Abstract
Tibetans are well adapted to the hypoxic environments at high altitude, yet the molecular mechanism of this adaptation remains elusive. We reported comprehensive genetic and functional analyses of EPAS1, a gene encoding hypoxia inducible factor 2α (HIF-2α) with the strongest signal of selection in previous genome-wide scans of Tibetans. We showed that the Tibetan-enriched EPAS1 variants down-regulate expression in human umbilical endothelial cells and placentas. Heterozygous EPAS1 knockout mice display blunted physiological responses to chronic hypoxia, mirroring the situation in Tibetans. Furthermore, we found that the Tibetan version of EPAS1 is not only associated with the relatively low hemoglobin level as a polycythemia protectant, but also is associated with a low pulmonary vasoconstriction response in Tibetans. We propose that the down-regulation of EPAS1 contributes to the molecular basis of Tibetans’ adaption to high-altitude hypoxia.
Keywords: high altitude, Tibetans, hypoxia, genetic adaptation, EPAS1, transgenic mice
Introduction
The genetic adaptation of Tibetans to high-altitude environments at the Qinghai-Tibetan plateau (average elevation: 4,000 m) represents a remarkable case of natural selection during recent human evolution (Beall et al. 2004; Wu and Kayser 2006; Bigham and Lee 2014). Previous genome-wide scans have identified many candidate genes that may contribute to this adaptation, including two genes (EPAS1 and EGLN1) that are involved in the HIF (hypoxia inducible factor) pathway, which showed the strongest signals of selective sweeps in Tibetans (Beall et al. 2010; Bigham et al. 2010; Simonson et al. 2010; Yi et al. 2010; Peng et al. 2011; Xu et al. 2011). Both genes harbor sequence variants highly enriched in Tibetans (50–80%), but absent or rare (<5%) in lowlanders (Peng et al. 2011; Xiang et al. 2013; Huerta-Sanchez et al. 2014; Lorenzo et al. 2014).
EPAS1 encodes HIF-2α, and EGLN1 encodes prolyl 4-hydroxylase 2 (PHD2). Under normoxia, EGLN1 performs its oxygen-dependent hydroxylase function and triggers degradation of HIF proteins (HIF-1α and HIF-2α). Under hypoxia, the hydroxylase activity of EGLN1 is suppressed, resulting in the accumulation of HIFs that can transactivate hundreds of downstream target genes and induce diverse physiological responses, for example, erythropoiesis (Hu et al. 2003). It was reported that the Tibetan version of EGLN1 carries two Tibetan-specific missense mutations (Xiang et al. 2013), which cause increased HIF degradation under hypoxia that revokes the HIF-mediated augmentation of erythropoiesis, and protects Tibetans from polycythemia (red cell overproduction) at high altitudes (Lorenzo et al. 2014). Another study showed that the Tibetan EGLN1 variant might be a loss of function allele, leading to augmented HIF activation to facilitate adaptation to high altitude (Song et al. 2014). However, how the Tibetan version of EPAS1 works is still unclear. Unlike EGLN1 variants, all identified EPAS1 variants are located in the noncoding regions (mostly in introns), suggesting that they may affect EPAS1’s regulation at the transcriptional level (Peng et al. 2011). We conducted a series of experiments to understand the functional effects of the Tibetan-enriched EPAS1 variants and reveal the underlying molecular mechanism.
Result
Identification of EPAS1 Adaptive Variant Candidates
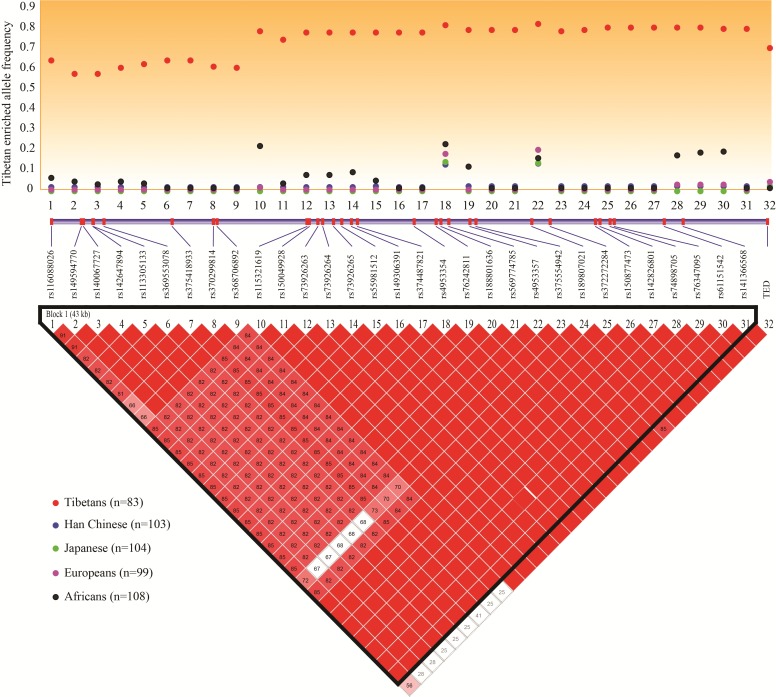
Using published genome data (Peng et al. 2011; Lu 2016), we identified a total of 204 variants across the entire gene region of EPAS1 (97.0 kb) in Tibetans (refer to the “Material and Methods” section for details), and 180 of them are shared with the lowland reference populations (Han Chinese, Japanese, Europeans and Africans from the 1000 Genomes Project). It is expected that Darwinian positive selection would drive enrichment of the adaptive alleles in Tibetans, but not in lowlanders. To search for functional variants, we chose EPAS1 variants showing the largest allelic divergence (FST >0.50) between high-altitude Tibetans and worldwide lowlanders. We obtained 32 EPAS1 candidate variants for further analysis (supplementary table S1, Supplementary Material online), including the recently reported 3.4-kb Tibetan-enriched deletion (TED) 80-kb downstream of EPAS1 (Lou et al. 2015). The map of linkage disequilibrium (LD) among the 32 candidate variants fits the expectation of an extended Tibetan-specific haplotype resulting from selective sweep (fig. 1). Notably, TED showed a strong LD with the downstream EPAS1 SNPs (D′ > 0.85) (fig. 1).
Fig. 1.
Map of linkage disequilibrium among the 32 EPAS1 sequence variants (lower panel) with the largest allelic divergence (FST > 0.5) between high altitude Tibetans and low altitude populations (upper panel).
To further evaluate the evolutionary and functional roles of the 32 candidate variants, we next conducted two haplotype-based selection tests, the iHS test and the XP-EHH test (Voight et al. 2006; Szpiech and Hernandez 2014), and the results confirmed that the Tibetan-specific haplotype had undergone a strong selective sweep (supplementary table S1, Supplementary Material online). Furthermore, we performed multiple functional predictions, including cross-species sequence conservation (measured by GREP ++), transcription factor binding site (TFBS), splicing motif, H3K4Me1/H3K4Me3 sites and DNAase-I hypersensitive site. The results suggest that many variants are located within the predicted functional elements (supplementary table S1, Supplementary Material online). We speculate that there might be multiple functional variants and they form a Tibetan-specific haplotype to regulate EPAS1’s transcription. We previously showed that this Tibetan-specific EPAS1 haplotype was prevalent in Tibetans (72%), rare in Han Chinese (2.2%), and absent in other global populations (Peng et al. 2011).
EPAS1 Adaptive Allele at rs149594770 Weakens Promoter Activity
Among the 32 candidate variants, we selected one variant (rs149594770) located in a H3K4Me1 peak (peak score = 62.8) with the highest sequence conservation among all tested variants (GERP ++ score = 4.94). The A allele of rs149594770 is prevalent (presumably adaptive) in Tibetans (57.2%), but rare in lowlanders (0.5% in Han Chinese, 3.2% in Africans, and absent in Japanese and Europeans).
We first used EMSA (electrophoretic mobility shift assay) to test if the two alleles at rs149594770 have different transcription factor binding affinity. The result showed that the Tibetan-enriched “A” allele had a weaker binding affinity to Hela nuclear extracts compared with the wild-type “T” allele, and this was further confirmed by the competitor probe assay (fig. 2A). We next tested with the luciferase reporter gene assay using 293T cells, and the results clearly indicated weaker promoter activity of the Tibetan-enriched “A” allele compared with the wild-type “T” allele (fig. 2B). We observed the same pattern when repeating the experiment using Hela cells (supplementary fig. S1, Supplementary Material online). As the 32 candidate variants are tightly linked, this result implies that the Tibetan-specific haplotype is likely functional.
Fig. 2.
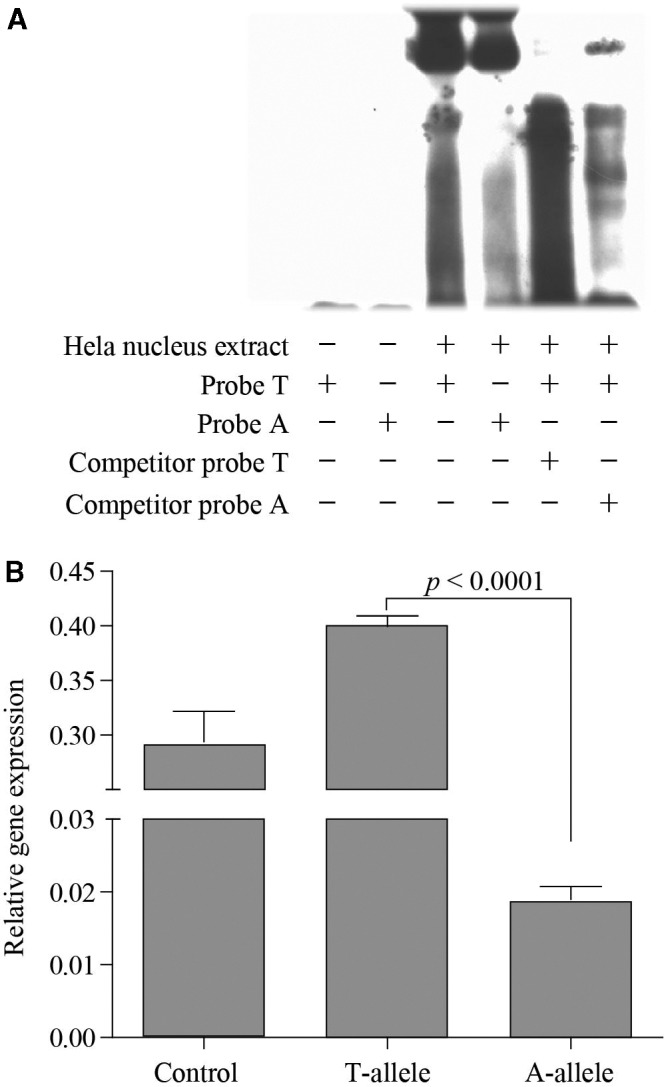
In vitro functional test of the EPAS1 variant (rs149594770) using an electrophoretic mobility shift assay (A) and a luciferase reporter gene assay (B). The two-tailed t test was used for statistical assessment.
Tibetan-Specific EPAS1 Haplotype Down-Regulates Expression
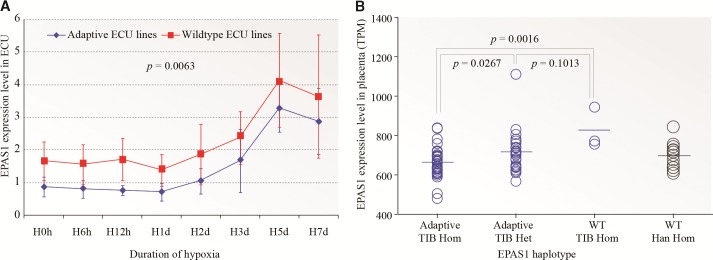
To reveal the transcriptional regulation of the Tibetan-specific EPAS1 haplotype, we conducted an in vitro analysis using endothelial cells derived from Tibetan umbilical cords (ECU) because endothelial cells are responsive to hypoxia and selectively express EPAS1 (Tian et al. 1997). By genotyping a subset (10 variants) of the candidate variants, we were able to obtain three wild-type haplotype homozygotes from 105 Tibetan umbilical cords, and we paired them with three adaptive haplotype homozygotes and conducted tests using a hypoxic assay (supplementary table S2, Supplementary Material online). Cells were subject to prolonged hypoxic conditions (1% oxygen) for 7 days and the expression level of EPAS1 was determined by qPCR at eight time points (0 h, 6 h, 12 h, 1 day, 2 days, 3 days, 5 days and 7 days). As expected the expression of EPAS1, being a hypoxia inducible factor, started to rise at Day-2 and kept increasing till Day-7, consistent with previous observations (Hu et al. 2003). Importantly, the adaptive ECU line showed a relatively lower EPAS1 expression compared with the wild-type ECU line (fig. 3A), which was confirmed by two replicate experiments though the magnitudes varied (supplementary fig. S2, Supplementary Material online). When pooling data from all three replicates, we detected a significant down-regulation of the adaptive ECU lines (P = 0.0063, one-way ANOVA). On an average, there was a 31.3% reduction of EPAS1 expression in the adaptive ECU lines compared with the wild-type controls over the 7-day period. These results suggest that the EPAS1 adaptive haplotype down-regulates EPAS1’s expression under hypoxic conditions. Consistently, a previous report also observed a decreased EPAS1 mRNA expression in Tibetan lymphocytes (Petousi and Robbins 2014).
Fig. 3.
Changes of EPAS1 expression in Tibetan umbilical endothelial cells during prolonged hypoxic treatment (A) and in term Tibetan placentas (B). The adaptive ECU lines are homozygotes of the adaptive EPAS1 haplotype, and the wildtype ECU lines are homozygotes of the wildtype EPAS1 haplotype. TIB Hom, Tibetan homozygote; TIB Het, Tibetan heterozygote; Han Hom, Han Chinese homozygote; WT, wild-type. The one-way ANOVA and two-tailed t tests were conducted for panel A and panel B, respectively.
Furthermore, we tested 63 term placenta samples from Tibetans living in Lhasa (elevation: 3,680 m). The fetal section of villus parenchyma was analysed with RNAseq. We classified the Tibetan placenta samples into three haplotype groups, the wild-type haplotype homozygotes (WHH, n = 3), the heterozygotes (HET, n = 26) and the adaptive haplotype homozygotes (AHH, n = 34). Consistent with the results of the endothelial cells, the AHHs showed a significantly lower EPAS1 expression compared with the WHHs (27.1% reduction on an average), and the HETs fell in between (fig. 3B). In comparison, we also analysed 14 Han Chinese placentas from Kunming of southwestern China (elevation: 1,800 m), and these Han WHHs showed relatively low EPAS1 expression, similar with the Tibetan AHHs and HETs (fig. 3B), indicating that most high-altitude Tibetans have a similar level of EPAS1 expression in the placenta as the lowlander Han Chinese, a reflection of Tibetans’ adaptation at altitude. In addition, the single variant analysis also showed significant association with EPAS1’s expression (supplementary table S3, Supplementary Material online). Thus, both endothelial cell and placenta data suggest a down-regulation of EPAS1 expression in Tibetans, which may serve as the molecular basis of adaptation to high-altitude hypoxia.
EPAS1+/− Mice Show Blunted Hypoxic Responses
To dissect how EPAS1’s down-regulation leads to the adaptive physiological changes in Tibetans, we utilized heterozygous EPAS1 knockout mice (EPAS1+/−) as these mice have ∼50% reduction of intact EPAS1 transcripts (Brusselmans et al. 2003). It is known that EPAS1 not only works as a hypoxia inducible factor, but it is also involved in embryonic development because homozygous EPAS1 knockout mice (EPAS1−/−) either died of cardiac failure at mid-gestation or of severe respiratory failure as neonates (Tian et al. 1998; Compernolle et al. 2002). Interestingly, the EPAS1+/− mice were viable and healthy, and they were indistinguishable from their wild-type (WT) littermates under normoxia (Tian et al. 1998). Under long-term severe hypoxia, the EPAS1+/− mice performed better than the WT mice, and showed blunted hypoxic responses (Brusselmans et al. 2003).
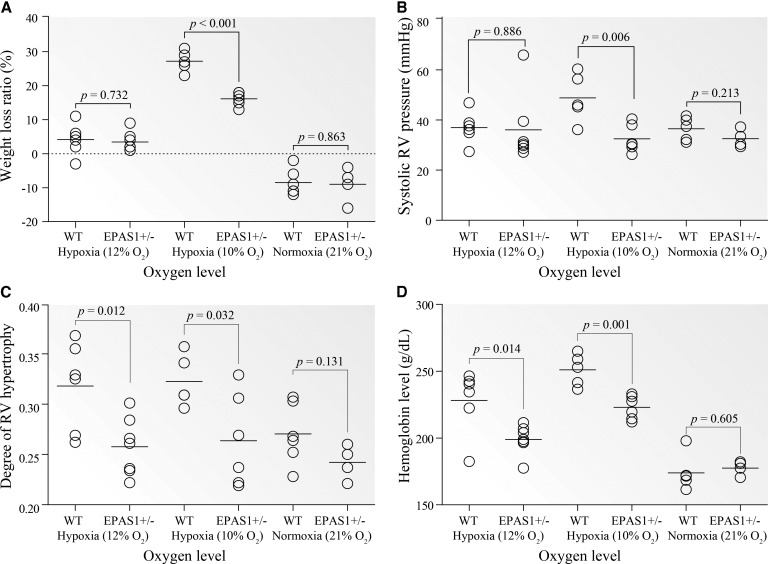
We conducted the experiment by housing both EPAS1+/− and WT mice in a hypoxic chamber (Coy Laboratory Products, MI) and tested their physiological responses to prolonged normobaric hypoxia (4 weeks at 12% O2 and 10% O2, equivalent to the oxygen levels at ∼4,000 m and ∼5,000 m, respectively). The experiment under normoxia (21% O2) was performed as reference. Consistent with the previous data (Brusselmans et al. 2003), under 10% O2, the EPAS1+/− mice indeed performed better than the WT mice, reflected by less weight loss, lower hemoglobin concentration, lower right ventricle (RV) pressure and less RV hypertrophy (P < 0.05, two-tailed t test). Similar differences were also observed under 12% O2 though less significant, while no difference was detected under normoxia (fig. 4). In particular, during prolonged hypoxia, the WT mice experienced vascular remodeling with thick-walled vessels in the lung, but this did not occur in the EPAS1+/− mice (supplementary fig. S2, Supplementary Material online). Hence, the data indicates that a 50% reduction of EPAS1 expression causes blunted responses to prolonged hypoxia in the EPAS1+/− mice, mirroring the situation in Tibetans.
Fig. 4.
In vivo tests of EPAS1+/− and WT mice under prolonged hypoxia. (A) Weight loss percentage; (B) systolic RV pressure; (C) degree of RV hypertrophy; and (D) hemoglobin concentration. The degree of RV hypertrophy was determined by the weight ratio of right ventricle to heart. Mice were kept in a hypoxic chamber for 4 weeks at two oxygen levels (7 EPAS1+/− vs. 6 wild-type mice at 12%, and 6 EPAS1+/− vs. 5 wild-type mice at 10%). The data at normoxia (4 EPAS1+/− vs. 5 wild-type mice at 21% O2) was collected as reference. WT, wild-type mice.
To reveal the underlying molecular mechanisms of the observed physiological changes in mice, we next conducted RNAseq for five types of tissues (lung, heart, liver, kidney, and spleen; six EPAS1+/− mice vs. four WT mice at 10% O2). As expected, the EPAS1+/− mice showed a 53.8–59.7% reduction of intact EPAS1 transcripts compared with the WT mice in all five tissues, and the highest EPAS1 expression was observed in the lung (supplementary fig. S4, Supplementary Material online). We identified a total of 4,494 differentially expressed genes (DEGs) between EPAS1+/− and WT mice in at least one tissue (supplementary data set S1, Supplementary Material online), an indication of the large impact of EPAS1 on downstream gene regulation. Interestingly, most of the DEGs (79.3%) were tissue-specific (supplementary fig. S5A, Supplementary Material online), suggesting a tissue-differential regulation by EPAS1.
The liver and the kidneys have the largest numbers of DEGs (1,997 and 1,523 genes, respectively) (supplementary table S4, Supplementary Material online) with functional enrichment in protein transportation, energy metabolism, oxidation–reduction process, heme/iron-ion binding and others, reflecting mostly different transitions from oxidative to glycolytic metabolism under hypoxia between EPAS1+/− and WT mice (Semenza 2012). The spleen has 880 DEGs (supplementary table S4, Supplementary Material online), most of which are involved in immune responses to hypoxia (Palazon et al. 2014). Notably, the most significant KEGG pathway in the spleen is hematopoietic cell lineage (adjusted P = 2.29e − 10) (supplementary table S5, Supplementary Material online), which is consistent with the known role of EPAS1 in erythropoiesis under hypoxia (Hu et al. 2003). In contrast, the lung and the heart have the least number of DEGs (742 and 520 genes, respectively) with one enriched functional category (extracellular space) for lung and none for heart (supplementary table S4, Supplementary Material online).
To further establish the molecular connections between EPAS1’s down-regulation and the adaptive physiological changes in the EPAS1+/− mice, we checked a set of 473 gene candidates with known functions in hypoxic responses and/or related to the physiological traits (Simonson et al. 2010; Lu 2016). Of those, 127 genes (26.8%) were DEGs (supplementary table S6, Supplementary Material online). Similar with the aforementioned pattern, most of the DEGs (78.0%) were tissue-specific (supplementary fig. S5B, Supplementary Material online). As a known target gene of EPAS1 and a negative mediator of EPAS1-regulated genes, HIF-3α showed a consistent down-regulation across all tissues (3.3-to 6.8-fold reduction) (supplementary table S7, Supplementary Material online), which may help fine-tune the overall hypoxic responses of the EPAS1+/− mice under hypoxia (Hatanaka et al. 2009).
In the regulation of erythropoiesis, the kidney serves as the most important oxygen sensor and responds to systemic hypoxia with a rapid increase in erythropoietin (EPO) production by renal interstitial fibroblast-like cells, and EPAS1 is the main regulator of hypoxic EPO induction (Haase 2006). Although we observed a significantly lower Hb level in the EPAS1+/− mice than that in the WT mice (fig. 4D), we did not detect an EPO expression difference in the kidneys (P > 0.05, two-tailed t test). This was likely caused by the observed low expression of EPO (0–2.18 TPM in the kidney and none in other tissues) as the kidney samples for RNAseq probably only contained a small number of EPO-producing renal cells. We measured EPO concentration (at protein level) in the blood, and we observed a higher EPO level in the WT mice compared with the EPAS1+/− mice under hypoxia, consistent with the pattern of EPAS1 expression difference. No EPO level difference was detected under normoxia (supplementary fig. S6A, Supplementary Material online).
Notably, ACE (angiotensin I converting enzyme), a key blood pressure regulator was significantly down regulated in the lungs and the heart (33% and 66% reductions, respectively) (supplementary table S7, Supplementary Material online). This explains the relatively low pulmonary arterial pressure (PAP, measured as RV pressure) and less RV hypertrophy of the EPAS1+/− mice since the ACE knockout mice displayed a low blood pressure (Krege et al. 1995), and increased cardiac ACE augments cardiac hypertrophy (Tian et al. 2004). In contrast, the kidneys showed an up-regulation of ACE, which could be explained by the reported independent effects of ACE on maintenance of normal blood pressure and renal functions (Kessler et al. 2003). Consistently, VEGFC (vascular endothelial growth factor C), a gene involved in chronic hypoxic pulmonary hypertension (Partovian et al. 1998) was also down regulated in the lungs (62% reduction) (supplementary table S7, Supplementary Material online). Additionally, a recent study showed that the vasodilatory apelin (a vasodilatory peptide encoded by APLN) signaling was down regulated in the PHD2 (encoded by EGLN1) deficient mice (Kapitsinou et al. 2016). In the EPAS+/− mice, we observed significant up-regulation of APLN (logFC = −0.771, P = 2.32e-4) compared with the wild-type mice, implying that the vasodilatory apelin signaling may also contribute to the observed low pulmonary pressure of the EPAS1+/− mice under prolonged hypoxia.
Finally, two nitric oxide synthases (iNOS and eNOS, also called NOS2 and NOS3) were differentially expressed with an up-regulation of iNOS in the heart and a down-regulation of eNOS in the lungs, the liver and the kidneys (supplementary table S7, Supplementary Material online). In mice, both iNOS and eNOS are the target genes of EPAS1 and they produce nitric oxide (NO), an important vasodilator (Ho et al. 2012). We measured the blood NO concentration and we did not see difference between the EPAS1+/− and WT mice (P > 0.05, two-tailed t test) either under normoxia or under hypoxia (supplementary fig. S6B, Supplementary Material online), which seems to contradict the previous observation that high-altitude Tibetans had much higher levels of blood NO levels compared with lowlanders (Erzurum et al. 2007).
High-Altitude Tibetans Have Similar Placenta Expression Profiles as Lowlander Han Chinese
The placenta is the key organ of maternal–fetal oxygen exchange, and EPAS1 is highly expressed in the placenta as an important gene for fetal development (Tian et al. 1998). We analysed the transcriptomes of 63 high altitude Tibetan and 14 low altitude Han Chinese term placentas. As aforementioned, the 63 Tibetan placentas were classified into three groups based on their EPAS1 haplotypes (3 WHHs, 26 HETs and 34 AHHs). We first compared the Tibetan AHHs with the Han controls. Only 276 DEGs (135 up-regulated and 141 down-regulated) were identified and no enriched functional categories or pathways were detected (supplementary table S8, Supplementary Material online). When using a stringent statistical cutoff (corrected P < 0.01), no DEGs remained. We also compared the Tibetan HETs with the Han controls, and no DEGs were identified, nor in the comparison between the Tibetan AHHs and HETs. This suggests that high-altitude Tibetans have in general similar placenta transcriptional profiles as lowlander Han Chinese, a reflection of adaptation at altitude. Due to the small sample size of the Tibetan WHHs (n = 3), they were excluded from the analysis.
EPAS1 Variants Are Associated with Multiple Physiological Traits of Tibetans
We conducted a survey of physiological traits among 508 unrelated Tibetan individuals from Bange of Tibet (elevation: 4,700 m) to test whether the EPAS1 variants are responsible for the known adaptive traits of Tibetans. We measured three physiological parameters, including the Hb concentration, the systolic pulmonary arterial pressure (PAP) and the degree of blood oxygen saturation (SaO2). As expected, males had a higher Hb level compared with females. We also observed a between-sex difference for SaO2, but not for PAP (supplementary fig. S7, Supplementary Material online). Because the elevation at Bange is among the highest altitude regions in Tibet with permanent residents, we observed a relatively large portion of Tibetans showing high Hb (≥ 200 g/l, 23.4% in males and 4.8% in females) and PAP (≥ 30 mmHg, 43.9% in males and 54.7% in females), which implied that even for Tibetans, an elevation of 4,700 m might be a big challenge.
We genotyped five EPAS1 variants (rs149594770, rs370299814, rs73926265, rs1888801636 and TED) in the 508 Tibetans. As a reference, we also genotyped two EGLN1 variants (rs186996510 and rs12097901). The result of association is listed in table 1. Consistent with previous observations (Beall et al. 2010; Yi et al. 2010; Peng et al. 2011; Xiang et al. 2013), all tested variants were significantly associated with the hemoglobin levels in Tibetans. On an average, each EPAS1 variant could explain 2.87% variance of Hb, and the percentages were 1.91% for TED and 1.44% for the two EGLN1 variants (table 1). For example, the adaptive alleles of rs149594770 and rs73926265 accounted for 6.15 and 12.65 g/l deductions of hemoglobin levels, respectively (supplementary fig. S8, Supplementary Material online). This result suggests that the Tibetan versions of EPAS1 and EGLN1 contribute to the relatively low Hb level of high-altitude Tibetans.
Table 1.
Results of Genetic Association Analysis of Three Physiological Traits in Bange Tibetans.
| Trait | Gene | SNP ID | Male (n=219) |
Female (n=289) |
All (n=508) |
||||
|---|---|---|---|---|---|---|---|---|---|
| Beta | EMP′ | Beta | EMP′ | Beta | EMP″ | R2 (%) | |||
| Hb | EGLN1 | rs12097901 | −12.19 | 2.8E-04 | −4.11 | 0.07 | −7.41 | 8.9E-05 | 2.11 |
| rs186996510 | −5.95 | 0.02 | −2.05 | 0.27 | −3.79 | 0.02 | 0.77 | ||
| EPAS1 | rs149594770 | −4.40 | 0.06 | −3.72 | 0.03 | −4.04 | 6.7E-03 | 1.40 | |
| rs370299814 | −5.27 | 0.04 | −2.60 | 0.20 | −3.80 | 0.02 | 1.33 | ||
| rs73926265 | −10.82 | 1.9E-04 | −8.00 | 9.0E-04 | −9.47 | 1.0E-06 | 4.93 | ||
| rs188801636 | −8.44 | 4.3E-03 | −7.90 | 9.7E-03 | −8.14 | 1.2E-04 | 3.83 | ||
| / | TED | −4.88 | 0.08 | −5.19 | 8.0E-03 | −5.04 | 1.2E-03 | 1.91 | |
| PAP | EGLN1 | rs12097901 | −1.30 | 0.48 | −0.53 | 0.70 | −0.77 | 0.44 | 0.23 |
| rs186996510 | 1.11 | 0.46 | −1.59 | 0.19 | −0.59 | 0.75 | 0.27 | ||
| EPAS1 | rs149594770 | 1.36 | 0.44 | −2.83 | 6.5E-03 | −1.20 | 0.12 | 0.68 | |
| rs370299814 | 1.30 | 0.42 | −1.37 | 0.24 | −0.32 | 0.86 | 0.11 | ||
| rs73926265 | −0.32 | 0.86 | −3.08 | 0.03 | −1.99 | 0.04 | 0.99 | ||
| rs188801636 | −0.41 | 0.86 | −2.21 | 0.18 | −1.33 | 0.41 | 0.20 | ||
| / | TED | 1.74 | 0.20 | −0.36 | 0.64 | 0.41 | 0.86 | 0.08 | |
| SaO2 | EGLN1 | rs12097901 | −0.34 | 0.75 | −0.29 | 0.86 | −0.29 | 0.46 | 0.05 |
| rs186996510 | 0.48 | 0.59 | 0.05 | 1.00 | 0.27 | 0.56 | 0.17 | ||
| EPAS1 | rs149594770 | −0.92 | 0.07 | 0.26 | 0.86 | −0.27 | 0.56 | 0.09 | |
| rs370299814 | −1.14 | 0.07 | −0.70 | 0.15 | −0.89 | 0.04 | 0.87 | ||
| rs73926265 | −1.27 | 0.05 | 0.09 | 0.86 | −0.61 | 0.26 | 0.33 | ||
| rs188801636 | −1.01 | 0.17 | −0.07 | 0.86 | −0.58 | 0.32 | 0.50 | ||
| / | TED | −0.79 | 0.27 | 0.24 | 0.75 | −0.22 | 0.52 | 0.05 | |
Note.—All presented P values (EMP′ and EMP″) were subject to multiple test corrections with age (EMP′) or age and sex (EMP″). The significant P values (P < 0.05) are highlighted. R2 (%) indicates the percentage of the explained variance of a trait by an EPAS1 variant. The two nonsynonymous SNPs of EGLN1 cause amino acid changes from aspartic acid to glutamic acid (rs186996510) and from cysteine to serine (rs12097901). Hb, hemoglobin concentration; PAP, systolic pulmonary arterial pressure; SaO2, degree of blood oxygen saturation.
We observed a significant association of two EPAS1 variants (rs149594770 and rs73926265) with PAP (corrected P <0.05) (table 1). The adaptive alleles tended to have a lower PAP compared with the wild-type alleles (supplementary fig. S9, Supplementary Material online), explaining the well-known low hypoxic pulmonary vasoconstriction response in Tibetans (Wu and Kayser 2006). Compared with Hb, there was less PAP variance explained by the EPAS1 variants (e.g., 0.68% by rs149594770 and 0.99% by rs73926265) (table 1).
Lastly, we observed a weak association of one EPAS1 variant (rs370299814, corrected P = 0.04) with SaO2. But the presumably adaptive allele (enriched in Tibetans) at rs370299814 showed a lower SaO2 level compared with the wild-type allele (supplementary fig. S10, Supplementary Material online). The same trend was observed for the other EPAS1 variants including TED although it was statistically not significant (corrected P > 0.05) (table 1). This result implies that EPAS1 may not be the gene contributing to the reported better SaO2 of Tibetans compared with Han newcomers at altitude (Beall et al. 2004; Wu and Kayser 2006).
Discussion
When lowlanders stay at high altitudes, hypobaric hypoxia induces erythropoiesis, mainly triggered by the accumulation of HIF-2α and the downstream transactivation of EPO production. The prolonged exposure of lowlanders to hypobaric hypoxia usually leads to overproduction of red cells (polycythemia), which increases the risk of heart attack, stroke, and fetal loss during pregnancy (Semenza 2012). In contrast, Tibetans are genetically protected from polycythemia. It was speculated that the Tibetan genotype associated with absence of an erythrocytotic response to hypoxia may encode reduced HIF-2α activity along with other alterations that increase metabolic efficiency (Semenza 2012). Here, we provided evidence that the functional change of EPAS1 as an adaptive strategy for hypoxia occurred at the transcription level, where the EPAS1 adaptive variants caused a reduced EPAS1 expression under hypoxia compared with the wild-type controls. Based on the data from Tibetan endothelial cells and placentas, the reduced levels of EPAS1 expression were 31.3% and 27.1%, respectively, a significant reduction that serves as the molecular basis for blunted hypoxic responses in Tibetans. It should be noted that in the reporter gene assay, there was over a 10-fold reduction of promoter activity with the Tibetan-enriched “A” allele of rs149594770. Although the trend is the same as those in human endothelial cells and placentas, this magnitude may not reflect the in vivo situation as 293T and Hela cells were used.
Although both HIF-1α and HIF-2α can induce EPO production during hypoxia, HIF-2α is dominant in renal EPO-regulation, as HIF-2α expression in the kidney co-localizes with EPO-producing renal cells (Haase 2006). This regulation mechanism explains why the EPAS1 adaptive variants have the greatest impact on Hb among the tested physiological parameters in Tibetans (table 1). Accordingly, the EPAS1+/− mice (50% reduction) showed a similar Hb regulation with Tibetans (fig. 4). Interestingly, EGLN1, the other key adaptive gene has a similar strategy as EPAS1 does, but works at the post-translational level. The Tibetan version of EGLN1 carries two amino acid mutations (Xiang et al. 2013), resulting in a stronger enzyme activity and an increased degradation of HIFs under hypoxic conditions (Lorenzo et al. 2014). Taken together, current evidence suggests that the Tibetan versions of EPAS1 and EGLN1 push toward the same direction for blunted responses to hypoxia (Bigham and Lee 2014), and they both reduce the amount of HIF-2α in Tibetans under hypoxic conditions—one at transcriptional level and the other at post-translational level—leading to a decreased HIF-mediated augmentation of erythropoiesis that protects Tibetans from high-altitude polycythemia (Lorenzo et al. 2014).
Besides a relatively low hemoglobin level, Tibetans also exhibit other adaptive traits, including sustained high resting-ventilation, low hypoxic pulmonary vasoconstriction response and better arterial oxygen saturation (Wu and Kayser 2006). We showed that two EPAS1 variants were significantly associated with a low PAP in Tibetans (table 1). The mouse data provided a possible molecular mechanism of how EPAS1 affects hypoxic pulmonary vasoconstriction response. We observed a significant down-regulation of ACE and VEGFC in the heart and the lungs of the EPAS1+/− mice compared with the WT mice (supplementary table S7, Supplementary Material online). Being a key regulator of normal blood pressure, ACE generates the vasoconstricting peptide angiotensin II and inactivates the vasodilating peptide bradykinin (Erdos 1990). ACE knockout mice exhibit low blood pressure (Krege et al. 1995), and over-expression of ACE augments cardiac hypertrophy in transgenic rats (Tian et al. 2004). VEGF is a target gene of EPAS1 and plays an important role in angiogenesis. In rats, the up-regulation of VEGF is involved in vascular remodeling associated with the development of pulmonary hypertension (Partovian et al. 1998). Hence, the mouse data suggest that the Tibetan version of EPAS1 likely contributes to low PAP of Tibetans through a down-regulation of ACE and VEGFC.
Both iNOS and eNOS are the target genes of EPAS1, and they produce NO, an important molecule for vasodilation (Coulet et al. 2003; Schodel et al. 2011). It was reported that in comparison to sea level controls, Tibetans had >10-fold-higher circulating concentrations of bioactive NO products (Erzurum et al. 2007), which may explain the improved blood flow and low PAP in Tibetans. However, in mice, iNOS and eNOS showed an opposite pattern with an up-regulation of iNOS and a down-regulation of eNOS in the EPAS1+/− mice. We did not detect a blood NO concentration difference between the EPAS1+/− mice and the WT mice. The observed higher blood NO concentration of high-altitude Tibetans could be caused merely by altitude difference because the previous report did not test NO levels in lowlanders living at high altitude, nor in Tibetans living at sea level (Erzurum et al. 2007). Thus, further experiments are required to determine if NO plays a role in blood flow and pulmonary vasoconstriction response of Tibetans.
The Tibetan versions of EPAS1 and EGLN1 do not seem to account for the reported better SaO2 in Tibetans compared with Han new comers at altitude (Wu and Kayser 2006), although a major autosomal dominant gene was previously inferred to explain the difference of the SaO2 levels in Tibetan women (Beall et al. 2004). Current evidence suggests that Tibetans have a better SaO2 level than lowlander Han Chinese during submaximal and maximal exercise, but no difference was detected at rest (Weitz and Garruto 2007). In this study, we measured SaO2 of Tibetans at rest, and we observed a weak negative association. Hence, whether SaO2 serves as one of the adaptive traits of Tibetans and what gene is responsible remains unanswered.
In view of fetal development, although the EPAS1 expression reduction can cause regulatory changes in many downstream genes, it may not have a great impact on placenta and fetal development. We observed similar transcriptional profiles in the placentas of high-altitude Tibetans and lowlander Han Chinese. Also, the EPAS1+/− mice with a 50% expression reduction turned out to be developmentally normal (Tian et al. 1998). Alternatively, since the oxygen exchange of the developing fetus depends on maternal blood circulation, the genetic background of the mother instead of the fetus is presumably more influential to fetal development as previous data showed that Tibetan women with high oxygen saturation had more surviving children than women with low oxygen saturation (Beall et al. 2004).
Finally, Tibetans are likely the earliest human populations who adapted to high altitude environments because their permanent settlement in the Qinghai-Tibetan plateau was dated as early as 30,000 years ago based on genetic data (Shi et al. 2008; Qi et al. 2013). Previous studies indicated that the onset of EPAS1 and EGLN1 selection occurred at different times, ∼18,000 years ago for EPAS1 and ∼8,000 years ago for EGLN1, which coincide the two proposed migratory waves into Qinghai-Tibetan Plateau during Upper Paleolithic and early Neolithic times (Qi et al. 2013). Regardless of whether the Tibetan version of EPAS1 was introduced by introgression from archaic humans (Huerta-Sanchez et al. 2014), the selection on EPAS1 likely represents the initial Paleolithic adaptation of Tibetan ancestors at altitude, and the Tibetan version of EGLN1 was probably introduced later during early Neolithic due to the expansion of the earliest Neolithic agriculturalists who originated in the Upper Yellow River basin of China ∼8,500 years ago (Qi et al. 2013). This two-episode selection model accounts for the prevalence of the adaptive versions of EPAS1 and EGLN1 in current Tibetan populations.
Materials and Methods
Collection of Physiological Data and Blood Samples of Tibetans
Physiological data and blood samples were collected from unrelated Tibetans permanently residing at Bange county of Nagqu prefecture (n = 508, 219 males and 289 females, 37.41 ± 3.8 years old) with an elevation 4,700 m. Biologically related individuals were excluded from this study. Written informed consents for all participants were obtained. For physiological parameters, we collected hemoglobin concentration (Hb), level of arterial oxygen saturation (SaO2) and systolic pulmonary artery pressure (PAP), which are among the key adaptive physiological traits in Tibetans (Wu and Kayser 2006). Hb was measured using a HemoCue Hb 201+ analyzer (Angelholm, Sweden). SaO2 was measured at forefinger tip with a hand-held pulse oximeter (Nellcor NPB-40, CA) at rest, and the fingertip was cleaned with alcohol swab before measurement. Venous blood was collected from each individual for Hb measurement and DNA extraction. For echocardiographic measurements, standard parasternal, apical, and subcostal two-dimensional views and color flow-directed pulsed wave Doppler of transvalvular flows and continuous wave Doppler of the tricuspid regurgitant jet flow were performed on fully rested subjects in the supine or left lateral decubitus position using a portable ultrasound system (Vivid I, GE Healthcare) equipped with a 1.5–3.6 MHz M4S-RS matrix sector array transducer. The peak systolic tricuspid regurgitation pressure gradient (TRPG) as an estimate of the systolic pulmonary artery pressure was calculated using the simplified Bernoulli equation: TRPG = 4 × TRVmax2, where TRVmax is the maximal velocity of the tricuspid regurgitant jet. All measurements were made in triplicates, and results are presented as means and the studies were evaluated blindly as to the source of individual recordings. The protocol of this study was approved by the Institutional Review Board of the Kunming Institute of Zoology, Chinese Academy of Sciences.
Selection of Candidate EPAS1 Variants
We analysed the entire 93-kb gene region of EPAS1. The 3.4-kb deletion (TED) 80-kb downstream of EPAS1 was also included. A total of 83 unrelated Tibetan individuals were analysed with 50 of them from the genome-wide SNP data (by Affymetrix Genome-wide Human SNP Array 6.0) (Peng et al. 2011) and 33 of them from genome sequencing (by Illumina HiSeq X10) (Lu 2016). Among the 83 unrelated Tibetans, we identified a total of 204 EPAS1 sequence variants. After compared with four lowlander populations from the 1000 Genomes Project, including 103 Han Chinese from Beijing (CHB), 104 Japanese (JPT), 99 Europeans (CEU) and 108 Africans (YRI) (http://www.1000genomes.org), we obtained 180 EPAS1 shared variants for further analysis.
Locus specific FST was calculated between Tibetans and the four lowlander populations following the previous method (Weir and Cockerham 1984). We also calculated the FST value of TED (Tibetan enriched deletion) by treating TED as a bi-allelic variant.
For iHS and XP-EHH tests, due to the high rate of missing genotype in the genome array data, we only used the genome sequencing data (30× genome coverage) of the 33 unrelated Tibetan individuals (Lu 2016). The missing genotypes were rare (0.92%) and retrieved by imputation. The iHS scores were calculated for all variants using Selscan (Szpiech and Hernandez 2014) based on the phased haplotypes. Phasing was conducted using Beagle 4.1 (Browning and Browning 2007), and only loci whose ancestral alleles were known with certainty and a minor allele frequency (MAF) ≥ 0.05 were included in the analysis (Voight et al. 2006). The XP-EHH analysis was used to detect the extended haplotypes resulting from positive selection in Tibetans (Sabeti et al. 2007). We computed XP-EHH scores using Selscan (Szpiech and Hernandez 2014) based on the phased haplotypes of Tibetans and CHB (as the reference population). The XP-EHH score at each SNP was standardized by the mean XP-EHH and the standard deviation over the genome. An absolute iHS or XP-EHH value of >2.0 is considered a signal of selection.
For functional prediction of the candidate EPAS1 variants, we measured the conservation score for each variant using GERP (Genome Evolutionary Rate Profiling) (http://mendel.stanford.edu/SidowLab/downloads/gerp/). The GERP ++ method was used (Davydov et al. 2010) to calculate site-specific RS scores and to discover evolutionarily constrained elements. Positive scores represent a substitution deficit (i.e., fewer substitutions than the average neutral site) and thus indicate that a site may be under evolutionary constraint. Positive scores scale with the level of constraint, such that the greater the score, the greater the level of evolutionary constraint inferred to be acting on that site. Negative scores indicate that a site is probably evolving neutrally. For calculating the GERP value of TED, we took the average of the entire 3.4 kb (3,400 sites) region.
Functional enrichment analysis of the candidate variants was conducted using CADD (Combined Annotation Dependent Depletion) (http://krishna.gs.washington.edu/download/CADD/v1.3/1000G_phase3_inclAnno.tsv.gz), which contains a list of 114 annotation categories for all variants in 1000G Phase 3. Five annotation categories were used in this study, including maximum ENCODE H3K4 methylation level (EncH3K4Me1), maximum ENCODE H3K4 trimethylation level (EncH3K4Me3), P value (PHRED-scale) of DNase I hypersensitivity evidence for open chromatin (EncOCDNasePVal), number of different overlapping ChIP transcription factor binding sites (TFBS), and ACCEPTOR or DONOR of the closest splice site (Dst2SplType). The H3K4Me1 value indicates the maximum ENCODE H3K4 methylation level (the maximum value observed across the 16 ENCODE cell lines at a given position), where modification of histone proteins is suggestive of enhancer and, to a lesser extent, other regulatory activity. The H3K4Me3 value indicates the maximum ENCODE H3K4 trimethylation level (the maximum value observed across the 16 ENCODE cell lines at a given position), where modification of histone proteins is suggestive of promoters. The DNase I hypersensitivity track indicates where chromatin is hypersensitive to cutting by the DNase enzyme, which has been assayed in a large number of cell types. Regulatory regions tend to be DNase-sensitive, and promoters are particularly DNase-sensitive. Dnase P indicates P value (PHRED-scale) of Dnase evidence for open chromatin. The transcription factor binding site (TFBS) is indicated by the number of different overlapping ChIP transcription factor binding sites (supplementary table S1, Supplementary Material online). In addition, Splice indicates that the closest splice site of the tested variant is either ACCEPTOR or DONOR (supplementary table S1, Supplementary Material online).
Endothelial Cell Hypoxic Assay
Tibetan umbilical cords were obtained from 105 normal, full-term pregnancies at the People’s Hospital of Lhasa, Tibetan Autonomous Region of China. Written informed consent was obtained from each subject. The human umbilical vein endothelial cells (HUVECs) were isolated from the veins of umbilical cord by the well-established method (Jaffe et al. 1973). In brief, cords were separated from placentas within 2 h after delivery. The umbilical vein was cannulated and washed through with sterile phosphate buffered saline (PBS), supplemented with 100 units/ml penicillin, 0.1 mg/ml streptomycin and 5.6 µg/ml amphotericin B in order to remove any blood clots. It was then infused with 1 mg/ml (125 U/ml) of type I collagenase (Gibco) in Ca2+ and Mg2+ free PBS, and incubated at 37 °C for 15 min. After incubation, the HUVECs were flushed from the vein vessel together with the collagenase solution. The obtained HUVECs were cultured on flasks coated with 1% gelatin. The cells were grown in complete medium (Medium199 supplemented with 10% FBS, 20 mmol/l HEPES (pH 7.4), 1 ng/ml recombinant human fibroblast growth factor (bFGF), 1 ng/ml endothelial growth factors (EGF), and 100 units/ml penicillin, 0.1 mg/ml streptomycin) (Gibco) at 37ºC in 5% CO2, and the medium was changed every 2–3 days. About 7∼10 days, the primary culture cells were confluent in the flask. For subculture, cells were detached and harvested with 0.05% trypsin-EDTA solution, then neutralized with FBS (Gibco). When the cells were cultured to passage-3, they were cryopreserved with cell freezing medium (NCS + 10% DMSO) in liquid nitrogen.
All hypoxia experiments were performed in passage-5 cells. The HUVECs were divided into two groups (adaptive vs. nonadaptive EPAS1 haplotype). We conducted three paired experiments, that is, ECU-A01 versus ECu-W01, ECU-A02 versus ECU-W02, and ECU-A03 versus ECU-W03. Each cell line has two replication dishes. Cells were placed in a conventional 37 °C humidified incubator (Thermo-Forma, Model 3141) with automated gas control of 1% O2, 5% CO2, balanced with N2. The oxygen concentration of the hypoxic incubator was constantly monitored by a real-time O2 sensor with 0.1% sensitivity. Cells were harvested at different time points, including 0 h, 6 h, 12 h, 24 h (1 day), 48 h (2 days), 72 h (3 days), 120 h (5 days) and 168 h (7 days). For each dish, three parallel quantitative real-time PCR were performed to determine the expression level of EPAS1, therefore, there are six data points for each cell line.
RNAseq Data Generation and Analysis
Paired-end reads were generated from Illumina HiSeq 2500 platform. We followed a previous protocol to remove low quality reads (Yan et al. 2013), where reads with adapters or with >10% ambiguous bases or with >50% low quality bases (Q-score ≤ 5) were removed. On an average, we generated 24.5 million paired-end reads for each sample. The RNAseq data was deposited in an open-access archive (Genome Sequence Achieve at http://gsa.big.ac.cn) and the accession numbers are PRJCA000268 and PRJCA000269.
Pre-processed RNA-Seq reads were mapped to the genome using split-mapping aligner STAR (release 2.4.1a) (Dobin et al. 2013), and the resulting bam files were feed to RSEM (version 1.2.11) (Li and Dewey 2011) to estimate gene expression abundance. To handle reads mapped to multiple locations, RSEM takes mapping uncertainty into account with a generative model and the EM algorithm to achieve accurate abundance estimates at both the isoform and gene levels (Li and Dewey 2011). The mouse genome sequence was downloaded from Ensembl (release 81) (Cunningham et al. 2015), and the human genome was downloaded from Gencode release 19 (Harrow et al. 2012). To detect differentially expressed genes, we used a python framework HTSeq (Anders et al. 2015) to count the number of reads mapped to each gene, and then applied the voom method embedded in the R package limma (Law et al. 2014), which is a Bayesian method accounting for mean–variance relationship. Genes with multiple test corrected P value <0.05 were determined as differentially expressed.
Since the second exon of EPAS1 is replaced by a nonmouse-origin frame-shifting sequence in the EPAS1 knockout copy, it might be possible that the inactivated transcript decays quickly and cannot be detected, leading to similar expression levels across all exons. To explore this possibility, we inspected the number of reads mapped to exon2 of EPAS1 and its flanking exon1 and exon3 in both knockout and wildtype mice, and found similar expression level of exon1 and exon3 as well as a clear reduction of reads mapped to exon2 in knockout mice compared with wildtype, indicating the inactivated transcript retains to be measured. Moreover, we noticed that the estimate of active EPAS1 transcripts would be biased if we include reads mapped to inactivated copies. To correct this, we counted reads mapped to exon1, 2 and 3 of EPAS1, and calculated the average ratio of exon2/exon1 and exon2/exon3 for each sample, the adjusted EPAS1 expression estimate is then obtained by multiplying the ratio with the uncorrected estimate.
Electrophoretic Mobility Shift Assay and Luciferase Reporter Assay
To test the predicted functional of SNP rs149594770, we performed electrophoretic mobility shift assay (EMSA). The double-stranded probes were obtained through boiling and gradual cooling of the complementary single-strand oligonucleotides. The nucleotide sequences of the double-stranded oligonucleotides with either T or A allele are:
T allele: 5′-TCTTTAGGGTGAGCTGACCTTTCCAGATGGAGCAGGGAAAT-3
A allele: 5′-TCTTTAGGGTGAGCTGACCTATCCAGATGGAGCAGGGAAAT-3′.
DNA binding reactions were premixed with a 200-fold molar excess of unlabeled competitors at room temperature for 20 min, followed by another 20 min of incubation when adding the labeled probes. After incubation, samples were separated on a native 6% polyacrylamide gel, and then rapidly transferred to a positive nylon membrane, UV-cross-linked, probed with streptavidin-HRP conjugate and incubated with the substrate.
For luciferase reporter assay, the 100-bp synthetic single-strand oligonucleotides were annealed to form double strands consisting of the corresponding genotypes (TT and AA) for rs149594770 flanked by restriction sites. The fragments were then cloned into the multiple cloning site of the pGL3-promoter vector (Promega). All constructs were validated by sequencing to make sure no de novo mutation was introduced. The reporter vectors containing either T allele or A allele were co-transfected into HEK293T and HeLa cells, respectively, together with a reference vector (pRL-TK vector). HeLa and HEK293T cells were grown in Gibco Dulbecco’s Modified Eagle’s Medium (Gibco) supplemented with 10% fetal bovine serum (HyClone). Lipofectamine 2000 (Invitrogen) was used in transient transfection. After a 36-h incubation, we collected the cell lysates and measured luciferase activity using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI). The relative light units were measured using a luminometer. The mean values of three independent experiments were used.
Prolonged Hypoxic Experiments in Mice
Eight-week-old mice (littermates; mixed-background Swiss/129Sv) were weighted and placed in a polycarbonate hypoxic chamber (Coy Laboratory Products). The oxygen level in the chamber is controlled automatically by balancing the air with N2. After exposure to prolonged hypoxia for the indicated period, mice were first equilibrated by room air for 1 h in order to avoid acute vasomotor responses, and were then anesthetized with urethane (1.4 mg/g). The closed chest method was used to measure cardiac function. A 1.2-F pressure catheter (Transonic Scisense Inc) was used to measure right ventricle (RV) pressure. RV pressure and heart rate were recorded continuously (MP150, BIOPAC Systems). Arterial blood was collected in heparinized glass capillaries (50 µl) and immediately analysed for hemoglobin (Hb) concentration. Hb concentration was determined using a spectrophotometer (B-Hemoglobin, HemoCue). The degree of RV hypertrophy was determined by the weight ratio of right ventricle to heart.
The measurement of predominant species of NO2− and NO3− in plasma were employed to reveal the level of NO synthesis, and the NO level in plasma is almost all reflected by the level of nitrate because the nitrite level in plasma is only ∼1% of nitrate. We adopted a chemiluminescence method to measure the concentration of NO using a nitric oxide analyzer following the protocol provided by the manufacture (Sievers model 280, GE Analytical Instruments, Boulder, CO). The plasma sample was treated with precooling ethanol (2 folds) to eliminate proteins. The calibration curve was constructed by plotting the peak area versus concentration of the standard NaNO3 solution (pre-treated with 2 folds ethanol). Sample injection was carried out using a Hamilton syringe with 5-in long needle, and 30 µl plasma sample was injected for each sample. Two injections were measured for each sample and the average value of the two measurements was taken. To eliminate the noise of ethanol, a baseline correction was conducted before sample injection and measurement.
For HE staining, the mice were perfused with phosphate-buffered saline by cardiac perfusion techniques, and 4% PFA was used for tissue fixation. Sections of lungs (12 µm) were used for HE staining. The slides were treated at 60 °C for 30 min and then were rinsed with water. After washing, the slides were stained with hematoxylin for 3 min. After staining with hematoxylin, slides were washed sufficiently with water, immersed in 95% ethanol for 5 s, and counterstained with eosin for 10 s. The slides were dehydrated in a gradient concentration of ethanol and dehydrated with xylene for 3–5 min each. Finally, the slides were mounted with neutral balsam.
Genetic Association Analysis
The genetic association analysis of three key physiological traits, including hemoglobin level, pulmonary arterial pressure and degree of blood oxygen saturation, was performed using PLINK 1.07 (Purcell et al. 2007). Based on the EPAS1 expression analysis of endothelial cells and placentas, we adopted the additive genetic model in the association analysis. Gender and age were treated as covariates when applicable. For multiple test correction, we used a permutation test (100,000 times for each test) to generate empirical P values.
Statistical Analysis and Plot
Unless specified, all statistical analyses were performed using R (https://www.R-project.org). Multiple test correction was performed using the Benjamini–Hochberg approach (Benjamini and Hochberg 1995).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
B.S., X.Q., C.C. and T.W. conceived the project. X.Q., C.C., Ouzhuluobu, Y.P., Y.H., H.Z., Bianbazhuoma, K. X., S.L., Yangla, Dejiquzong, Baimakangzhuo, Duojizhuoma, Yongyue Pan, Cirenyangji, Baimayangji, Gonggalanzi and Bianba collected the samples; Y.P., X.Q., Y.H., H.Z., D.Y., K.X. collected the data; L.Y., Y.H., X.Z., S.B., P.S., Basang, Ciwangsangbu, L.L. and H.S provided technical support during the experiments; B.S., X.Q. Q.Z., Y.H., S.X. and H.C. conducted data analysis; B.S., X.Q., Y.P., Y.H. and Q.Z. wrote the article.
Supplementary Material
Acknowledgments
We are grateful to all of the volunteers in this study. We would like to thank Dr Akylbek Sydykov (Justus Liebig University of Giessen, Germany) for further confirmation of echocardiographic data in Bange population. We also thank Dr Peter Carmeliet (Katholieke Universiteit Leuven, Belgium) for kindly providing the genetically engineered mice. This study was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB13010000), the National Natural Science Foundation of China (91231203, 91631306, 31321002, 31401064, 31671329, 31371269, 31460287 and 31525014), the National 973 Program of China (2012CB518202), the State Key Laboratory of Genetic Resources and Evolution (GREKF15-05, GREKF16-04), the Zhufeng Scholar Program of Tibetan University, and the Program of Shanghai Academic Research Leader (16XD1404700).
References
- Anders S, Pyl PT, Huber W.. 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall CM, Cavalleri GL, Deng L, Elston RC, Gao Y, Knight J, Li C, Li JC, Liang Y, McCormack M, et al. 2010. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci U S A. 107:11459–11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall CM, Song K, Elston RC, Goldstein MC.. 2004. Higher offspring survival among Tibetan women with high oxygen saturation genotypes residing at 4,000 m. Proc Natl Acad Sci U S A. 101:14300–14304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J R Stat Soc B Methodol. 57:289–300. [Google Scholar]
- Bigham A, Bauchet M, Pinto D, Mao X, Akey JM, Mei R, Scherer SW, Julian CG, Wilson MJ, Lopez Herraez D, et al. 2010. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 6:e1001116.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham AW, Lee FS.. 2014. Human high-altitude adaptation: forward genetics meets the HIF pathway. Genes Dev. 28:2189–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL.. 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 81:1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusselmans K, Compernolle V, Tjwa M, Wiesener MS, Maxwell PH, Collen D, Carmeliet P.. 2003. Heterozygous deficiency of hypoxia-inducible factor-2alpha protects mice against pulmonary hypertension and right ventricular dysfunction during prolonged hypoxia. J Clin Invest. 111:1519–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compernolle V, Brusselmans K, Acker T, Hoet P, Tjwa M, Beck H, Plaisance S, Dor Y, Keshet E, Lupu F, et al. 2002. Loss of HIF-2alpha and inhibition of VEGF impair fetal lung maturation, whereas treatment with VEGF prevents fatal respiratory distress in premature mice. Nat Med. 8:702–710. [DOI] [PubMed] [Google Scholar]
- Coulet F, Nadaud S, Agrapart M, Soubrier F.. 2003. Identification of hypoxia-response element in the human endothelial nitric-oxide synthase gene promoter. J Biol Chem. 278:46230–46240. [DOI] [PubMed] [Google Scholar]
- Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. 2015. Ensembl 2015. Nucleic Acids Res. 43:D662–D669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S.. 2010. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol. 6(12):e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdos EG. 1990. Angiotensin I converting enzyme and the changes in our concepts through the years. Lewis K. Dahl memorial lecture. Hypertension 16:363–370. [DOI] [PubMed] [Google Scholar]
- Erzurum SC, Ghosh S, Janocha AJ, Xu W, Bauer S, Bryan NS, Tejero J, Hemann C, Hille R, Stuehr DJ, et al. 2007. Higher blood flow and circulating NO products offset high-altitude hypoxia among Tibetans. Proc Natl Acad Sci U S A. 104:17593–17598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase VH. 2006. Hypoxia-inducible factors in the kidney. Am J Physiol Renal Physiol. 291:F271–F281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. 2012. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka M, Shimba S, Sakaue M, Kondo Y, Kagechika H, Kokame K, Miyata T, Hara S.. 2009. Hypoxia-inducible factor-3alpha functions as an accelerator of 3T3-L1 adipose differentiation. Biol Pharm Bull. 32:1166–1172. [DOI] [PubMed] [Google Scholar]
- Ho JJ, Man HS, Marsden PA.. 2012. Nitric oxide signaling in hypoxia. J Mol Med (Berl) 90:217–231. [DOI] [PubMed] [Google Scholar]
- Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC.. 2003. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol. 23:9361–9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Sanchez E, Jin X, Asan Bianba Z, Peter BM, Vinckenbosch N, Liang Y, Yi X, He M, Somel M, et al. 2014. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512:194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe EA, Nachman RL, Becker CG, Minick CR.. 1973. Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J Clin Invest. 52:2745–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitsinou PP, Rajendran G, Astleford L, Michael M, Schonfeld MP, Fields T, Shay S, French JL, West J, Haase VH.. 2016. The endothelial prolyl-4-hydroxylase domain 2/hypoxia-inducible factor 2 axis regulates pulmonary artery pressure in mice. Mol Cell Biol. 36:1584–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler SP, de SSP, Scheidemantel TS, Gomos JB, Rowe TM, Sen GC.. 2003. Maintenance of normal blood pressure and renal functions are independent effects of angiotensin-converting enzyme. J Biol Chem. 278:21105–21112. [DOI] [PubMed] [Google Scholar]
- Krege JH, John SW, Langenbach LL, Hodgin JB, Hagaman JR, Bachman ES, Jennette JC, O’Brien DA, Smithies O.. 1995. Male-female differences in fertility and blood pressure in ACE-deficient mice. Nature 375:146–148. [DOI] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK.. 2014. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15:R29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo FR, Huff C, Myllymaki M, Olenchock B, Swierczek S, Tashi T, Gordeuk V, Wuren T, Ri-Li G, McClain DA, et al. 2014. A genetic mechanism for Tibetan high-altitude adaptation. Nat Genet. 46:951–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou H, Lu Y, Lu D, Fu R, Wang X, Feng Q, Wu S, Yang Y, Li S, Kang L, et al. 2015. A 3.4-kb copy-number deletion near EPAS1 is significantly enriched in high-altitude Tibetans but absent from the Denisovan sequence. Am J Hum Genet. 97:54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Lou H, Yuan K, Wang X, Wang Y, Zhang C, Lu Y, Yang X, Deng L, Zhou Y, et al. 2016. Ancestral origins and genetic history of Tibetan highlanders. Am J Hum Genet. 99:580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazon A, Goldrath AW, Nizet V, Johnson RS.. 2014. HIF transcription factors, inflammation, and immunity. Immunity 41:518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partovian C, Adnot S, Eddahibi S, Teiger E, Levame M, Dreyfus P, Raffestin B, Frelin C.. 1998. Heart and lung VEGF mRNA expression in rats with monocrotaline- or hypoxia-induced pulmonary hypertension. Am J Physiol. 275:H1948–H1956. [DOI] [PubMed] [Google Scholar]
- Peng Y, Yang Z, Zhang H, Cui C, Qi X, Luo X, Tao X, Wu T, Ouzhuluobu Basang, et al. 2011. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol Biol Evol. 28:1075–1081. [DOI] [PubMed] [Google Scholar]
- Petousi N, Robbins PA.. 2014. Human adaptation to the hypoxia of high altitude: the Tibetan paradigm from the pregenomic to the postgenomic era. J Appl Physiol (1985) 116:875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC.. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Cui C, Peng Y, Zhang X, Yang Z, Zhong H, Zhang H, Xiang K, Cao X, Wang Y, et al. 2013. Genetic evidence of Paleolithic colonization and Neolithic expansion of modern humans on the Tibetan Plateau. Mol Biol Evol. 30:1761–1778. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449:913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schodel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, Mole DR.. 2011. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood 117:e207–e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. 2012. Hypoxia-inducible factors in physiology and medicine. Cell 148:399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Zhong H, Peng Y, Dong YL, Qi XB, Zhang F, Liu LF, Tan SJ, Ma RZ, Xiao CJ, et al. 2008. Y chromosome evidence of earliest modern human settlement in East Asia and multiple origins of Tibetan and Japanese populations. BMC Biol. 6:45.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, Bai Z, Lorenzo FR, Xing J, Jorde LB, et al. 2010. Genetic evidence for high-altitude adaptation in Tibet. Science 329:72–75. [DOI] [PubMed] [Google Scholar]
- Song D, Li LS, Arsenault PR, Tan Q, Bigham AW, Heaton-Johnson KJ, Master SR, Lee FS.. 2014. Defective Tibetan PHD2 binding to p23 links high altitude adaption to altered oxygen sensing. J Biol Chem. 289:14656–14665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpiech ZA, Hernandez RD.. 2014. selscan: an efficient multithreaded program to perform EHH-based scans for positive selection. Mol Biol Evol. 31:2824–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Hammer RE, Matsumoto AM, Russell DW, McKnight SL.. 1998. The hypoxia-responsive transcription factor EPAS1 is essential for catecholamine homeostasis and protection against heart failure during embryonic development. Genes Dev. 12:3320–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, McKnight SL, Russell DW.. 1997. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 11:72–82. [DOI] [PubMed] [Google Scholar]
- Tian XL, Pinto YM, Costerousse O, Franz WM, Lippoldt A, Hoffmann S, Unger T, Paul M.. 2004. Over-expression of angiotensin converting enzyme-1 augments cardiac hypertrophy in transgenic rats. Hum Mol Genet. 13:1441–1450. [DOI] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK.. 2006. A map of recent positive selection in the human genome. PLoS Biol. 4:e72.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC.. 1984. Estimating F-statistics for the analysis of population-structure. Evolution 38:1358–1370. [DOI] [PubMed] [Google Scholar]
- Weitz CA, Garruto RM.. 2007. A comparative analysis of arterial oxygen saturation among Tibetans and Han born and raised at high altitude. High Alt Med Biol. 8:13–26. [DOI] [PubMed] [Google Scholar]
- Wu T, Kayser B.. 2006. High altitude adaptation in Tibetans. High Alt Med Biol. 7:193–208. [DOI] [PubMed] [Google Scholar]
- Xiang K, Ouzhuluobu Peng Y, Yang Z, Zhang X, Cui C, Zhang H, Li M, Zhang Y, Bianba, et al. 2013. Identification of a Tibetan-specific mutation in the hypoxic gene EGLN1 and its contribution to high-altitude adaptation. Mol Biol Evol. 30:1889–1898. [DOI] [PubMed] [Google Scholar]
- Xu S, Li S, Yang Y, Tan J, Lou H, Jin W, Yang L, Pan X, Wang J, Shen Y, et al. 2011. A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol Biol Evol. 28:1003–1011. [DOI] [PubMed] [Google Scholar]
- Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, Yan J, et al. 2013. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 20:1131–1139. [DOI] [PubMed] [Google Scholar]
- Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, et al. 2010. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329:75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.