Abstract
DNA methylation contributes to gene and transcriptional regulation in eukaryotes, and therefore has been hypothesized to facilitate the evolution of plastic traits such as sociality in insects. However, DNA methylation is sparsely studied in insects. Therefore, we documented patterns of DNA methylation across a wide diversity of insects. We predicted that underlying enzymatic machinery is concordant with patterns of DNA methylation. Finally, given the suggestion that DNA methylation facilitated social evolution in Hymenoptera, we tested the hypothesis that the DNA methylation system will be associated with presence/absence of sociality among other insect orders. We found DNA methylation to be widespread, detected in all orders examined except Diptera (flies). Whole genome bisulfite sequencing showed that orders differed in levels of DNA methylation. Hymenopteran (ants, bees, wasps and sawflies) had some of the lowest levels, including several potential losses. Blattodea (cockroaches and termites) show all possible patterns, including a potential loss of DNA methylation in a eusocial species whereas solitary species had the highest levels. Species with DNA methylation do not always possess the typical enzymatic machinery. We identified a gene duplication event in the maintenance DNA methyltransferase 1 (DNMT1) that is shared by some Hymenoptera, and paralogs have experienced divergent, nonneutral evolution. This diversity and nonneutral evolution of underlying machinery suggests alternative DNA methylation pathways may exist. Phylogenetically corrected comparisons revealed no evidence that supports evolutionary association between sociality and DNA methylation. Future functional studies will be required to advance our understanding of DNA methylation in insects.
Keywords: DNA methylation, whole genome bisulfite sequencing, social behavior, phylogenetic comparisons, molecular evolution.
Introduction
DNA methylation is recognized as an important DNA modification, which aids in structural integrity and proper regulation of the genome for many species. In animals DNA methylation typically occurs at CG sites, established de novo by DNA methyltransferase 3 (DNMT3), and maintained by the maintenance methyltransferase DNMT1 (Goll and Bestor 2005; Cheng and Blumenthal 2008; Kim et al. 2008). Homologous to DNMT1 and DNMT3 is DNMT2: a tRNAAsp DNA methyltransferase, which does not contribute to the DNA methylome (Goll et al. 2006). DNA methylation in insects is restricted to the transcribed regions of genes (Feng et al. 2010; Lyko et al. 2010; Bonasio et al. 2012). This is in contrast to DNA methylation in mammals, which is found throughout the genome except at CpG islands near promoters of genes (Ehrlich et al. 1982; Suzuki and Bird 2008; Cedar and Bergman 2009). As in insects, DNA methylation is found within genes of plants, but in contrast to insects it predominantly occurs in transposons and other repetitive elements (Niederhuth et al. 2016). Despite CG DNA methylation conservation across the tree of life (Feng et al. 2010; Zemach et al. 2010; Huff and Zilberman 2014), the understanding of its contribution to genome function and complex traits is limited. In insects, DNA methylation is implicated in behavioral plasticity and social behavior, especially eusociality in Hymenoptera (ants, bees, wasps, and sawflies) (Yan et al. 2014; Yan et al. 2015). Comparisons between pairs of species that are social or are not social, and between pairs of castes within a single species of the order Hymenoptera, support the association between social behavior and DNA methylation (Elango et al. 2009; Bonasio et al. 2012; Patalano et al. 2015). Although others have observed little association between social behavior and DNA methylation (Libbrecht et al. 2016; Standage et al. 2016). Overall, comparative epigenomics in insects has been taxonomically piecemeal, and examining potential influences on insect sociality beyond eusocial traits have yet to be performed.
Here we investigate patterns of DNA methylation, its underlying mechanism and its relationship to social behavior across a taxonomically diverse sampling of insects. We first define the extent that DNA methylation occurs in insects. We tested for the presence/absence of DNA methylation using whole genome bisulfite sequencing (WGBS) and utilized previously published data to generate a dataset comprising of 41 species of insects from six orders, including three orders where eusociality has evolved. To expand our taxonomic sampling and cover insect orders and species where sequencing has not yet been done, we documented the presence/absence of DNA methylation using in 123 species of insects from 11 orders. The measure relies on natural, spontaneous deamination of methylated cytosines to thymines, and robustly recovers presence/absence of DNA methylation (see Materials and Methods). Next we tested the hypothesis that the presence/absence of DNMT1 and DNMT3 explains the patterns of DNA methylation observed across insects. Finally, we test the hypothesis that DNA methylation is associated with the evolution of sociality, more broadly defined (Wilson 1971; Costa 2006) across insects and independent of phylogenetic history. We tested our hypothesis at two broad levels of sociality: (1) the evolution of sociality along a spectrum from solitary to eusocial and (2) the evolution of any level of sociality. To reduce error in how social behavior was classified we further tested our hypothesis within Hymenoptera only.
We found widespread presence but diverse patterns of DNA methylation across the insect tree of life. This is in part supported by the presence/absence of DNA methyltransferases, including duplication of DNMT1 in some groups of insects and lack of DNMT3 in others. However, the presence of DNA methylation in insects is associated with the presence of the maintenance rather than the de novo DNA methyltransferase. We found no association between sociality and DNA methylation in insects. Instead we suggest that DNA methylation plays a role in a broader, ubiquitous function or has diverse roles. Phylogenetic comparative tests that take into account evolutionary history and nonindependence of species are a first step in identifying function of DNA methylation in insects. Functional tests are now needed to elucidate the role of DNA methylation in insects, and this study will facilitate the identification of species suitable for such experimentation.
Results and Discussion
DNA Methylation Levels Are Highly Variable across Insects
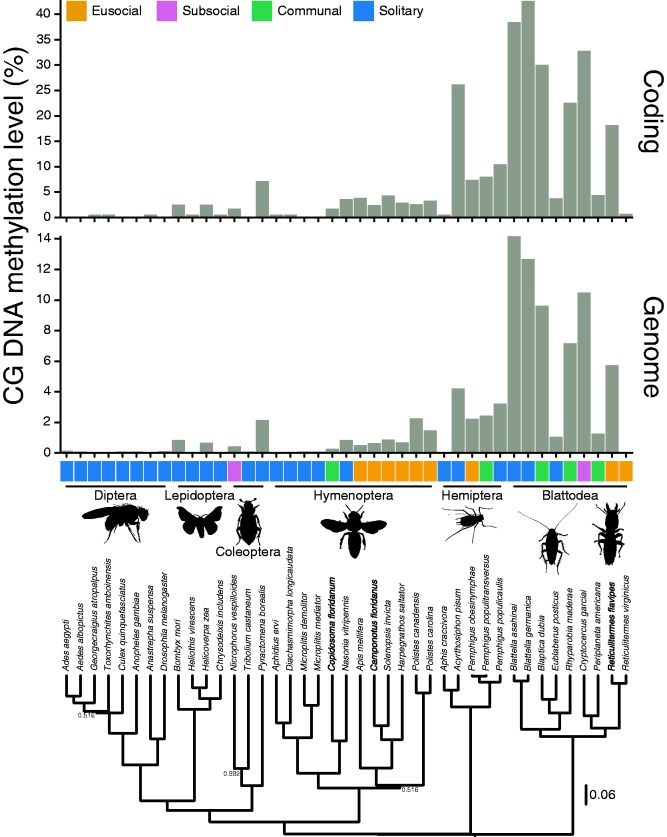
Estimates of DNA methylation from WGBS from 41 species from six orders (Blattodea [n = 9], Hemiptera [n = 5], Hymenoptera [n = 12], Coleoptera [n = 3], Lepidoptera [n = 4], and Diptera [n = 8]) revealed diverse levels of DNA methylation. DNA methylation was found in all insect orders except Diptera and in insects that are social and solitary (fig. 1 and supplementary table S1, Supplementary Material online). DNA methylation levels within coding regions are higher than genome-wide levels for all species, which suggests DNA methylation is predominantly found in coding regions across insects (fig. 1 and supplementary table S1, Supplementary Material online). Higher levels of DNA methylation within coding regions compared with genome-wide might partially reflect highly conserved protein-coding genes in insects, which have the highest levels of DNA methylation (Glastad et al. 2014). DNA methylation levels within the commonly studied hymenopteran (Kronforst et al. 2008; Elango et al. 2009; Bonasio et al. 2012; Patalano et al. 2015; Rehan et al. 2016) are some of the lowest compared with other species sampled (fig. 1 and supplementary table S1, Supplementary Material online). Additionally, several species of Hymenoptera (Aphidius ervi, Microplitis demolitor and Microplitis mediator) are completely absent or have extremely reduced levels of DNA methylation (fig. 1 and supplementary table S1, Supplementary Material online). This loss or extreme reduction in DNA methylation is similar to what is observed in the eusocial species Polistes dominula (Hymenoptera) (Standage et al. 2016). Levels of DNA methylation genome-wide and within coding regions were overall higher in the order Blattodea (fig. 1). Genome-wide and coding levels of DNA methylation were highest in the solitary cockroaches in the genus Blattella, B. asahinai and B. germanica, respectively. Furthermore, drastic differences in levels of DNA methylation were observed between the highly eusocial species Reticulitermes flavipes and R. virginicus within Blattodea (fig. 1). Conversely, similar levels of DNA methylation were observed between closely related Pemphigus spp. of aphids (Hemiptera) with differing social behaviors (fig. 1). Additionally, the solitary species Acyrthosiphon pisum had the highest levels of DNA methylation within the Hemiptera. Overall, DNA methylation levels were variable in insects, with some species’ levels reaching those observed in plants (Feng et al. 2010; Zemach et al. 2010; Niederhuth et al. 2016; Takuno et al. 2016), where its contribution to complex traits still remains largely unknown.
Fig. 1.
WGBS reveals extensive variation of DNA methylation in insects. (A) Genomic levels of DNA methylation in insects ranges from zero (all Diptera examined) to ∼14% (Blattella asahinai). Higher ranges are observed for coding regions; zero (all Diptera examined) to ∼42% (Blattella germanica). Overall, levels are highest in Blattodea, and do not always associate with social species. (B) A species tree constructed from nuclear and mitochondrial loci, which was used in Phylogenetic Generalized Least Squares (PGLS) analysis. Results from this analysis revealed that there is no correlation between social behavior and DNA methylation (table 1). Values at nodes are posterior probabilities <0.95; all blank nodes have ≥0.95 posterior probability.
DNA Methylation Is Present across the Insect Tree of Life
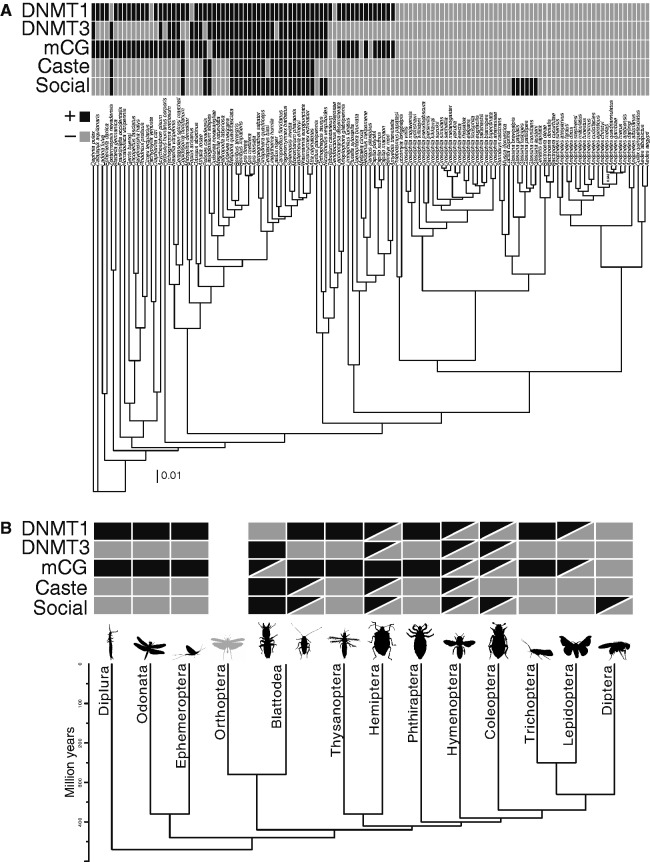
To taxonomically expand on insect orders and species, the distribution of of 123 insect species from 11 orders were investigated for signatures of DNA methylation. The bimodal distribution of values is a robust estimator for the simpler question of presence/absence of DNA methylation in insect genomes. When compared with estimates of DNA methylation from WGBS, accurately estimates presence/absence of DNA methylation in 18/18 (100%) species (see Materials and Methods, and supplementary table S1, Supplementary Material online). We again found that DNA methylation was identified in insect species from all orders except Diptera (fig. 2). Species belonging to the order Diptera comprised ∼46% (57/124) of the total number of species investigated, which was the largest number of species sampled from one order. The absence of bimodal distribution from members of both the Nematocera and Brachycera sub-orders suggests that DNA methylation was lost early on in dipteran evolution. Furthermore, stochastic mapping supports a single loss of DNA methylation at the base of Diptera (supplementary fig. S1A, Supplementary Material online). Other losses of DNA methylation in Microplitis demolitor (Hymenoptera), Lasioglossum albipes (Hymenoptera), Tribolium castaneum (Coleoptera), Dendroctonus ponderosae (Coleoptera), Melitaea cinxia (Lepidoptera), and Danaus plexippus (Lepidoptera) are species/lineage-specific (supplementary fig. S1A, Supplementary Material online). However, denser taxonomic sampling may reveal some of these losses to be shared events. The evolution of DNA methylation is deeply rooted in Hexapoda, going as far back as the Ordovician and the divergence of Diplura (∼489.84–436.68 Ma; Misof et al. 2014), but is most likely older (Feng et al. 2010; Zemach et al. 2010; Huff and Zilberman 2014).
Fig. 2.
Patterns of DNA methylation and social behavior across the insect tree of life. Relationships of 123 insect species, and outgroups Catajapyx aquilonaris (Dipluran) and Daphnia pulex (Crustacea) investigated with DNA methyltransferases, sociality, division of labor and DNA methylation scored as a binary (presence/absence) trait. The tree was constructed from 58 nuclear protein coding loci, and was used in Pagel’s test for evolutionary dependence. All nodes except one had posterior probability of 1.00, which is indicated on the phylogeny. (B) A chronogram of insect order relationships with sociality, division of labor and DNA methylation scored as binary (presence/absence) trait. The chronogram was modified from Misof et al. 2014. For (A) and (B), traits are represented as shaded boxes above each species or order. Half-filled boxes indicate the trait is variable within the corresponding order.
Evolution of DNMT1, DNMT2 and DNMT3, and DNA Methylation
DNMT1 is found in all orders of insects investigated in this study except Diptera (fig. 3A and supplementary table S2, Supplementary Material online). Only DNMT2 is found in Diptera, which reflects the lack of DNA methylation in the genomes of all species belonging to this order (fig. 2B and supplementary table S1, Supplementary Material online). The loss of DNA methylation co-occurred with the loss of DNMT1 and DNMT3 in Diptera ∼206.28–107.26 Ma (Misof et al. 2014) (supplementary fig. S1, Supplementary Material online).
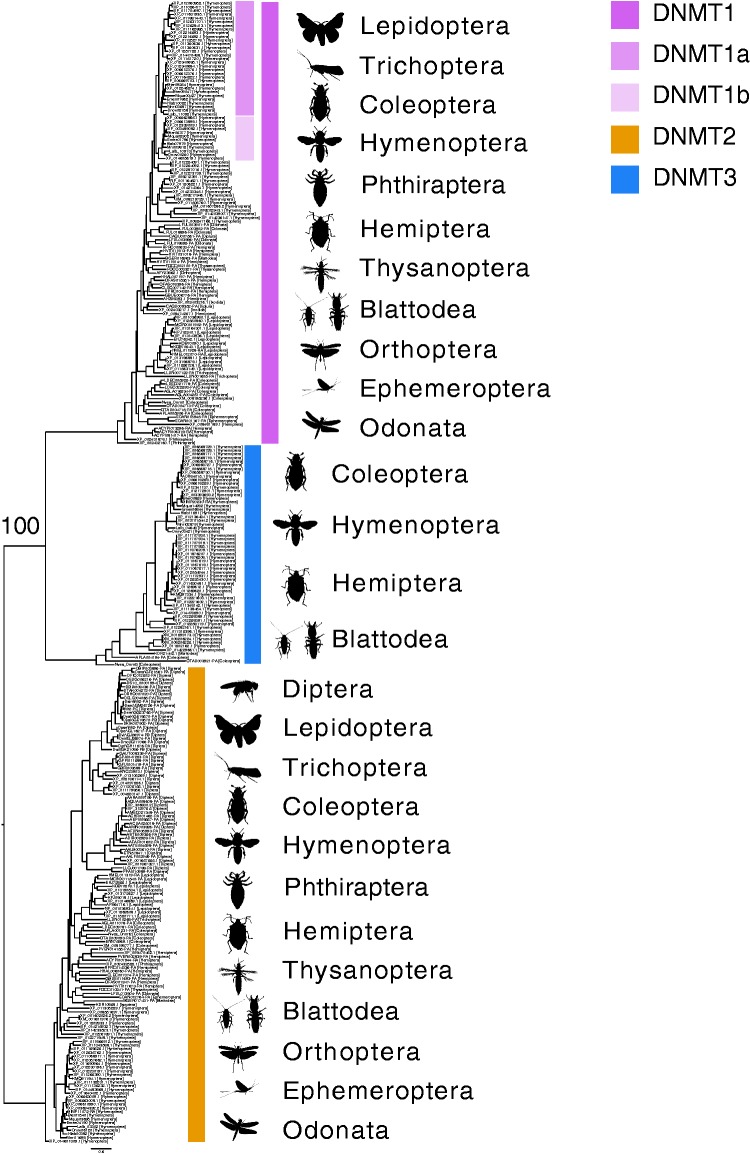
Fig. 3.
Evolution of DNMT1, 2 and 3 across Insecta and other Arthropoda. Relationships of DNMT1, 2, and 3 in insects, Diplura and Ixodida (Arachnida). DNMT2 can be found in all insect orders investigated, while DNMT1 and 3 are more order-poor. The insect order for each sequence is provided in square brackets following the GenBank or genome annotation accession number. The tree was rooted to DNMT2.
Only DNMT1 and DNMT2 are found in lepidopteran species (fig. 3A and supplementary table S2, Supplementary Material online). This observation supports the hypothesis that DNMT3 was lost from species belonging to this order between ∼177.99–116.45 Ma (Misof et al. 2014). Also, the sister order to Lepidoptera, Trichoptera, only possesses DNMT1 and DNMT2. Therefore, the timing of this loss of DNMT3 may be older and shared by Diptera, Lepidoptera and Trichoptera. However, Trichoptera is represented by a single species in our data and this loss might be species/lineage-specific. Despite missing DNMT3 from assembled transcriptomes or genomes—as in the lepidopteran Bombyx mori—evidence for DNA methylation is still observed from WGBS (supplementary table S1, Supplementary Material online) data and from distributions of values (supplementary table S1 and fig. S2, Supplementary Material online) (Xiang et al. 2010). Similarly, 2/7 Coleoptera and 9/10 Hemiptera only possess DNMT1 and DNMT2, and DNA methylation is expected to be present based on bimodality of (supplementary table S1, Supplementary Material online).
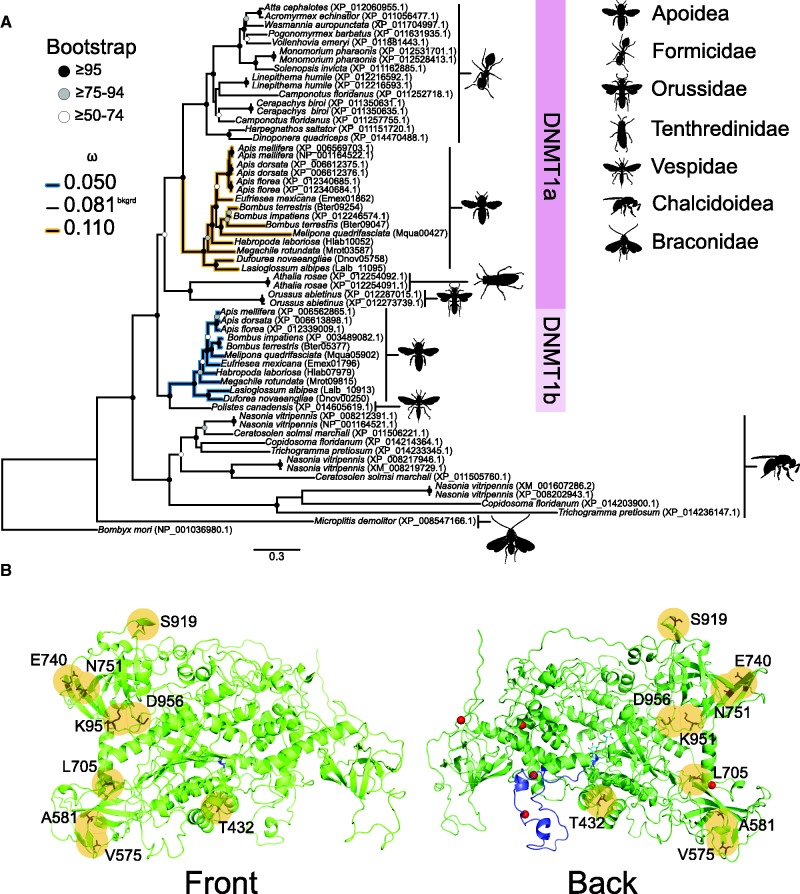
Several duplications were observed in the DNMT1 clade, but none coincide with presence or absence of DNA methylation or sociality. A duplication event shared by many hymenopteran gave rise to what is referred to as DNMT1a and DNMT1b (fig. 4). Differences in codon and amino acid alignment tree topology suggest all hymenopterans or bees and ants shared the duplication, respectively (fig. 4A and supplementary fig. S3, Supplementary Material online). The amino acid alignment tree topology has higher node support than the codon alignment, which provides stronger support for the duplication event being shared by the superfamilies Apoidea (bees) and family Formicidae (ants) and places the timing for this duplication following their divergence, ∼145 Ma (Branstetter et al. 2016) (fig. 4A and supplementary fig. S3, Supplementary Material online). Species relationships suggest a loss of DNMT1b in Formicidae. Whereas DNMT1a and DNMT1b can both be found in the superfamily Apoidea (families Apidae, Halictidae, and Megachilidae). Chalcid wasps (family Chalcidoidea) experienced at least two superfamily-specific duplication events, as suggested by the monophyletic groups containing Nasonia vitripennis (fig. 4A). Additional species-specific duplications—in-paralogs—were observed in orders Hemiptera and Lepidoptera (fig. 3), but these could represent allelic variation of DNMT1.
Fig. 4.
Divergent nonneutral evolution of DNMT1a and DNMT1b in Apoidea. (A) Hypothesized relationships among DNMT1 in Hymenoptera suggests a duplication event shared by the superfamilies Apoidea (bees) and family Formicidae (ants) gave rise to what is referred to as DNMT1a and DNMTb (supplementary fig. S3, Supplementary Material online). DNMT1b appears to have been lost from Formicidae (ants), whereas both DNMT1a and DNMT1b were retained in Apoidea (families Apidae, Halictidae, and Megachilidae). Divergent selection between DNMT1a and DNMT1b in Apoidea suggests the former is under relaxed purifying selection and the latter is under purifying selection. Bombyx mori (Lepidoptera) was used to root the tree, and was excluded from PAML analyses. (B) Several sites in DNMT1b were identified as under positive selection (yellow circles), with one site (T432) in the CXXC zinc finger domain, and three (V575, A581, S919) in the BAH domain. The crystal structure of Apis mellifera DNMT1b was predicted from Mus musculus DNMT1 using the program HHpred with default settings (Hildebrand et al. 2009). Details including red spheres, dark and light blue colouring specify Zn2+ ions, CXXC domain and s-adenosyl-l-homocysteine, the secondary product from the DNA methylation reaction it performs, according to Mus musculus DNMT1, respectively. Bootstrap support values are characterized as shaded circles. dN/dS (ω) values are for the most preferred branch model.
Gene duplication is a source of genetic novelty and functional divergence, which can be characterized by nonneutral evolution (Lynch and Conery 2000). Divergent selection is observed between DNMT1a and DNMT1b in Apoidea (bees), with DNMT1a having experienced relaxed purifying selection and DNMT1b purifying selection (fig. 4). Furthermore, positively selected sites were identified in DNMT1b, and these include one site (T432) in the CXXC zinc finger domain, and three (V575, A581, S919) in the BAH domain (fig. 4). DNMT1 paralogs might maintain DNA methylation differently, including efficiency or rate, and spatially and/or temporally. The contribution of each paralog to the maintenance of the DNA methylome is unknown, and functional studies are required to address the fates of these duplicated genes.
DNMT3 is the most order-poor of the DNA methyltransferases, and was only identified in species belonging to Blattodea, Coleoptera, Hemiptera, and Hymenoptera (fig. 3A and supplementary table S2, Supplementary Material online). Multiple lineage-specific (n = 9) and several larger shared (n = 5) losses of DNMT3 are expected to have occurred based on stochastic mapping (supplementary fig. S1, Supplementary Material online). For species with annotated genomes, DNMT3 is not always accompanied by the presence of DNMT1 (supplementary table S2, Supplementary Material online). Given the scarcity of DNMT3 in insects, a positive correlation between the presence of DNA methylation and DNMT1, and lack of a correlation between DNA methylation and DNMT3 (supplementary fig. S1 and table S3, Supplementary Material online), suggests DNMT3 may be dispensable for DNA methylation or DNMT1 compensates for DNMT3. However, DNMT1 is missing protein domains typically associated with metazoan de novo DNA methyltransferases. For example, DNMT1 does not contain a PWWP domain (PF00855), which interacts with DNA and histone lysine modified nucleosomes (Qiu et al. 2002; Qin and Min 2014). PWWP domain containing proteins exist (e.g., Bombyx mori), however none of these proteins contain a C-5 cytosine-specific DNA methylase domain (PF00145), which is required for DNA methylation (Bestor and Verdine 1994; Goll and Bestor 2005; Cheng and Blumenthal 2008). Based on these observations two possible alternative mechanisms explain the presence of DNA methylation in lieu of typical enzymes: (1) de novo DNA methylation does not occur via a DNMT-like protein and (2) DNA methylation is not reprogrammed during embryogenesis and is robustly maintained by DNMT1 during each cell replication. However, alternative mechanisms might exist, and functional tests of DNA methylation mechanisms in insects warrant further investigation.
Sociality in Insects Is Not Evolutionarily Associated with DNA Methylation
DNA methylation has been proposed to control many aspects of sociality, especially in Hymenoptera, including behavior expressed in social interactions, caste determination, and learning and memory (reviewed in Li-Byarlay 2016). Other investigators have suggested that empirical evidence for caste-specific DNA methylation in Hymenoptera is weak (Libbrecht et al. 2016). However, relatedness of species has not been considered in previous studies. The most powerful comparative tests of association control for nonindependence of species relationships (Felsenstein 1985).
We found DNA methylation in insects exhibiting the complete range of social behavior based on modality of (supplementary table S1, Supplementary Material online). This includes insects classified as solitary, communal (group living), subsocial (parental care), and eusocial (cooperative brood care, common nest site, overlapping generations, and reproductive castes) (Wilson 1971; Costa 2006). The order Hymenoptera contains species that are solitary or eusocial; however, evidence for DNA methylation based on did not always co-occur with eusociality, and vice versa (supplementary table S1, Supplementary Material online). Moreover, all species examined within the order Lepidoptera are solitary, yet 8/10 showed evidence for DNA methylation based on the distribution of , respectively (supplementary table S1, Supplementary Material online). Similarly, 3/4 solitary coleopteran species were expected to have DNA methylation based on bimodality of . Six Diptera in the genus Glossina have adenotrophic viviparity, a unique type of parental care in insects where larvae hatch, develop, and are fed through special glands within the female (International Glossina Genome Initiative 2014). Yet Glossina spp. do not have DNA methylation based on the distribution of . Together this supports the hypothesis for the evolution of adenotrophic viviparity in Glossina spp. occurring after the loss of DNA methylation. This is contrary to the burying beetle Nicrophorus vespilloides, which has DNA methylation based on WGBS and (Cunningham et al. 2015), and also provides parental care through provisioning developing offspring (Walling et al. 2008) (supplementary table S1, Supplementary Material online). The presence of DNA methylation across insect diversity suggests a more ubiquitous function, which may include transcriptional gene regulation and alternative splicing (Lyko et al. 2010; Shukla et al. 2011; Li-Byarlay et al. 2013; Yearim et al. 2015). Alternatively, diverse roles of DNA methylation may exist.
If sociality reflects DNA methylation, we would expect a gain in DNA methylation to result in either a gain of social behavior or a predisposition to evolve social behavior. Thus, if sociality reflects DNA methylation we would expect a phylogenetically corrected correlation between these two traits. We do not find this. Sociality—including division of labor—is not evolutionary dependent on DNA methylation (table 1 and supplementary tables S3 and S4, Supplementary Material online). This lack of dependence holds true when DNA methylation is categorized as a discrete or continuous trait, and sociality is categorized into multiple classes or as presence/absence (figs. 1 and2A;table 1). Although there are differences in transition rates between social behavior and DNA methylation states, they are not dependent on one another (table 1 and supplementary fig. S4, Supplementary Material online). For example, transition from social to solitary occurs at higher rate compared with transitions from solitary to social when DNA methylation is present (supplementary fig. 2A, Supplementary Material online). Furthermore, based on Akaike information criterion (AIC), an independent model of evolution for DNA methylation and social behavior is preferred (table 1). Social behavior can be qualitatively classified in several ways (Wilson 1971; Costa 2006), which can potentially affect comparisons to presence/absence of DNA methylation. Thus, to limit any confounding effect of social behavior classification methods we performed Pagel’s test with only hymenopteran. This analysis further showed that these traits are not dependent when only the 33 species within Hymenoptera are considered (table 1). This strongly supports a lack of a correlation, including in insects highly recognized as social. As dipterans composed a large fraction of species investigated in this study and do not have DNA methylation (figs. 1 and2), we were concerned that they may be biasing our analyses. However, when all species within Diptera are removed we similarly recover a lack of dependence between sociality and DNA methylation (table 1). Together this evidence supports the hypothesis that sociality and DNA methylation are not evolutionarily dependent.
Table 1.
Sociality and DNA Methylation Are Not Evolutionary Dependent.
| Method | Taxa | n | Trait x | Trait y | Preferred Model | P value |
|---|---|---|---|---|---|---|
| PGLS | Insecta | 41 | DNA methylationC | Eusocial | λfree | 0.445 |
| Subsocial | 0.130 | |||||
| Communal | 0.580 | |||||
| Solitary | 0.820 | |||||
| Insecta | 41 | DNA methylationG | Eusocial | λfree | 0.442 | |
| Subsocial | 0.097 | |||||
| Communal | 0.539 | |||||
| Solitary | 0.880 | |||||
| Pagel’s test | Insectaa | 125 | DNA methylation | Sociality | Independent | 0.721 |
| Insectaa | 125 | DNA methylation | Caste | Independent | 0.659 | |
| Insectaa | 125 | DNA methylation | DNMT1 | Dependent | 0.005 | |
| Insectaa | 125 | DNA methylation | DNMT3 | Independent | 0.277 | |
| Hymenoptera | 35 | DNA methylation | Sociality | Independent | 0.344 | |
| Hymenoptera | 35 | DNA methylation | Caste | Independent | 0.425 | |
| Diptera– | 68 | DNA methylation | Sociality | Independent | 0.748 | |
| Diptera– | 68 | DNA methylation | Caste | Independent | 0.946 |
Note.—Output from PGLS using the phylogeny and traits from figure 1 and Pagel’s test for evolutionary dependence using the phylogeny and traits from figure 2A. For PGLS, a free-model of phylogenetic signal () was preferred over a model of zero (independence) and a model of one (random, Brownian motion) based on a likelihood ratio test (LRT). Preference for a free-model of suggests dependence among species’ trait values of DNA methylation and social behavior due to their phylogenetic relationship. The dependent model of trait evolution is not preferred based on a LRT, with DNA methylation and DNMT1 being the exception. Dependence suggests the evolution of social behavior is not reliant on the evolution of DNA methylation. For both tests, P values represent the significance of correlations between trait x and y. P values for PGLS are given for the preferred model.
Catajapyx aquilonaris (Dipluran) and Daphnia pulex (Crustacea) included the following: Ccoding and Ggenome.
Conclusion
We have conducted the largest phylogenetic investigation of DNA methylation in insects. Our investigation suggests that DNA methylation is too variable in insects to support a single, common role in regulating social traits. DNA methylation is found in all insect orders except Diptera, but the level of DNA methylation is fluid and phylogeny does not predict the pattern. Furthermore, the gain, loss, and duplication of DNMT1 suggests that there may be novel functions in different species, and justifies further studies using RNAi or CRISPR/Cas9 to mutate DNA methyltransferases (Gilles et al. 2015). Finally, we found no association between level or presence of DNA methylation and the presence of sociality. Although this does not preclude a role for DNA methylation in regulating caste in eusocial insects, one possibility is that DNA methylation in most insects that display sociality is a downstream consequence of upstream environment, genetic or hormonal factors (Kapheim et al. 2015; Wallberg et al. 2016). For now, the universal role or diverse roles of DNA methylation in insects remains elusive, and is likely to remain so until functional tests are made to advance our understanding of this evolutionarily conserved and important DNA modification.
Materials and Methods
Tissue Collection and DNA Extraction
All samples were collected from established laboratory colonies with the following exceptions. Cryptocercus garciai was collected from northeast Georgia by Brian Forschler, Polistes carolina were collected from the campus of the University of Georgia by K.J.V.
DNA extractions were performed on freshly sacrificed insects with the exception of three species of aphids (Pemphigus obesinymphae, P. populicaulis, and P. populitransversus), which were ethanol preserved. For samples likely to contain significant gut microbial contamination (Blattodea), guts were dissected and discarded. Approximately 10 mg of material was frozen in liquid nitrogen, ground with a pestle. Samples were then extracted with the Qiagen DNEasy Mini kit following the manufacturer’s instructions for animal tissues.
Behavior Classification
Sociality can be classified numerous ways (Costa 2006) but most classifications share a hierarchical ordering based on the addition of derived and specialized social traits occurring in family groups (Rehan and Toth 2015). We therefore adopt a classification scheme that is most common, incorporating four levels: solitary, communal, subsocial, and eusocial. This collapses variation that may occur within the eusocial species (i.e., species that may switch back and forth) but captures the most common classifications (Wilson 1971; Costa 2006). Species with individuals of the same generation that use the same composite nest site but do not cooperate in brood care were classified as communal. Species that exhibit only parental care or cooperative brood rearing were classified as subsocial. Species that share a common nest site, exhibit cooperative rearing of young, at some point have reproductive division of labor with sterile or less fecund workers, and have overlap of generations are considered eusocial. Species lacking any one of the traits listed above were categorized as solitary. For binary classification of social behavior (i.e., social vs. solitary classification), eusocial, subsocial and/or communal species were classified as social and all others were categorized as solitary.
Whole-Genome Bisulfite Sequencing and Levels of DNA Methylation
MethylC-seq libraries were prepared according to the following protocol (Urich et al. 2015). Sequencing data for Acyrthosiphon pisum, Aedes aegypti, Aedes albopictus, Anopheles gambiae, Apis mellifera, Blattella germanica, Camponotus floridanus, Copidosoma floridanum, Culex quinquefasciatus, Dinoponera quadriceps, Drosophila melanogaster, Harpegnathos saltator, Microplitis demolitor, Nasonia vitripennis, Nicrophorus vespilloides, Polistes canadensis, Solenopsis invicta, Tribolium castaneum was aligned to their respective genome assembly using the methylpy pipeline (Schultz et al. 2015) (supplementary table S1, Supplementary Material online). Blattella asahinai was aligned to its sister species Blattella germanica. Similarly, Microplitis mediator was aligned to its sister species Microplitis demolitor. Dinoponera quadriceps was removed from figure 1 and associated analyses because of possible genomic sequence contamination with other insect species. Contamination was based on the unplaced genomic scaffold harboring cytochrome c oxidase I (COI) (KQ473275.1), which has a best BLASTn hit to the dipteran Megaselia scalaris. Further phylogenetic analysis of the COI locus revealed that it is more closely related to Diptera than Hymenoptera COI (supplementary fig. S5, Supplementary Material online). Other loci and genomic scaffolds may be contaminated, but were not investigated. Species for MethylC-seq were chosen to maximize taxonomic breadth based on previously published insect DNA methylomes. Thus, species within the same genus with published methylomes were often limited to a single representative. However, a priori knowledge of closely related species with differences in social behavior was taken into consideration. Overall, the species chosen for MethylC-seq represent phylogenetically deep and shallow switches for social behavior. Other species, including Locusta migratoria and Schistocerca gregaria, were excluded because no genome annotation exists on GenBank.
In brief, reads were trimmed of sequencing adapters using Cutadapt (Martin and Marcel 2011), and then mapped to both a converted forward strand (cytosines to thymines) and converted reverse strand (guanines to adenines) using bowtie (Langmead et al. 2009). Reads that mapped to multiple locations, and clonal reads were removed. Weighted DNA methylation was calculated for CG sites by dividing the total number of aligned methylated reads by the total number of methylated plus un-methylated reads. Slight differences in levels of DNA methylation can be found between this and previously published studies (Bonasio et al. 2012). These differences are shared by all studies and are most likely due to technical and methodological differences. The majority of these differences are negated within this study due to the use of identical methods applied to all samples, including mapping of MethylC-Seq reads, categorization of coding sequences, and estimates of DNA methylation levels. Thus, levels of DNA methylation are comparable within this study. Furthermore, the discrete categorization of DNA methylation is not affected by slight differences between different social castes or individuals of a population, as genome-wide reductions or gains in DNA methylation are not observed (Elango et al. 2009). For the remaining species, genome-wide levels of DNA methylation were estimated using FASTmC and the—animal model (Bewick et al. 2015). All sequencing data can be accessed from Gene Expression Omnibus (GEO) using accession GSE83497.
CG DNA methylation in insects is enriched in coding sequences (see Lyko et al. 2010; Beeler et al. 2014) and a strong correlation between genome-wide and within coding sequence levels of CG DNA methylation is observed (supplementary fig. S6, Supplementary Material online). Therefore, using genome-wide levels of CG DNA methylation estimated from FASTmC, we were able to extrapolate levels of CG DNA methylation within coding sequence for species without sequenced genomes (supplementary table S1, Supplementary Material online).
Measurement of and Tests for Bimodality
is a metric of CpG dinucleotides normalized by G and C nucleotide content (GC content) and length (bp) of a specific region of interest (e.g., a transcript or protein coding gene) (Elango et al. 2009; Kocher et al. 2013). Due to spontaneous deamination of methylated cytosines, genes that are hypermethylated are expected to have a lower value than hypomethylated genes. Thus, in a mixture of genes that are methylated and low to un-methylated, a bimodal distribution of values is expected. Conversely, a unimodal distribution is suggestive of a set of genes that are mostly low to un-methylated. We therefore used this metric to expand our sampling of insect species for the presence of DNA methylation in 11 orders: Odonata (n = 1), Ephemeroptera (n = 1), Blattodea (n = 2), Thysanoptera (n = 1), Hemiptera (n = 9), Phthiraptera (n = 1), Hymenoptera (n = 33), Coleoptera (n = 7), Trichoptera (n = 1), Lepidoptera (n = 11), and Diptera (n = 57). The value for each gene within 124 total transcriptomes (supplementary table S1, Supplementary Material online) or gene annotations was defined as:
| (1) |
where , , and are the frequencies of CpG dinucleotides, C nucleotides, and G nucleotides, respectively, estimated from each gene of length () in bp. Only exonic sequences of a gene were considered when estimating .
The modality of distributions was tested using Gaussian mixture modeling (mixtools v1.0.4). Two modes were modeled for each distribution, and the subsequent means and 95% confidence interval (CI) of the means were compared with overlapping or nonoverlapping CI’s signifying unimodality or bimodality, respectively. Based on the largest set of genomes and WGBS data to date, Gaussian mixture modeling using mixtools identified 18/18 (100%) species correctly for presence/absence of DNA methylation within coding regions. Thus, modality tested with Gaussian mixture modeling is a robust and accurate predictor of DNA methylation.
Phylogenetic Comparative Methods
The R package phytools (Revell 2011) was used for all phylogenetic comparative methods. Two tests of correlated evolution were conducted: (1) Pagel’s (Pagel 1994) method for detecting correlated evolution of two binary traits and (2) Phylogenetic generalized least squares analysis. Briefly, Pagel’s method uses a continuous-time Markov model to simultaneously estimate transition rates in pairs of binary characters on a phylogeny. These rates are then used to test whether an independent or dependent model of evolution is preferred using the likelihood ratio test (LRT). DNA methylation was categorized into un-methylated or methylated based on the distribution of , unimodal or bimodal, respectively. Sociality was categorized into social (eusocial, subsocial, and communal) or solitary binary traits. Similarly, the division of labor trait was categorized into species with (caste+) or without (caste–) castes. DNMT1 and DNMT3 were categorized as presence/absence in each species. Pagel’s test was performed on DNA methylation versus sociality, DNA methylation versus division of labor, DNA methylation versus DNMT1 and DNA methylation versus DNMT3. Phylogenetic generalized least squares (PGLS) was used to correlate continuously categorized estimates of DNA methylation generated through WGBS and alignment to reference assemblies (methylpy), or nonreferenced based methods (FASTmC) to discretely coded social traits (eusocial, subsocial, communal, and solitary). BEAST 2 (Bouckaert et al. 2014) was used to estimate a multilocus coalescent tree, which was used to control for relatedness of species (nonindependence) for both comparative tests. For (1), the phylogenetic tree was estimated from a subset of previously identified orthologous protein coding loci (58 of the >1400 described in Misof et al. 2014). These loci were chosen because each contained <5% missing species for 123 insects, and outgroups Catajapyx aquilonaris (Diplura) and Daphnia pulex (Crustacea) based on best BLASTp hit to a set of core species (Acyrthosiphon pisum, Anopheles gambiae, Acromyrmex echinatior, Bombyx mori, Drosophila melanogaster, Apis mellifera, Ixodes scapularis, Nasonia vitripennis, Pediculus humanus, Tribolium castaneum, Zootermopsis nevadensis, Daphnia pulex). Loci were aligned using PASTA (Mirarab et al. 2015), and Gblocks was used to identify sections of conserved protein coding sequence (Castresana 2000). For (2), a smaller number of loci were used due to the obscurity of species and available data on Genbank. Loci used for both trees can be found in supplementary table S5, Supplementary Material online. For both trees, the program Tracer (http://tree.bio.ed.ac.uk/software/tracer/) was used to assess stationarity and effective sample size (ESS; ≥100) of the Markov Chain Monte Carlo (MCMC) chains.
DNA Methyltransferase (DNMT) Phylogeny and Evolution
DNA methyltransferases (DNMT1, DNMT 2, and DNMT3) were curated from 124 insect species through homology searches using BLASTp and previously identified DNMT1, DNMT2, and DNMT3 proteins in Apis mellifera, Bombyx mori (DNMT1 only), Drosophila melanogaster (DNMT2 only), and Nicrophorus vespilloides. A series of alignment and phylogenetic estimation steps were conducted to eliminate partial and poor sequences, which can affect alignment and subsequently the topology of phylogeny. Protein sequences were aligned using the program PASTA, and back-translated using the CDS sequence to generate an in-frame codon alignment. RAxML was used to generate the phylogeny with 1000 rapid bootstrap replicates and the GTR + G model of nucleotide substitution (Stamatakis 2014). The phylogeny was rooted to the DNMT2 clade in the program FigTree (http://tree.bio.ed.ac.uk/software/figtree/) and exported for stylization. An identical method was used to generate the Hymenoptera-specific DNMT1 gene tree. Nine iterations of alignment and phylogeny construction were performed removing sequences on long branches, which is indicative of low sequence homology. For species with assembled transcriptomes or gene annotations, additional DNA methyltransferases were identified by Interproscan (Jones et al. 2014), and filtering those sequences with a DNA methylase domain (PF00145). These DNA methylase domain-containing sequences were then subjected to the previously mentioned homology searches using BLASTp.
Codon Analysis
Similar methodology as described above was used to construct phylogenetic trees for testing hypotheses on the rates of evolution in a phylogenetic context. However, the program Gblocks was used to identify conserved amino acids in codon alignments. The parameters for Gblocks were kept at the default settings, except we allowed for 50% gapped positions. The program Phylogenetic Analysis by Maximum Likelihood (PAML) was used to test branches (branch test) and sites along branches (branch-site test) for deviations from the background rate of molecular evolution (dN/dS; ω) and for deviations from the neutral expectation, respectively (Yang 2007). Positively selected sites were determined by Bayes Empirical Bayes (BEB) score of ≥0.95. Branches tested and a summary of each test can be found in supplementary table S6, Supplementary Material online.
Stochastic Mutational Mapping
A stochastic mutational map describes the historical pattern of states along a phylogeny or genealogy (Huelsenbeck et al. 2003). Furthermore, stochastic mutational mapping uses an MCMC approach to sample character histories from their posterior probability distribution. This method was used to estimate the ancestral state at each node, the occurrence and timing of different states, and the timing of changes of DNA methylation and DNA methyltransferases along the multilocus coalescent tree. Stochastic mutational mapping with 1,000 simulations was implemented in the R package phytools (Revell 2011). A one-rate transition matrix was used, which only allowed for loss of DNA methylation or DNA methyltransferases.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
The study was conceived by A.J.B. and R.J.S. All authors were involved in the experimental design. K.J.V. and A.J.M. curated samples. K.J.V. and A.J.M. classified social behavior for insects used in this study. A.J.B. performed all analyses, with contribution from K.J.V. to the phylogenetic analyses. A.J.B. wrote the article with contributions made by all authors.
Supplementary Material
Acknowledgments
We thank Lauren A. Eserman and Ben L. S. Furman for phylogenetic comparative advice, and William T. Jordan for crystal structure visualization. We thank Patrick Abbot, Gaelen Burke, Brian Forschler, Sarah E. Sander, Coby Schal, Kathrin Stanger-Hall, and Michael Strand for tissue and/or insect specimens. We also thank Patrick T. Griffin, Libby McKinney, and Nick A. Rohr for library preparation, and the Georgia Genomics Facility (GGF) for sequencing. Computational resources were provided by the Georgia Advanced Computing Resource Center (GACRC). We thank the i5K initiative and the Baylor College of Medicine Human Genome Sequencing Center (BCM-HGSC) for the use of pilot data. This work was supported by funding from the Office of the Vice President for Research at the University of Georgia to RJS, and by NIH award F32GM109750 to KJV.
Footnotes
Associate editor: Nadia Singh
References
- Beeler SM, Wong GT, Zheng JM, Bush EC, Remnant EJ, Oldroyd BP, Drewell RA. 2014. Whole-genome DNA methylation profile of the jewel wasp (Nasonia vitripennis). G3 4:383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor TH, Verdine GL. 1994. DNA methyltransferases. Curr Opin Cell Biol. 6:380–389. [DOI] [PubMed] [Google Scholar]
- Bewick AJ, Hofmeister BT, Lee K, Zhang X, Hall DW, Schmitz RJ. 2015. FASTmC: a suite of predictive models for non-reference-based estimations of DNA methylation. G3 6:447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Li Q, Lian J, Mutti NS, Jin L, Zhao H, Zhang P, Wen P, Xiang H, Ding Y, et al. 2012. Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr Biol. 22:1755–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 10:e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branstetter MG, Danforth BN, Pitts JP, Faircloth BC, Ward PS, Buffington ML, Gates MW, Kula RR, Brady SG. 2016. Phylogenomic analysis of ants, bees and stinging wasps: improved taxon sampling enhances understanding of hymenopteran evolution. Biorxiv doi: 10.1101/068957. [DOI] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. 2009. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 10:295–304. [DOI] [PubMed] [Google Scholar]
- Cheng X, Blumenthal RM. 2008. Mammalian DNA methyltransferases: a structural perspective. Structure 16:341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa JT. 2006. The other insect societies. Cambridge, MA: Harvard University Press. [Google Scholar]
- Cunningham CB, Ji L, Wiberg RA, Shelton J, McKinney EC, Parker DJ, Meagher RB, Benowitz KM, Roy-Zokan EM, Ritchie MG, et al. 2015. The genome and methylome of a beetle with complex social behavior, Nicrophorus vespilloides (Coleoptera: Silphidae). Genome Biol Evol. 7:3383–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C. 1982. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 10:2709–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elango N, Hunt BG, Goodisman MAD, Yi SV. 2009. DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proc Natl Acad Sci U S A. 106:11206–11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1985. Phylogenies and the comparative method. Am Nat. 125:1–15. [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. 2010. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A. 107:8689–8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles AF, Schinko JB, Averof M. 2015. Efficient CRISPR-mediated gene targeting and transgene replacement in the beetle Tribolium castaneum. Development 142:2832–2839. [DOI] [PubMed] [Google Scholar]
- Glastad KM, Hunt BG, Goodisman MAD. 2014. Evolutionary insights into DNA methylation in insects. Curr Opin Insect Sci. 1:25–30. [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. 2005. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 74:481–514. [DOI] [PubMed] [Google Scholar]
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. 2006. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311:395–398. [DOI] [PubMed] [Google Scholar]
- Hildebrand A, Remmert A, Biegert A, Söding J. 2009. Fast and accurate automatic structure prediction with HHpred. Proteins 77:128–132. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Nielsen R, Bollback JP. 2003. Stochastic mapping of morphological characters. Syst Biol. 52:131–158. [DOI] [PubMed] [Google Scholar]
- Huff JT, Zilberman D. 2014. Dnmt1-independent CG methylation contributes to nucleosome positioning in diverse eukaryotes. Cell 156:1286–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Glossina Genome Initiative 2014. Genome sequence of the tsetse fly (Glossina morsitans): vector of African trypanosomiasis. Science 344:380–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapheim KM, Pan H, Li C, Salzberg SL, Puiu D, Magoc T, Robertson HM, Hudson ME, Venkat A, Fischman BJ, et al. 2015. Social evolution. Genomic signatures of evolutionary transitions from solitary to group living. Science 348:1139–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Samaranayake M, Pradhan S. 2008. Epigenetic mechanisms in mammals. Cell Mol Life Sci. 66:596–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher SD, Li C, Yang W, Tan H, Yi SV, Yang X, Hoekstra HE, Zhang G, Pierce NE, Yu DW. 2013. The draft genome of a socially polymorphic halictid bee, Lasioglossum albipes. Genome Biol. 14:R142.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronforst MR, Gilley DC, Strassmann JE, Queller DC. 2008. DNA methylation is widespread across social Hymenoptera. Curr Biol. 18:R287–R288. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libbrecht R, Oxley PO, Keller L, Kronauer DJC. 2016. Robust DNA methylation in the clonal raider ant brain. Curr Biol. 26:391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Byarlay H, Li Y, Stroud H, Feng S, Newman TC, Kaneda M, Hou KK, Worley KC, Elsik CG, Wickline SA, et al. 2013. RNA interference knockdown of DNA methyl-transferase 3 affects gene alternative splicing in the honey bee. Proc Natl Acad Sci U S A. 110:12750–12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Byarlay H. 2016. The function of DNA methylation marks in social insects. Front Ecol Evol. 4: [Google Scholar]
- Lyko F, Foret S, Kucharski R, Wolf S, Falckenhayn C, Maleszka R. 2010. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 8:e1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. 2000. The evolutionary fate and consequences of duplicate genes. Science 90:1151–1155. [DOI] [PubMed] [Google Scholar]
- Martin M, Marcel M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10–12. [Google Scholar]
- Mirarab S, Nguyen N, Guo S, Wang LS, Kim J, Warnow T. 2015. PASTA: ultra-large multiple sequence alignment for nucleotide and amino-acid sequences. J Comput Biol. 22:377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misof B, Liu S, Meusemann K, Peters RS, Donath A, Mayer C, Frandsen PB, Ware J, Flouri T, Beutel RG, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science 346:763–767. [DOI] [PubMed] [Google Scholar]
- Niederhuth CE, Bewick AJ, Ji L, Alabady M, Kim KD, Page JT, Li Q, Rohr NA, Rambani A, Burke JM, et al. 2016. Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 17:194.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M. 1994. Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proc R Soc Lond B. 255:37–45. [Google Scholar]
- Patalano S, Vlasova A, Wyatt C, Ewels P, Camara F, Ferreira PG, Asher CL, Jurkowski TP, Segonds-Pichon A, Bachman M, et al. 2015. Molecular signatures of plastic phenotypes in two eusocial insect species with simple societies. Proc Natl Acad Sci U S A. 112:13970–13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Min J. 2014. Structure and function of the nucleosome-binding PWWP domain. Trends Biochem Sci. 39:536–547. [DOI] [PubMed] [Google Scholar]
- Qiu C, Sawada K, Zhang X, Cheng X. 2002. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nat Struct Biol. 9:217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehan SM, Glastad KM, Lawson SP, Hunt BG. 2016. The genome and methylome of a subsocial small carpenter bee, Ceratina calcarata. Genome Biol Evol. 8:1401–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehan SM, Toth AL. 2015. Climbing the social ladder: the molecular evolution of sociality. Trends Ecol Evol. 30:426–433. [DOI] [PubMed] [Google Scholar]
- Revell LJ. 2011. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 3:217–223. [Google Scholar]
- Schultz MD, He Y, Whitaker JW, Hariharan M, Mukamel EA, Leung D, Rajagopal N, Nery JR, Urich MA, Chen H, et al. 2015. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 523:212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. 2011. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479:74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standage DS, Berens AJ, Glastad KM, Severin AJ, Brendel VP, Toth AL. 2016. Genome, transcriptome and methylome sequencing of a primitively eusocial wasp reveal a greatly reduced DNA methylation system in a social insect. Mol Ecol. 25:1769–1784. [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. 2008. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 9:465–476. [DOI] [PubMed] [Google Scholar]
- Takuno S, Shohei T, Jin-Hua R, Gaut BS. 2016. Evolutionary patterns of genic DNA methylation vary across land plants. Nat Plants 2:15222.. [DOI] [PubMed] [Google Scholar]
- Urich MA, Nery JR, Lister R, Schmitz RJ, Ecker JR. 2015. MethylC-seq library preparation for base-resolution whole-genome bisulfite sequencing. Nat Protoc. 10:475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallberg A, Pirk CW, Allsopp MH, Webster MT. 2016. Identification of multiple loci associated with social parasitism in honeybees. PLoS Genet. 12:e1006097.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling CA, Stamper CE, Smiseth PT, Moore AJ. 2008. The quantitative genetics of sex differences in parenting. Proc Natl Acad Sci. U S A. 105:18430–18435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson EO. 1971. The insect societies. Cambridge, MA: Belknap Press. [Google Scholar]
- Xiang H, Zhu J, Chen Q, Dai F, Li X, Li M, Zhang H, Zhang G, Li D, Dong Y, et al. 2010. Single base-resolution methylome of the silkworm reveals a sparse epigenomic map. Nat Biotechnol. 28:516–520. [DOI] [PubMed] [Google Scholar]
- Yan H, Bonasio R, Simola DF, Liebig J, Berger SL, Reinberg D. 2015. DNA methylation in social insects: how epigenetics can control behavior and longevity. Annu Rev Entomol. 60:435–452. [DOI] [PubMed] [Google Scholar]
- Yan H, Simola DF, Bonasio R, Liebig J, Berger SL, Reinberg D. 2014. Eusocial insects as emerging models for behavioural epigenetics. Nat Rev Genet. 15:677–688. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yearim A, Gelfman S, Shayevitch R, Melcer S, Glaich O, Mallm JP, Nissim-Rafinia M, Cohen AH, Rippe K, Meshorer E, et al. 2015. HP1 is involved in regulating the global impact of DNA methylation on alternative splicing. Cell Rep. 10:1122–1134. [DOI] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. 2010. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328:916–919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.