksson T.. 2002. Phylogenetic relationships of Geum (Rosaceae) and relatives inferred from the nrITS and trnL-trnF regions. Syst Bot. 27:303–317. [Google Scholar]
- Spooner DM. 2016. Species delimitations in plants: lessons learned from potato taxonomy by a practicing taxonomist. J Syst Evol. 54:191–203. [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Talent N, Dickinson TA.. 2005. Polyploidy in Crataegus and Mespilus (Rosaceae, Maloideae): evolutionary inferences from flow cytometry of nuclear DNA amounts. Can J Bot. 83:1268–1304. [Google Scholar]
- The Angiosperm Phylogeny Group. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20. [Google Scholar]
- Töpel M, Antonelli A, Yesson C, Eriksen B.. 2012. Past climate change and plant evolution in western North America: a case study in Rosaceae. PLoS One 7:e50358.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vamosi JC, Dickinson TA.. 2006. Polyploidy and diversification: a phylogenetic investigation in Rosaceae. Int J Plant Sci. 167:349–358. [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, et al. 2010. The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet. 42:833–839. [DOI] [PubMed] [Google Scholar]
- Verde I, Abbott AG, Scalabrin S, Jung S, Shu S, Marroni F, Zhebentyayeva T, Dettori MT, Grimwood J, Cattonaro F, et al. 2013. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet. 45:487–494. [DOI] [PubMed] [Google Scholar]
- Wang H, Moore MJ, Soltis PS, Bell CD, Brockington SF, Alexandre R, Davis CC, Latvis M, Manchester SR, Soltis DE.. 2009. Rosid radiation and the rapid rise of angiosperm-dominated forests. Proc Natl Acad Sci U S A. 106:3853–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, et al. 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehr WC, Hopkins DQ.. 1994. The Eocene orchards and gardens of Republic, Washington. Wash Geol. 22:27–34. [Google Scholar]
- Wen J, Egan A, Dikow R, Zimmer E.. 2015. Utility of transcriptome sequencing for phylogenetic inference and character evolution In: Hörandl E, Appelhans M, editors. Next-generation sequencing in plant systematics. Koenigstein, Germany: Koeltz Scientific Books; p. 51–91. [Google Scholar]
- Wendel JF, Doyle JJ.. 1998. Phylogenetic incongruence: window into genome history and molecular evolution In: Soltis DE, Soltis PS, Doyle JJ, editors. Molecular systematics of plants II: DNA sequencing. Boston: Kluwer; p. 265–296. [Google Scholar]
- Wolfe JA, Gregory-Wodzicki KM, Molnar P, Mustoe G.. 2003. Rapid uplift and then collapse in the Eocene of the Okanagan? evidence from paleobotany. Geological Association of Canada, Mineralogical Association of Canada, Society of Economic Geologists, Joint Annual Meeting; Vancouver, Canada. p. abstract 533.
- Wu J, Wang Z, Shi Z, Zhang S, Ming R, Zhu S, Khan MA, Tao S, Korban SS, Wang H, et al. 2013. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 23:396–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Moore MJ, Brockington SF, Soltis DE, Wong GK-S, Carpenter EJ, Zhang Y, Chen L, Yan Z, Xie Y, et al. 2015. Dissecting molecular evolution in the highly diverse plant clade Caryophyllales using transcriptome sequencing. Mol Biol Evol. 32:2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachos J, Pagani M, Sloan L, Thomas E, Billups K.. 2001. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292:686–693. [DOI] [PubMed] [Google Scholar]
- Zachos JC, Dickens GR, Zeebe RE.. 2008. An early Cenozoic perspective on greenhouse warming and carbon-cycle dynamics. Nature 451:279–283. [DOI] [PubMed] [Google Scholar]
- Zeng L, Zhang Q, Sun R, Kong H, Zhang N, Ma H.. 2014. Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat Commun. 5:4956.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetter R, Hofmann CC, Draxler I, de Cabrera JD, Vergel MM, Vervoorst F.. 1999. A rich Middle Eocene microflora at Arroyo de los Mineros, near Cañadón Beta, NE Tierra del Fuego Province, Argentina. Abh Geol Bundesanst. 56:436–460. [Google Scholar]
- Zhang N, Zeng L, Shan H, Ma H.. 2012a. Highly conserved low-copy nuclear genes as effective markers for phylogenetic analyses in angiosperms. New Phytol. 195:923–937. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Chen W, Sun L, Zhao F, Huang B, Yang W, Tao Y, Wang J, Yuan Z, Fan G, et al. 2012b. The genome of Prunus mume. Nat Commun. 3:1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Jiang XW, Zuo YJ, Liu XL, Chin SW, Haberle R, Potter D, Chang ZY, Wen J.. 2016. Multiple events of allopolyploidy in the evolution of the racemose lineages in Prunus (Rosaceae) based on integrated evidence from nuclear and plastid data. PLoS One 11:e0157123.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu ZM, Gao XF, Fougere-Danezan M.. 2015. Phylogeny of Rosa sections Chinenses and Synstylae (Rosaceae) based on chloroplast and nuclear markers. Mol Phylogen Evol. 87:50–64. [DOI] [PubMed] [Google Scholar]
- Zimmer E, Wen J.. 2015. Using nuclear gene data for plant phylogenetics: progress and prospects II. Next-gen approaches. J Syst Evol. 53:371–379. [Google Scholar]
- Zimmer EA, Wen J.. 2013. Using nuclear gene data for plant phylogenetics: progress and prospects. Mol Phylogen Evol. 66:539–550. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.