Abstract
New genes arise through a variety of mechanisms, including the duplication of existing genes and the de novo birth of genes from noncoding DNA sequences. While there are numerous examples of duplicated genes with important functional roles, the functions of de novo genes remain largely unexplored. Many newly evolved genes are expressed in the male reproductive tract, suggesting that these evolutionary innovations may provide advantages to males experiencing sexual selection. Using testis-specific RNA interference, we screened 11 putative de novo genes in Drosophila melanogaster for effects on male fertility and identified two, goddard and saturn, that are essential for spermatogenesis and sperm function. Goddard knockdown (KD) males fail to produce mature sperm, while saturn KD males produce few sperm, and these function inefficiently once transferred to females. Consistent with a de novo origin, both genes are identifiable only in Drosophila and are predicted to encode proteins with no sequence similarity to any annotated protein. However, since high levels of divergence prevented the unambiguous identification of the noncoding sequences from which each gene arose, we consider goddard and saturn to be putative de novo genes. Within Drosophila, both genes have been lost in certain lineages, but show conserved, male-specific patterns of expression in the species in which they are found. Goddard is consistently found in single-copy and evolves under purifying selection. In contrast, saturn has diversified through gene duplication and positive selection. These data suggest that de novo genes can acquire essential roles in male reproduction.
Keywords: de novo gene, Drosophila, fertility, sperm, sexual selection
Introduction
Genomes acquire new genes through a variety of mechanisms, including gene duplication, retrotransposition, and horizontal gene transfer (Chen et al. 2013). While such newly evolved genes can be important for various biological processes, many function in reproduction. Gene duplication events are well documented for reproductive proteins in a range of organisms (Vacquier et al. 1997; Karn et al. 2008; Meslin et al. 2012), with many examples in Drosophila (Saudan et al. 2002; Wagstaff and Begun 2005, 2007; Dorus et al. 2008; Findlay et al. 2008; Sirot et al. 2014). For instance, genes arising from duplication events are known to play roles in sperm competitive ability (Nurminsky et al. 1998; Yeh et al. 2012), transcriptional and post-transcriptional regulation during spermatogenesis (Ding et al. 2010; Sartain et al. 2011) and the ability of the paternally derived chromatin to undergo proper mitotic division upon fertilization (Yasuda et al. 1995; Loppin et al. 2005).
While gene duplication is the best-characterized process for creating new genes, emerging evidence suggests that the de novo evolution of genes from noncoding sequence may also be an important source of evolutionary innovation (Chen et al. 2010; Carvunis et al. 2012; Silveira et al. 2013; Wissler et al. 2013; McLysaght and Hurst 2016). Like duplicate genes, de novo genes are often expressed in reproductive tissues (Wu et al. 2011; Reinhardt et al. 2013; Palmieri et al. 2014). One of the first such genes to be characterized was the Mus musculus gene Poldi, which is believed to encode a nonprotein-coding RNA (Heinen et al. 2009). Knockout of this gene caused reduced testis size and decreased sperm swimming speed. In Drosophila, 7% of the annotated protein-coding genes in the D. pseudoobscura genome lack apparent orthologs and paralogs outside of the obscura group and thus potentially arose de novo (Palmieri et al. 2014), and nearly 250 putative de novo, testis-expressed genes were found in a sample of just six genomes drawn from a single population of D. melanogaster (Zhao et al. 2014). Such de novo genes also have a high rate of inactivating mutations, but male-biased expression is a significant predictor of retention after gene birth (Palmieri et al. 2014). Indeed, many other de novo genes show male-biased expression patterns (Begun et al. 2006; Levine et al. 2006; Metta and Schlotterer 2008; Zhou et al. 2008; Findlay et al. 2009). An analogous pattern is observed in rice and Arabidopsis, in which many de novo genes are expressed in the male gametophyte (Cui et al. 2015), and in primates, in which de novo genes are often testis-expressed (Guerzoni and McLysaght 2011; Wu et al. 2011).
The example of Poldi shows that de novo genes can impact phenotypes even if they do not encode proteins, but many of the de novo genes discovered to date are predicted to be protein coding. For such genes to evolve, two major evolutionary changes must occur in a nongenic region: The region must become reliably transcribed, and it must come to contain an open reading frame (ORF) that encodes a biologically useful polypeptide. Models can be constructed in which either transcription or protein-coding potential evolves first (McLysaght and Guerzoni 2015; Schlotterer 2015); in the former case, it is possible for de novo proteins to evolve from noncoding RNAs. These changes were initially believed to be difficult to evolve, but it is becoming clear that in Drosophila melanogaster (as well as other organisms), a high fraction of the genome is transcribed in at least some individuals, organs, and/or developmental stages (Brown et al. 2014; Chen et al. 2014; Neme and Tautz 2016). Furthermore, in D. melanogaster, there are nearly 175,000 potential ORFs in annotated intergenic or intronic regions that could encode polypeptides of >40 amino acids (Begun et al. 2006). An ORF that is transcribed and translated is termed a proto-gene (Carvunis et al. 2012), but the encoded polypeptide, even when biologically useful, is unlikely to arise in a fully optimized form. Indeed, data from yeast and flies suggest that de novo proteins differ from existing proteins in their length, amino acid composition, and degree of intrinsic disorder (Carvunis et al. 2012; Abrusan 2013; Zhao et al. 2014; Bitard-Feildel et al. 2015).
Many de novo genes in Drosophila that are retained by selection are expressed specifically in the male reproductive tract (Levine et al. 2006; Zhou et al. 2008). Kaessmann (2010) hypothesized that the requirements for transcription in the testes may be less stringent than in other tissues, and a recent analysis of two newly evolved, testis-specific retrogenes shows that the regulatory regions that induce testis expression can be quite short (Sorourian et al. 2014). However, work in the obscura group of Drosophila has shown that the youngest putative de novo genes (found in just one species) show less of an expression bias toward males than those found in several species (Palmieri et al. 2014). These data suggest that when a de novo gene is expressed in the testes, it has a higher chance of being retained by selection. Additionally, Zhao et al. (2014) found that sometimes only one or two nucleotide changes in the cis-regulatory region of a de novo gene separate a testis-expressed allele from a nontestis-expressed allele, implying that relatively few cis-regulatory changes would be needed for a de novo gene to become testis expressed. These studies imply that de novo genes may be well positioned to underlie lineage-specific changes in reproductive phenotypes. For example, the process of spermatogenesis may evolve rapidly between closely related species that show divergence in mating systems, levels of sperm competition, and/or patterns of sperm usage or storage (Ramm et al. 2014). Such divergence is indeed observed between related species of Drosophila (Pitnick et al. 1999; Markow and O'Grady 2005; Scharer et al. 2008).
While putative de novo genes have been identified in many species and often expressed in male reproductive organs, their functions remain largely unexplored. Previous functional studies in D. melanogaster have relied largely on whole-organism RNA interference (Chen et al. 2010; Reinhardt et al. 2013), rather than targeted knockdown (KD) in the male reproductive tract. These studies found that de novo genes are often essential for fitness because they are required for viability. Analysis of nonviable KD progeny showed that, for most genes tested, the pupal-to-adult transition was the most common stage of growth arrest. While these results are interesting, their interpretation is potentially problematic for two reasons. First, recent reports suggest that ∼25% of the lines from the RNAi collection used in those experiments can produce a dominant, pupal-lethal phenotype upon ubiquitous KD, regardless of the specific gene being targeted (Green et al. 2014; Vissers et al. 2016). Furthermore, Reinhardt et al. (2013) could not link these de novo genes’ patterns of expression—many of which are expressed primarily or exclusively in the testes (Chintapalli et al. 2007)—to the preadult viability phenotype observed in both sexes. Thus, the specific roles played by de novo genes in Drosophila male reproduction remain unknown. An additional limitation is that many genes termed de novo have not undergone fine-scale evolutionary analyses to pinpoint the exact circumstances of their origins (McLysaght and Hurst 2016; though see Reinhardt et al. 2013 for good examples of such analyses).
To circumvent these technical issues, we screened 11 putative de novo genes with testis-specific expression for functions in male reproduction using testis-specific RNA interference. We found two genes, which we named goddard and saturn, that had major effects on male fertility. KD of goddard blocked the production of mature sperm, while KD of saturn reduced sperm production and caused the sperm that were produced to be stored inefficiently in mated females. Both genes encode proteins with no detectable homology to other proteins from Drosophila and other taxa, and the proteins lack functional domains that would be consistent with membership in a gene family. These features, combined with other bioinformatic properties of the gene and protein sequences, suggest a de novo origin for each gene, though there remains some uncertainty about their origins because of difficulty in aligning the syntenic regions in non-Drosophila species. Within Drosophila, each gene is absent in a subset of species. In species that have goddard, it is present in single copy and has evolved under purifying selection. In contrast, saturn has undergone gene duplication, rearrangement, and adaptive evolution in different Drosophila lineages. Our findings suggest that sexual selection may be an important force that promotes the retention of recently born de novo genes.
Results
A Preliminary Screen for Essential De Novo Genes
Many potential de novo genes in D. melanogaster show testis-biased expression patterns (Chintapalli et al. 2007; Reinhardt et al. 2013). To investigate whether any such genes have become essential for male fertility, we conducted a preliminary RNAi screen of 11 candidate genes (supplementary fig. S1, Supplementary Material online). These genes were identified as potential de novo genes because of their bioinformatic classification as such in a previous study (Zhang et al. 2010) and/or because, at the time of their selection, their patterns of annotated orthologs on FlyBase suggested the genes’ existence in a limited subset of Drosophila species. We knocked down each gene with Bam-GAL4 (White-Cooper 2012), which drives RNAi expression in the spermatogonia and spermatocytes prior to the onset of meiosis. This preliminary screen had low power (see Materials and Methods), but it revealed two genes essential for male fertility, CG13477 and CG32141, as well as other genes with potential minor effects that await more detailed characterization (supplementary fig. S1, Supplementary Material online). We previously named male seminal fluid proteins essential for postmating responses after lunar modules of the Apollo space program (Findlay et al. 2014); we extend this convention here by naming these two new genes after icons in rocketry that were necessary for the Apollo missions. We refer to CG13477 as goddard, after Robert Goddard, the father of modern rocketry, and we denote CG32141 as saturn, after the Saturn V rocket.
Goddard and Saturn Are Required for Male Fertility through Effects on Sperm Production and Function
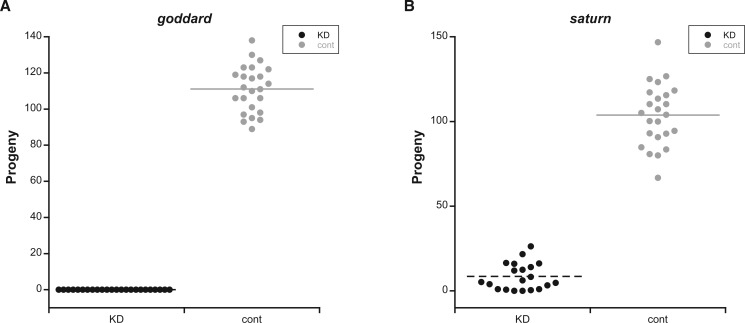
To characterize the fertility defects caused by goddard or saturn KD in greater detail, we first performed single-pair mating experiments using males generated with Bam-GAL4 and a variety of RNAi lines. Near-complete KD was verified by RT-PCR and occurred for all RNAi constructs described below (supplementary fig. S2, Supplementary Material online). We initially used "KK" RNAi lines (see Materials and Methods) that knocked down each gene effectively. KD or control males were mated individually to wild-type females, and then removed. These females were allowed to lay eggs for 4 days and were then discarded. We found that KD of goddard (fig. 1A) caused complete male sterility, while KD of saturn (supplementary fig. S3, Supplementary Material online) caused a significant, 90% reduction in male fertility (two-sample t-test, t = −8.92, df = 24.6, P < 0.0001). Because a fraction of KK lines have an additional transgene insertion that causes dominant phenotypes (Green et al. 2014), and because the KK line for saturn had two predicted off-target genes that were also expressed in the testes, we validated these results by using the available "GD" RNAi lines: One for goddard and two for saturn. These results confirmed the results of the KK lines. KD of goddard again caused complete sterility (supplementary fig. S3, Supplementary Material online). KD of saturn with either GD line caused a significant reduction in fertility, though one GD line had a stronger effect (line 41108: Two-sample t-test, t = −23.07, df = 32.5, P < 0.0001; fig. 1B) than the other (line 41107: Two-sample t-test, t = −5.73, df = 22.9, P < 0.0001; supplementary fig. S3, Supplementary Material online), likely due to differences in the degree of KD (supplementary fig. S2, Supplementary Material online). This difference in KD level could have been caused by different sites of transgene insertion and/or different sequences being used to trigger KD. We also tested the fertility of RNAi lines that targeted the genes identified as potential off-targets of the saturn KK line. We observed no significant fertility defects when each gene was knocked down with Bam-GAL4 (data not shown).
Fig. 1.
Goddard and saturn are required for male fertility. Number of progeny produced from single-pair matings between wild-type females and A) goddard knockdown (KD, n = 24) or control (cont, n = 24) males, or B) saturn KD (n = 20) or cont (n = 24) males. Both comparisons were analyzed by two-sample t-tests, with P < 0.0001 in both cases.
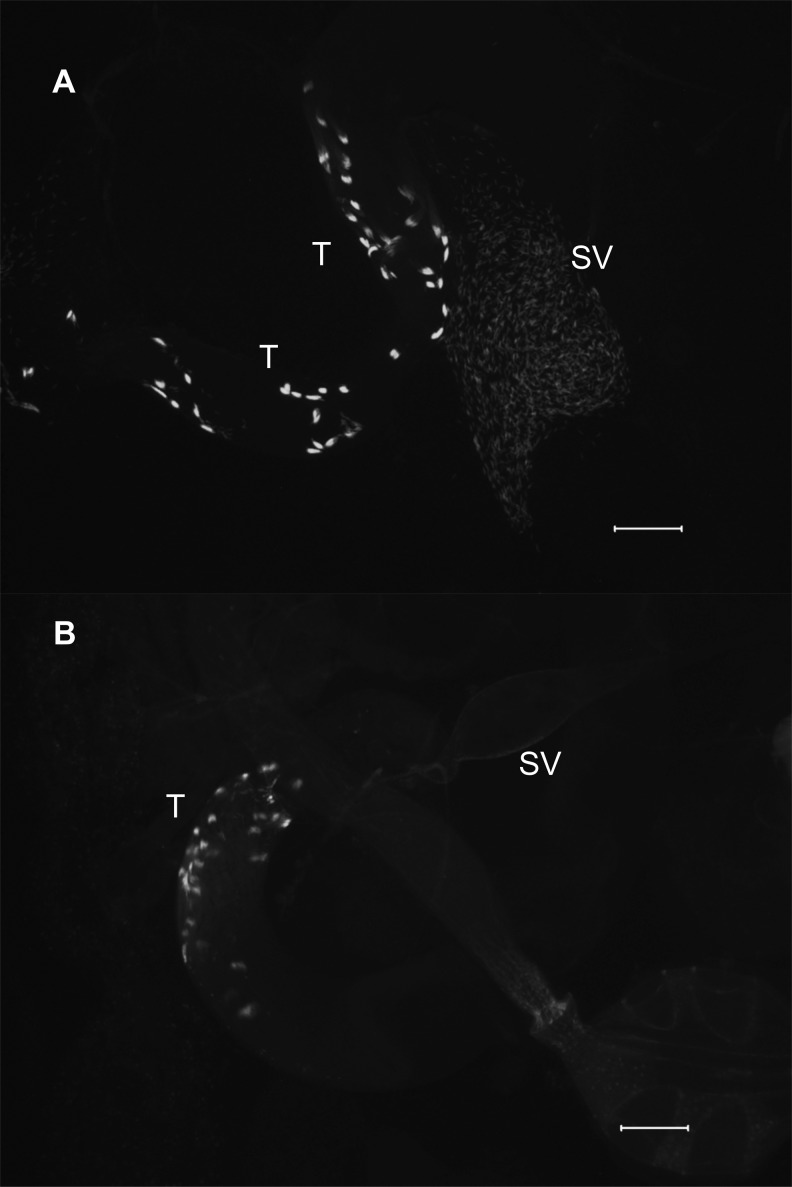
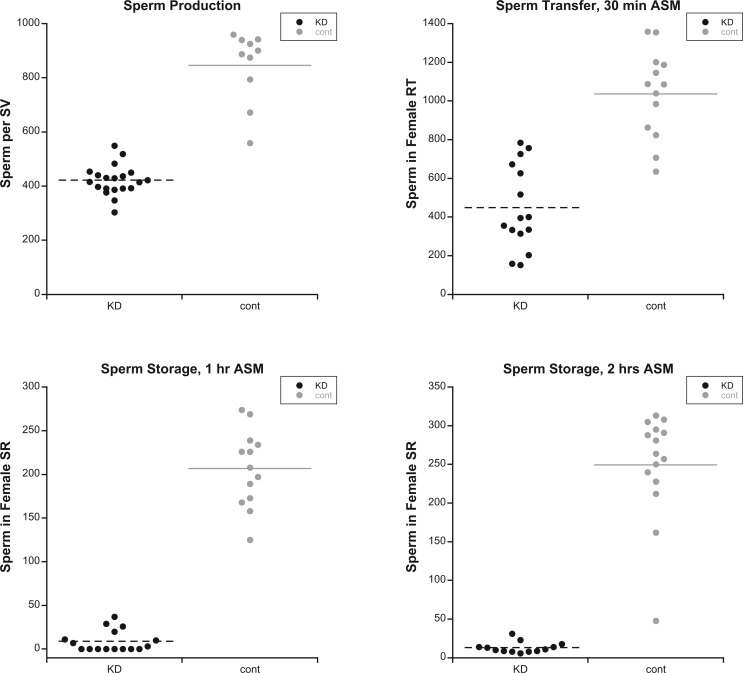
To investigate the cellular nature of the above fertility defects, we used the Mst35Bb-GFP marker (Manier et al. 2010) to label the nuclei of late-stage spermatids and individualized sperm in KD and control males. We first examined the seminal vesicles of KD and control males to evaluate levels of sperm production. Males knocked down for goddard showed no individualized sperm in their seminal vesicles (fig. 2), consistent with their complete sterility. Seminal vesicles of males knocked down for saturn contained only ∼45% as many mature sperm as controls (two-sample t-test: t = −9.69, df = 10.6, P < 0.0001; fig. 3A), indicating that saturn is required for efficient spermatogenesis. To evaluate whether these sperm showed normal motility, we dissected reproductive tracts from KD and control males, pierced the seminal vesicles and testes to release the sperm, and took a series of images that were made into a movie. At this resolution, we observed no qualitative difference in the motility of KD and control male sperm (supplementary videos S1 and S2, Supplementary Material online).
Fig. 2.
Knockdown of goddard prevents production of mature sperm. The Mst35Bb-GFP marker was used to label the nuclei of late-stage sperm bundles during spermiogenesis in the testes (T) and individualized sperm in the seminal vesicle (SV). Relative to controls (A), goddard KD males (B) show some sperm bundles but fail to produce any individualized sperm. Scale bar: 100 μm.
Fig. 3.
Males depleted for saturn show reduced sperm production and inefficient sperm storage. (A) Mature, individualized sperm present in the seminal vesicle (SV) of saturn knockdown (KD, n = 20 flies) and control (cont, n = 10) males. Each data point represents the average number of sperm per SV for one male. (B) Sperm present in the female uterus 30 min ASM with saturn KD (n = 15) or control (n = 13) males. (C) Sperm present in the female seminal receptacle (SR) 1 h ASM with saturn KD (n = 15) or control (n = 13) males. (D) Sperm present in the SR 2 hours ASM with saturn KD (n = 13) or control (n = 15) males. In all panels, KD values are significantly less than controls (two-sample t-tests, all P < 0.0001).
We next evaluated the ability of saturn KD sperm to be transferred to and stored within females’ reproductive tracts. KD or control males were mated to wild-type females, and females were flash frozen 30 min after the start of mating (ASM). At this time point, most sperm transferred from males are present in the female’s uterus, though small numbers may have already entered the seminal receptacle (SR) and spermathecae (the two types of sperm storage organ in D. melanogaster) (Avila and Wolfner 2009; Manier et al. 2010). We observed that KD males transferred significantly fewer sperm to females than controls (fig. 3B; two-sample t-test: t = −6.96, df = 25.1, P < 0.001). However, the difference in the number of sperm transferred between KD and control males was roughly proportional to the difference in the number of sperm in the seminal vesicles of KD and control males. A parsimonious explanation for this result is that the mechanics of sperm transfer—that is, male contractions and the motility of sperm in the male—are normal in KD males; males transfer fewer sperm because they produce fewer sperm.
Finally, we investigated the ability of KD and control male sperm to enter the SR, the primary sperm storage organ in D. melanogaster. At 1 and 2 h ASM, sperm from control males were found in the SR at typical levels, but sperm from KD males were present at significantly lower levels (fig. 3C and D; two-sample t-tests, 1 h: t = −14.82, df = 14.8, P < 0.0001; 2 h: t = −13.11, df = 14.3, P < 0.0001). Thus, while sperm from saturn KD males were present in the uterus at ∼40% of the level of controls, their rate of storage—and thus their availability for fertilization—was depressed much further.
Putative De Novo Origins of Goddard and Saturn
The previous classification of goddard and saturn as potential de novo genes was based on high-throughput bioinformatic methods. To more sensitively investigate each gene’s evolutionary origin, we next performed detailed evolutionary analyses. The primary criterion for defining a protein-coding gene as de novo evolved is that the gene arose from noncoding DNA sequence (McLysaght and Hurst 2016). The most definitive evidence that satisfies this criterion is to identify the syntenic region in outgroup species that lack the gene and show that orthologous sequence is present, but does not encode an ORF. Depending on the age of the gene and the availability of genome sequences from closely related species, however, such evidence can be difficult to obtain. In such cases, a gene may be classified as putatively de novo if it encodes a protein with no sequence similarity to other proteins (McLysaght and Hurst 2016). This lack of similarity is inconsistent with an origin via gene duplication or horizontal gene transfer, which thus implicates a de novo origin by exclusion (McLysaght and Hurst 2016). However, it is important to note that a duplicated or horizontally transferred gene that evolves rapidly could lose detectable similarity and thus be mistakenly classified as de novo (Moyers and Zhang 2015).
Goddard was previously identified as a de novo gene due to a lack of orthologs in divergent Drosophila species (Chen et al. 2010; Zhang et al. 2010). To confirm its phylogenetic distribution, we used BLAST searches of annotated proteins to identify goddard orthologs in the genomes of all melanogaster group species (D. melanogaster, simulans, sechellia, yakuba, erecta, and ananassae) and obscura group species (D. pseudoobscura and persimilis) (fig. 4A). In each species, goddard exists within a large intron of the well-conserved omega gene (fig. 4B), so we used this gene to identify the syntenic regions of more distantly related Drosophila species. No ortholog could be found in D. willistoni, because the syntenic region appears to have undergone extensive deletion and/or rearrangement. In D. virilis, the syntenic region was present, and LASTZ alignment detected three regions of microsynteny (supplementary file S1, Supplementary Material online) to D. melanogaster goddard. Inspection of this region revealed a potential ORF that could encode a polypeptide of up to 190 amino acids. While this ORF had no significant homology by BLASTP to the orthologs of goddard described above, it showed highly significant, full-length homology to a potential ORF that we detected in the syntenic region of D. mojavensis (supplementary file S1, Supplementary Material online). This D. mojavensis ORF, in turn, returned a marginal BLASTP hit to goddard of D. melanogaster (e-value = 3.27). We tested for transcription of these ORFs in each species by RT-PCR using primers located in the predicted ORF. Both species showed robust amplification when cDNA from whole male flies was used as the template, but little or no amplification when whole female cDNA or cDNA from males from whom the testes had been removed by dissection were used (supplementary file S1, Supplementary Material online). Thus, these ORFs show evidence of male- and testis-enriched expression, as does goddard (Chintapalli et al. 2007; Brown et al. 2014). Based on their genomic position, testis-enriched expression, and marginal sequence identity, these putative genes are likely highly divergent orthologs of goddard, thus confirming the importance of using methods beyond BLAST to identify the phylogenetic distribution of de novo genes (Moyers and Zhang 2015). Finally, we found evidence of a potential goddard ortholog in the equally divergent D. grimshawi by searching for an ORF with identity to the D. virilis ORF in the syntenic region. We identified such an ORF, and while it is shorter than the D. virilis ORF, the degree of sequence identity is high (supplementary file S1, Supplementary Material online).
Fig. 4.
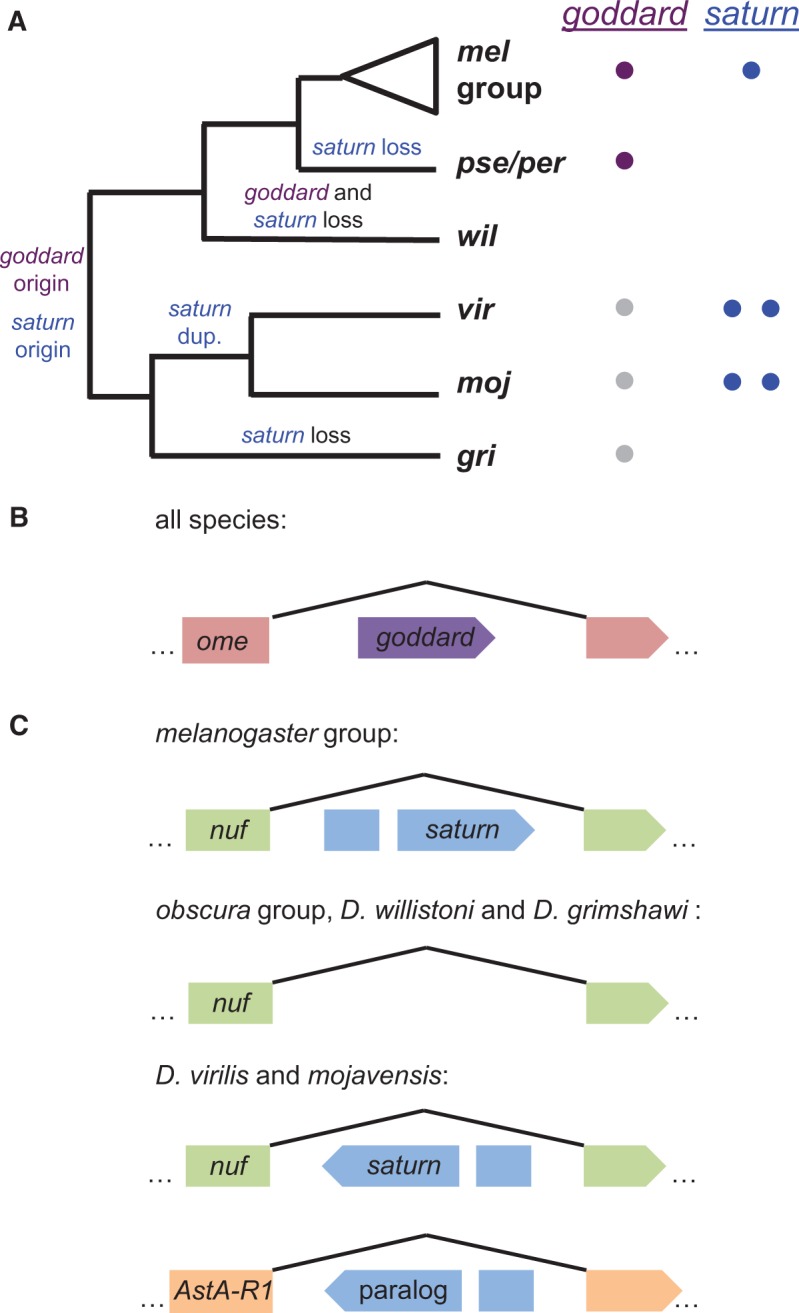
Evolutionary histories of goddard and saturn. (A) Copy numbers and inferred evolutionary histories of goddard and saturn across the Drosophila genus. Colored circles next to each species/group on the phylogeny indicate the number of copies of each gene identified in that taxon. Gray circles indicate putative, unannotated orthologs of goddard discovered based on syntenic location. (B) Goddard is located within an intron of the omega (ome) gene and is found in the same orientation in all species that have the gene. (C) Saturn gene structure in different groups of Drosophila species. Saturn is located within an intron of the nuclear fallout (nuf) gene in the melanogaster group and in D. virilis and mojavensis, though the orientation of saturn has been reversed in the latter species. Searches of the syntenic region in the obscura group and D. willistoni did not reveal any plausible saturn gene, and D. grimshawi had only partial remnants. In D. virilis and mojavensis, a saturn paralog is found in a large intron of the ortholog of the D. melanogaster gene, AstA-R1. Branch lengths in panel A and gene models in panels B and C are not to scale.
Given the presence of goddard in the most distantly diverged Drosophila species with sequenced genomes, we investigated the possibility that goddard exists in species outside of the Drosophila genus. Using the more highly conserved omega gene that harbors goddard, along with Exonerate searches of the goddard protein sequence against entire sequenced genomes, we searched for putative orthologs in the most closely related Dipteran species with a sequenced genome, the medfly Ceratitis capitata (Papanicolaou et al. 2016), as well as in two sequenced mosquito species, Anopheles gambiae and Aedes aegypti. Both methods failed to identify a possible goddard ortholog. The most parsimonious explanation for this pattern is that goddard arose de novo at the base of the Drosophila genus and was then lost in the D. willistoni lineage (fig. 4A). However, because the specific intron of the omega gene that harbors goddard in Drosophila could not be aligned to any of the genomes examined, we were unable to identify a noncoding orthologous region in an outgroup species, as required by McLysaght and Hurst (2016) to demonstrate a definitive de novo origin. Thus, we describe goddard as a putative de novo gene, though we cannot completely rule out the possibility that the gene could instead be a rapidly evolving member of a novel gene family.
Saturn (CG32141) was identified as a potential de novo gene because it initially had FlyBase-annotated orthologs only in the melanogaster group of species, though subsequent annotations of D. virilis identified a potential, more diverged copy. Like goddard, this gene is found within a large intron of another gene, nuclear fallout (nuf) (fig. 4C). We again used BLASTP to identify annotated copies of saturn in other species and used synteny to search for unrecognized orthologs. We found annotated orthologs in the melanogaster group species, as well as D. virilis and D. mojavensis. These latter orthologs were found in the same intron of the nuf gene, but their direction of transcription relative to nuf was reversed, suggesting a microinversion event (fig. 4C). To estimate the putative inversion breakpoints, we generated alignments of the respective nuf gene sequences using LASTZ. These alignments show a relatively close homology between the complete nuf gene sequences of D. melanogaster and both D. virilis and D. mojavensis. The genes align without major gaps from the 5′ to the 3′ end of nuf (supplementary fig. S4, Supplementary Material online). This good coverage of the alignments right next to the saturn coding sequence in D. melanogaster suggests that the inversion event was mostly restricted to the saturn coding sequence. Indeed, based on the position of the syntenic blocks found immediately adjacent to the inverted region, we estimate that the inversion breakpoints were no more than 350 bp away from the start and stop codons of saturn, suggesting that relatively little regulatory sequence was captured by the inversion.
Our BLAST searches also revealed one additional annotated protein in D. virilis and D. mojavensis that showed similar levels of identity to the D. melanogaster saturn protein, suggesting that saturn underwent gene duplication in the lineage leading to these species. The paralogous copy is located within an intron of the ortholog of the D. melanogaster gene AstA-R1. The paralog contains an intron in the same position as saturn, suggesting a duplication mechanism other than retrotransposition. However, we detected no evidence of conserved upstream or downstream sequences between the two paralogs in each species, leaving the mechanism of duplication unclear. Because AstA-R1 and nuf are unrelated genes, the duplication event likely involved only the saturn gene, rather than the larger gene in which it resides. The hypothesis that gene duplication preceded speciation in this lineage (fig. 4A) is also supported by a protein sequence tree that shows no clustering of within-species paralogs (supplementary fig. S5, Supplementary Material online).
We did not identify orthologs of saturn in the obscura group, D. willistoni, or D. grimshawi (fig. 4A and C). These results are similar to those of goddard in that they suggest an origin of the saturn gene at the base of the Drosophila phylogeny, followed by gene loss events in particular lineages (in this case, three independent events). The copy of saturn within the nuf intron is likely the ancestral copy, as we detected remnants of its protein-coding sequence in this syntenic regions of D. willistoni and D. grimshawi. Interestingly, saturn is found on chromosome 3L in D. melanogaster, which corresponds to the neo-X chromosome in D. pseudoobscura and D. willistoni. If the ancestral saturn gene in these species experienced meiotic sex chromosome inactivation (Kaiser and Bachtrog 2010) after the formation of the neo-X chromosome, the consequences of gene loss may have been ameliorated.
As with goddard above, we used Exonerate and a synteny-based analysis using the nuf intron to interrogate the medfly and mosquito genomes for potential orthologs. Exonerate identified no candidate orthologs in either species. The synteny analysis using the nuf intron was possible only in medfly (supplementary fig. S6, Supplementary Material online), since the relevant intron was not alignable to either mosquito genome. In medfly, the precise interval containing saturn had a very poor alignment quality. Scanning this region for ORFs revealed none resembling saturn. Nonetheless, because of the alignment difficulty, we conclude conservatively that saturn is a putative de novo gene. It is also possible that saturn encodes a member of a novel and rapidly evolving gene family, or that its sequence arose through the insertion of a sequence from an uncharacterized virus/mobile element, though this latter possibility was not supported by a search of known Drosophila transposable element families (J. Thomas and C. Feschotte, personal communication). We also note that the syntenic region of the AstA1 intron that harbors the saturn paralog was not alignable to the medfly or mosquito genomes.
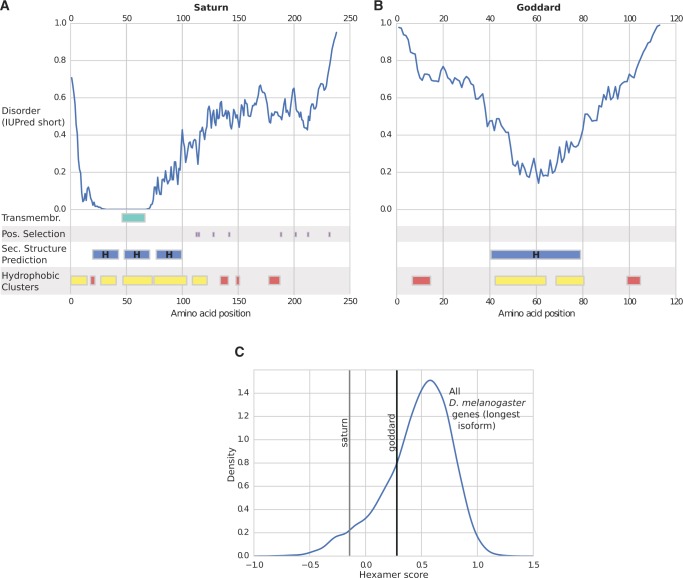
We next examined the sequences of goddard and saturn and the predicted biophysical properties of the proteins they encode. These analyses are largely consistent with predictions for de novo genes, but cannot positively confirm the mechanism of origination. First, neither the goddard protein nor the saturn protein shows detectable homology in either sequence-based (Marchler-Bauer et al. 2015) or predicted structure-based (Kelley et al. 2015) searches to any other annotated protein from Drosophila or any other taxa, which suggests neither belongs to a gene family or arose via horizontal gene transfer. Indeed, the most notable structural feature we could detect was a single predicted transmembrane domain in the saturn protein (fig. 5A).
Fig. 5.
Predicted biophysical and sequence properties of goddard and saturn. Line plots showing the likelihood of disorder (IUPred score) and schematic plots of other protein property predictions for saturn (A) and goddard (B). Hydrophobic clusters that do not match clusters from well-established globular proteins are highlighted in yellow, while those that do are shown in red (see Materials and Methods). Transmembrane domains, sites under positive selection, and predicted secondary structures (H: alpha helix) are also shown. (C) Hexamer score of saturn and goddard gene sequences compared with all D. melanogaster protein-coding genes. Hexamer scores are based on a comparison of in-frame hexamer usage of a coding sequence to a distribution of in-frame hexamer usage of a reference set of protein coding genes.
Second, the biophysical and other sequence properties of each protein include several features that are consistent with previous studies and theoretical expectations of de novo genes. We found both goddard and saturn proteins to contain large, intrinsically disordered stretches of sequence (fig. 5A and B). Compared with other D. melanogaster proteins, goddard and saturn exhibit high fractions of intrinsic disordered sequence (94th and 82nd percentiles, respectively; supplementary fig. S7, Supplementary Material online). Intrinsic disorder has previously been suggested to play an important role in de novo gene emergence (Carvunis et al. 2012; Zhao et al. 2014). Furthermore, a previous analysis of orphan domains in Drosophila found that younger domains contain clusters of hydrophobic residues with predicted folding patterns that differ from those of clusters commonly found in evolutionarily old, globular proteins (Bitard-Feildel et al. 2015). Applying this analysis to the goddard and saturn protein sequences, we found that most of these proteins’ hydrophobic clusters were also novel: They did not match any clusters from globular proteins (fig. 5A and B). However, this pattern did not deviate significantly from other D. melanogaster protein-coding genes (supplementary fig. S7, Supplementary Material online). Finally, we found that both genes show somewhat atypical patterns of hexamer usage bias patterns relative to most protein-coding genes in D. melanogaster (fig. 5C): Saturn’s hexamer usage is in the bottom 5% of all protein-coding genes, and goddard’s falls in the bottom 24%. Hexamer usage bias has previously been used to distinguish coding from noncoding sequence (Fickett and Tung 1992; Wang et al. 2013), and genes that arose recently from noncoding sequence may be predicted to have scores that are lower than average genes, but potentially increasing with gene age. However, the evolutionary dynamics of a gene’s hexamer score are as yet unclear. Specifically, it is unknown whether, and if so, how fast, the hexamer score of a de novo gene will evolve toward commonly observed levels. Consequently, a hexamer score that is somewhat divergent from the distribution of all protein-coding genes is consistent with de novo origination, but not proof of it.
Molecular Evolution
We used PAML to investigate the molecular evolution of goddard and saturn. We considered each gene’s evolution across two time frames: Within the melanogaster subgroup, represented here by six species estimated to have shared a common ancestor 3–4 Ma, and within the broader melanogaster group, represented here by 15–16 species estimated to have shared a common ancestor ∼15 Ma (Obbard et al. 2012). For each gene at each timescale, we estimated the overall dN/dS ratio (ω) across all alignable sites (PAML model M0), and we used the PAML sites tests (comparing models M7, M8, and M8a) to identify specific residues predicted to have evolved under recurrent positive selection (Yang et al. 2000; Swanson et al. 2003).
These tests revealed that goddard and saturn have evolved under somewhat different selective pressures. The goddard gene has evolved fairly slowly over both timescales (table 1). The calculated ω for all sites across the entire sequence (PAML model M0) was 0.21–0.25, and standard tests for positive selection on a subset of sites were nonsignificant (table 1). Within neutral model M7 and across both sets of species examined, six of the ten classes of sites had ω < 0.25, suggesting that most sites in the protein have evolved under purifying selection. This result is consistent with predictions that novel genes that acquire an essential function are more likely to evolve under purifying selection (Domazet-Loso and Tautz 2003). The observations of purifying selection across all species of the melanogaster group, and of patterns of substitution that preserve the full-length ORF, are consistent with goddard encoding a protein, rather than a noncoding RNA.
Table 1.
PAML-Based Tests for Positive Selection on Goddard.
| Data From Six Species of the melanogaster Subgroup: |
| Model M0 (uniform ω): ω = 0.25, ln L = −882.10, np = 11 |
| Model M7 (10 site classes, each with 0 ≤ ω ≤ 1): ln L = −879.18, np = 12 |
| Model M8 (10 site classes as in M7, and 1 class with ω ≥ 1): ln L = −879.18, np = 14, ω for extra class of sites = 1.00 (0.0% of sites) |
| Model M8a (10 site classes as in M7, and 1 class with ω = 1): ln L = −879.18, np = 13 |
| M7 vs. M8 LRT: χ2 = 0, df = 2, P = 1.00 |
| M8 vs. M8a LRT: χ2 = 0, df = 1, P = 1.00 |
| Data From 16 Species of the melanogaster Group: |
| Model M0 (uniform ω): ω = 0.21, ln L = −3038.97, np = 31 |
| Model M7 (10 site classes, each with 0 ≤ ω ≤ 1): ln L = −2979.22, np = 32 |
| Model M8 (10 site classes as in M7, and 1 class with ω ≥ 1): ln L = −2979.03, np = 34, ω for extra class of sites = 2.20 (0.7% of sites) |
| Model M8a (10 site classes as in M7, and 1 class with ω = 1): ln L = −2979.19, np = 33 |
| M7 vs. M8 LRT: χ2 = 0.38, df = 2, P = 0.83 |
| M8 vs. M8a LRT: χ2 = 0.32, df = 1, P = 0.57 |
NOTE.—ln L , log likelihood; LRT, likelihood ratio test; np, number of parameters.
In contrast, saturn has a complex evolutionary history. In addition to the gene duplication and inversion events described above, the protein-coding sequence has evolved rapidly. A protein sequence alignment for saturn showed that the N-terminal portion of the protein (amino acids 1–105 in D. melanogaster) could be aligned with high confidence across the melanogaster group (supplementary fig. S8, Supplementary Material online), excluding D. kikkawai. In contrast, the C-terminal end of the saturn protein (amino acids 106–238 in D. melanogaster) is characterized by repetitive, disordered sequences, rapid divergence, and indel variation (fig. 5A;supplementary fig. S8, Supplementary Material online), which prevented confident alignment outside of the melanogaster subgroup. Interestingly, analysis of the saturn sequences by the SBP algorithm (Kosakovsky Pond et al. 2006) revealed evidence for a putative recombination breakpoint in the 108th codon. Because of the alignment differences and the potential confounding effect of recombination on phylogenetic inference, we thus tested for selection on the N-terminus only in both the melanogaster group and subgroup, and tested the whole gene and the C-terminus only in the melanogaster subgroup (table 2; supplementary table S1, Supplementary Material online).
Table 2.
PAML-Based Tests for Positive Selection on Saturn.
| Data From Six Species of the melanogaster Subgroup: |
| Test of whole gene |
| Model M0 (uniform ω): ω = 0.90, ln L = −2405.30, np = 11 |
| Model M7 (10 site classes, each with 0 ≤ ω ≤ 1): ln L = −2391.89, np = 12 |
| Model M8 (10 site classes as in M7, and 1 class with ω ≥ 1): ln L = −2380.87, np = 14, ω for extra class of sites = 4.18 (14.0% of sites) |
| Model M8a (10 site classes as in M7, and 1 class with ω = 1): ln L = −2391.45, np = 13 |
| M7 vs. M8 LRT: χ2 = 22.04, df = 2, P < 0.0001 |
| M8 vs. M8a LRT: χ2 = 21.16, df = 1, P < 0.0001 |
| Sites identified under positive selection by BEB analysis with Pr ≥ 0.9 (numbers indicate the amino acid position in D. melanogaster): 111A, 113S, 115D, 128S, 142F, 188L, 201H, 212A, 231N. |
| Data From 15 Species of the melanogaster Groupa: |
| Test of N-terminus only |
| Model M0 (uniform ω): ω = 0.29, ln L = −2410.15, np = 29 |
| Model M7 (10 site classes, each with 0 ≤ ω ≤ 1): ln L = −2380.78, np = 30 |
| Model M8 (10 site classes as in M7, and 1 class with ω ≥ 1): ln L = −2380.78, np = 32, ω for extra class of sites = 1.00 (0.0% of sites) |
| Model M8a (10 site classes as in M7, and 1 class with ω = 1): ln L = −2380.78, np = 31 |
| M7 vs. M8 LRT: χ2 = 0.00, df = 2, P = 1.00 |
| M8 vs. M8a LRT: χ2 = 0.00, df = 1, P = 1.00 |
NOTE.—LRT, likelihood ratio test; np, number of parameters; BEB, Bayes Empircal Bayes analysis.
Only 15 species were used in this comparison because the identified ortholog from D. kikkawai could not be reliably aligned.
In the closely related melanogaster subgroup species, the full-length gene has a high, gene-wide ω of 0.90, near the dN/dS ≈ 1 expected for a neutrally evolving gene. Using the sites tests, we found that this high rate of evolution is driven at least in part by positive selection; selection model M8 fit the data significantly better than neutral models M7 and M8a, and nine sites in the C terminus were flagged as having a high posterior probability of having evolved adaptively. Upon partitioning the gene, the N terminus showed no evidence of positive selection and a lower, though still somewhat elevated, ω value. In contrast, the C terminus gave a highly significant signal of selection, with model M8 identifying the same nine sites as being under positive selection. In the broader melanogaster group, the rate of evolution of the N terminus was further reduced, with no evidence of sites under positive selection. This finding, combined with patterns of substitutions maintaining lengthy ORFs in all Drosophila species that possess the gene (supplementary fig. S8, Supplementary Material online), are consistent with a protein-coding gene. We could not formally test the C terminus in this group, but the difficulty of aligning this region suggests its continued, rapid evolution. This general pattern of rapid evolution for saturn is consistent with the recurrent observation of adaptive divergence in reproductive proteins (reviewed by Wilburn and Swanson 2016), and with observations that de novo genes show elevated rates of molecular evolution (Carvunis et al. 2012; Reinhardt et al. 2013; Palmieri et al. 2014).
Conservation of Gene Expression Patterns
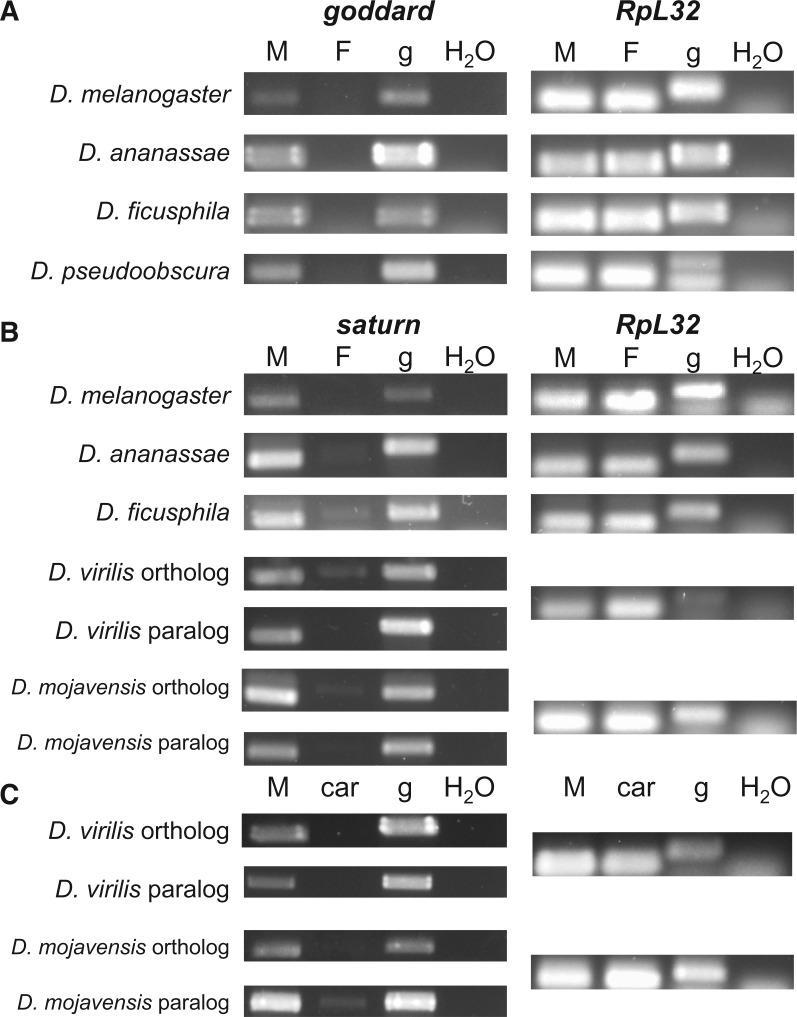
Given the different patterns of sequence evolution for goddard and saturn, as well as the copy number changes and inversion event for saturn, we next asked whether the expression patterns of these genes were conserved across the species in which they are found. Transcriptome profiling in D. melanogaster has shown that both genes are expressed in a testis-specific pattern in adult flies (Chintapalli et al. 2007; Brown et al. 2014) (supplementary fig. S9, Supplementary Material online). RNAseq data from D. pseudoobscura (Chen et al. 2014) showed that the goddard ortholog is also expressed testis-specifically. We investigated both genes’ expression patterns in additional species by performing RT-PCR on cDNA synthesized from RNA isolated from whole males or whole females. For goddard, we confirmed the gene’s male-specific pattern of expression in divergent members of the melanogaster group, D. ananassae and D. ficusphila, and in D. pseudoobscura (fig. 6A). These data, combined with those presented above for the putative orthologs in D. virilis and D. mojavensis (supplementary file S1, Supplementary Material online), suggest that goddard has maintained male-specific expression since its origin. Likewise, saturn orthologs showed male-specific, or strongly male-biased, expression in all species tested (fig. 6B). Interestingly, we observed weak RT-PCR amplification for saturn in females of D. ananassae, D. ficusphila, D. virilis, and D. mojavensis. These PCR products were found in repeated biological replicates and were consistently the size of genomic DNA, rather than spliced cDNA, yet they persisted even when extra DNase was used to remove genomic DNA from the RNA samples prior to cDNA synthesis (see Materials and Methods). We thus hypothesize that saturn is transcribed at low levels in females of these species, but perhaps the lack of an appropriate sex-specific splicing factor (Telonis-Scott et al. 2009) prevents the expression of the saturn protein.
Fig. 6.
Goddard and saturn show conserved, male-specific expression across a wide range of Drosophila species. RT-PCR was performed on goddard, saturn and a housekeeping control gene, RpL32, across a variety of species to assess the sex- and tissue-specificity of expression. Goddard (A) and saturn (B) show conserved male-specific expression patterns in divergent species spanning the Drosophila genus. Template DNA for PCR included M: whole male cDNA; F: whole female cDNA; g: genomic DNA; H2O: water (negative control). Faint bands can be seen in the female saturn lanes of some of the species, but these bands appear to be equal in size to the gDNA product, rather than the spliced, cDNA product. The presence and sizes of these bands were confirmed in multiple biological replicates, suggesting that they represent low levels of expression of an unspliced transcript in females. (C) RT-PCR performed on cDNA isolated from whole males (M) or male carcasses after the removal of the testes (car). Removal of the testes causes the loss of transcripts of saturn and its paralog, suggesting that all or most of the male-specific expression observed for these species in panel (B) is due to expression in the testes.
Both paralogs of saturn showed male-specific expression in D. virilis and D. mojavensis, in spite of the inversion and gene duplication events in this lineage (fig. 6B). To confirm that this pattern was due to expression in the testes, we compared RT-PCR amplification of cDNA isolated from whole males versus amplification of cDNA isolated from males from whom the testes had been removed by dissection. Removal of the testes resulted in the complete or near-complete removal of saturn transcripts (fig. 6C), confirming that saturn has retained its testis-specific expression pattern even in the face of gene duplication and inversion.
Discussion
Previous attempts to determine the functions of de novo genes in Drosophila (Chen et al. 2010; Reinhardt et al. 2013) have used ubiquitous RNA interference. These studies found that some de novo genes are essential for adult viability, though it remains an open question whether some of these results were caused by a dominant phenotypic artifact predicted for a fraction of the RNAi lines used (Green et al. 2014). However, even if they were not, the preadult lethality caused by ubiquitous KD prevented any investigation into how de novo genes affect adult male fertility. This question is important because of the testis-specific expression pattern shown for many de novo genes (Reinhardt et al. 2013; Palmieri et al. 2014; Zhao et al. 2014). By using tissue-restricted KD and multiple, independent RNAi lines, we investigated whether de novo genes expressed in the testes play essential roles in male reproduction. Following the de novo gene classification guidelines proposed by McLysaght and Hurst (2016), we characterized two putative de novo genes, goddard and saturn, that are each essential for male fertility. KD of goddard blocks production of mature sperm. KD of saturn causes reduced sperm production and a decrease in the likelihood of storage of the sperm that are transferred to the female during mating. While goddard has evolved under purifying selection, saturn has evolved rapidly in species closely related to D. melanogaster and has been both inverted and duplicated in more divergent species.
It will be important to determine the specific roles that goddard and saturn play during spermatogenesis. The Bam-GAL4 driver we used to knock down the genes acts primarily in the spermatogonia and spermatocytes, but not in postmeiotic stages or in somatic cells of the testis that support spermatogenesis (White-Cooper 2012). However, RNAi induced by this driver in premeiotic cells is known to persist after meiosis, as well. Microarray data on RNA isolated from mitotic, meiotic and postmeiotic cells in the testes detected transcripts from both genes throughout spermatogenesis, with slightly increased levels for each in meiotic and postmeiotic cells (Vibranovski et al. 2009). Thus, these proteins likely act in the germline, and not in somatic cells, but we cannot presently pinpoint the exact stage(s). In general, however, the phenotypes of male-sterile mutants are most likely to manifest at postmeiotic stages (Wakimoto et al. 2004). It also remains an open question whether either protein is present in mature sperm. Neither protein has been identified in the D. melanogaster sperm proteome (Wasbrough et al. 2010), but the lack of protein detection in shotgun mass spectrometry experiments is not strong evidence of absence. Notably, while no direct evidence currently exists to show that either gene encodes a protein, the low estimates of dN/dS for goddard and the N-terminus of saturn (tables 1 and 2), and the observation that indels between species occur in multiples of three and thus preserve the genes’ ORFs, strongly suggest that both genes encode proteins.
Whether the saturn protein, in particular, is incorporated into mature sperm and/or acts locally in the testes to regulate sperm production is interesting in light of the saturn KD phenotypes affecting both sperm production and sperm storage. One possibility is that saturn has multiple functions, one affecting sperm production and the another that influences sperm traits that are necessary for the sperms' ability to be stored. However, it is also possible that saturn is present only in the testes. In this scenario, the two effects of saturn KD could be explained if the reduction of expression alters spermatogenesis such that a reduced number of suboptimally functioning sperm are produced. Further investigation of this question will be best addressed with a null allele of saturn, given that RNAi greatly reduced, but did not fully eliminate, saturn transcripts (supplementary fig. S2, Supplementary Material online).
Due to the lack of homology to other proteins, the sequences of goddard and saturn provide few insights into their potential cellular functions. The only computationally predicted structural feature from either protein is the single potential transmembrane domain in the N-terminal portion of saturn, which is conserved across all orthologs and paralogs (fig. 5A;supplementary fig. S8, Supplementary Material online). We also note the high number of positively charged amino acids in the C-terminal region of saturn (e.g., 15 lysines found in amino acid positions 106–238), which are somewhat reminiscent of the lysine- and arginine-rich sperm nuclear basic proteins that contribute to the packaging of DNA in the highly condensed nucleus of mature sperm (Raja and Renkawitz-Pohl 2005; Rathke et al. 2007). Beyond these features, the sequences of goddard and saturn are broadly consistent with de novo origins, including their deviation from the mean hexamer frequency present in Drosophila protein coding genes and the high fraction of each protein predicted to be disordered (fig. 5).
While goddard and saturn have become essential for efficient sperm production in D. melanogaster, these genes arose in an ancestral species that must have already been producing functional sperm. Thus, hypotheses for how these putative de novo genes became established as bona fide genes must take into account both their initial nonessentiality and their current importance. One possibility is that each gene had a modest, but beneficial, effect on male reproduction when it was born. Even a slight positive effect on male fitness would be favored by sexual selection, allowing the gene to persist and potentially fix in an ancestral population, after which its sequence could become optimized. Other de novo genes have been observed to have this sort of modest benefit on male reproduction, such as Poldi in mice (Heinen et al. 2009), which appears to cause a modest increase in testis size, and the sdic gene cluster in flies (Yeh et al. 2012), the benefit of which is seen only under sperm competitive conditions. Upon fixation, the genes would then evolve to become essential, likely by becoming integrated into existing protein interaction networks. Data from yeast suggest that de novo genes may become essential fairly quickly (Abrusan 2013). However, in the cases of goddard and saturn, these genes have also been lost in certain lineages after their origins. One potential resolution to this discrepancy is that these genes might have been necessary in the ancestors of these species, but subsequent changes in each species’ male reproductive system (e.g., Snook and Karr 1998) rendered the genes dispensable. Furthermore, both genes are found on chromosome 3L in D. melanogaster, but this Muller element has become a neo-X chromosome in some other species, including D. pseudoobscura and D. willistoni. If genes on these neo-X chromosomes undergo meiotic sex chromosome inactivation (Kaiser and Bachtrog 2010), the consequences of deleterious mutations may be minimized, increasing the probability of gene loss. In any case, the observed pattern is consistent with a high loss probability of young genes (Palmieri et al. 2014).
The goddard and saturn genes show contrasting patterns of molecular evolution. Goddard is found in single copy and has evolved under purifying selection. This result is consistent with it evolving to become essential and gaining interacting partners shortly after gene birth, and with predictions that slow-evolving de novo genes are the most likely to be functionally important (Domazet-Loso and Tautz 2003). In contrast, the evolutionary history of saturn is dynamic. Two of the sequenced Drosophila species that diverged from the melanogaster group most anciently, D. virilis and D. mojavensis, now have two copies of the gene, while D. willistoni, D. grimshawi, and the obscura group species have no detectable copies. The gene’s conserved, male-specific expression across species suggests that saturn continues to play some role in male reproduction in each species, but in light of the gene’s rapid sequence divergence, it is possible that the orthologous copies have evolved to affect sperm production and function in their respective species in ways that differ from the effects of the D. melanogaster copy, perhaps due to differing sexual selection pressures. Furthermore, the testis-specific expression of the paralogous copies and the retention of testis-specific expression in the inverted orthologous copies suggests that only short upstream and downstream regulatory regions may be required for testis expression in Drosophila (Sorourian et al. 2014).
We relied initially on various types of alignment search tools to identify saturn and goddard orthologs, as these methods are commonly used in studies of de novo evolved genes (Neme and Tautz 2013; McLysaght and Hurst 2016). However, a recent study has demonstrated that, particularly for rapidly evolving genes, orthologs in distantly related species are sometimes not detected by the BLAST algorithm (Moyers and Zhang 2015). We thus supplemented BLAST-based analyses with the concurrent examination of syntenic regions. These efforts identified potential ORFs in D. virilis, mojavensis, and grimshawi that likely represent unannotated copies of goddard. We believe the most parsimonious explanation of these ORFs is that they are true goddard orthologs, but experienced extreme divergence after goddard’s likely de novo birth at the base of the Drosophila genus. The weak sequence identity detected between the translation of the D. mojavensis ORF and goddard in D. melanogaster, the conserved location in the omega intron, and the male- and testis-enriched expression of both ORFs, are consistent with this interpretation. An alternative, but less parsimonious, explanation would be two independent gene births in the same location.
We also note that it is possible that one or both of these genes may be present outside of the Drosophila genus and are simply undetectable by available methods. Likewise, it is possible these genes arose through horizontal gene transfer followed by divergence, rather than from noncoding sequence, since this is another source for newly evolved genes (Chen et al. 2013). However, no available evidence currently supports either hypothesis, and the proteins’ biophysical properties are consistent with de novo origins. Additionally, independent of their exact emergence mechanism, goddard and saturn represent two novel genes in the sense that no other genes with similar sequence exist. As such, the genes represent major evolutionary innovations that raise many of the same questions as de novo gene emergence in general.
An additional feature of both goddard and saturn is their genomic locations within large introns of well-conserved genes. One intriguing possibility raised by our results is that large introns of existing genes may represent hot spots for the birth of de novo genes, but this question should be addressed through a more systematic analysis of all putative de novo genes.
Putative de novo genes encode lineage-specific, male reproductive proteins in several Drosophila species (Begun et al. 2006; Levine et al. 2006; Findlay et al. 2009). RNA sequencing from the testes of several strains of the Drosophila Genetic Reference Panel suggests that de novo genes are born at a high rate in Drosophila and that numerous such genes are segregating in a current population (Zhao et al. 2014). Comparative data from the obscura group (Palmieri et al. 2014) show that many novel genes are expressed in the male reproductive system, and such expression correlates with their retention. These data, combined with our functional analyses presented here, suggest that de novo gene evolution may be an important mechanism that underlies the rapid evolution of male reproductive traits. The continued study of de novo genes is likely to yield insight into the genetic changes that underlie male evolutionary responses to interspecific differences in the structures and molecules of the female reproductive tract (e.g., Pitnick et al. 1999; Bono et al. 2011), changes in seminal fluid protein content (Findlay et al. 2008; Kelleher et al. 2009) and differences in sperm competition levels (Markow and O'Grady 2005).
Materials and Methods
A Preliminary Screen for De Novo Genes with Major Effects on Male Fertility
To rapidly screen for potential de novo genes with major effects on male fertility, we identified 11 genes with testis-enriched expression patterns (Chintapalli et al. 2007). All selected genes lacked identifiable protein domains and had either been denoted in a previous bioinformatic study as de novo (Zhang et al. 2010) or had, at the time of gene selection, patterns of annotated orthologs in FlyBase that suggested lineage-restriction. We obtained RNAi lines for these genes from the Vienna Drosophila RNAi Center and the Harvard Transgenic RNAi Project (TRiP) (see supplementary fig. S1, Supplementary Material online for the specific lines used) and used the Bam-GAL4 driver to induce KD in the testes. All lines reported here showed at least partial KD by RT-PCR as described below. To assess male fertility, we paired seven KD or control males with five Canton S females for 24 h. Males were then removed, and females were returned to the same vial for 72 additional hours of egg-laying. Females were then discarded, and the resulting progeny were counted. We included two replicate vials for each gene and report the mean number of progeny for each gene as a proportion of the progeny counts for genetically matched control males assayed simultaneously. Clearly, this screen is a blunt instrument for identifying genes with the most major effects on fertility; several other genes appeared to cause slightly attenuated fertility (supplementary fig. S1, Supplementary Material online), but these effects require more careful screening and quantification.
Flies and RNA Interference for Goddard and Saturn
We used first (GD)- and second (KK)-generation RNAi lines from the Vienna Drosophila RNAi Center (VDRC) to knock down goddard (CG13477) and saturn (CG32141) expression in the testes. We used GD stocks 38677 (goddard) and 41107 and 41108 (saturn), and KK stocks 109920 (goddard) and 105447 (saturn). GD stocks contain P-element inserted UAS-RNAi sequences at random genomic locations, while KK stocks contain distinct UAS-RNAi sequences inserted at an AttP site on chromosome II. For saturn, the GD and KK lines targeted nonoverlapping regions of the gene’s coding sequence, while the targeted regions were mostly overlapping for goddard (due to the gene’s short length). All lines were crossed to Bam-GAL4, UAS-Dicer2 to create KD flies. Control flies were generated by crossing the chromosome II AttP line without an RNAi insert (VDRC stock 60100) to Bam-GAL4, UAS-Dicer2.
The saturn KK stock #105447 has two predicted off-targets: Spps (CG5669) and Lrr47 (CG6098). FlyAtlas data (Chintapalli et al. 2007) showed that these genes are expressed in the testes (among other locations). However, we ruled out these genes as causing any part of the saturn phenotypes described here. First, RT-PCR of these transcripts in cDNA isolated from saturn KK KD males showed no reduction of Spps, though a partial reduction of Lrr47 was observed. Second, we obtained RNAi lines that specifically targeted each gene (GD stock 45300 for Spps, and KK stock 108096 for Lrr47), confirmed KD, and tested KD males for fertility defects with the Bam-GAL4 driver as above (data not shown). No defects were found. While we conducted fertility assays on each goddard and saturn RNAi line, we selected one line (that showed strong KD and lacked off-targets) for further functional experiments: Line 41108 for saturn and line 109920 for goddard.
Fly stocks were reared at room temperature with ambient lighting, while fly crosses and their progeny were maintained in 25° incubators with 12–12 h light–dark cycles. Flies were raised on standard molasses-cornmeal-yeast food with fresh yeast ad lib. Other species of flies were obtained from the Drosophila Species Stock Center (genome reference strains) and H. Malik.
Confirmation of RNAi KD
We evaluated KD using previously described procedures (Findlay et al. 2014). Briefly, we isolated RNA from whole knockdown and control males using TRIzol (Life Technologies), performed RQ1 DNase treatment (Promega), and synthesized cDNA using the SmartScribe kit (Clontech) and oligo-dT primers (Eurofins MWG Operon). This cDNA was then used in GoTaq (Promega) PCR to assay for gene expression, while substituting genomic DNA or water for template cDNA as controls. Amplification of RpL32 served as a KD control. The amount of RNA used for DNase treatment and cDNA synthesis was standardized between samples by quantification on a Nanodrop spectrophotometer (Thermo). PCR primers were designed to cause a size difference between cDNA and genomic DNA products when possible; primers and cycling conditions are available upon request. KD and control cDNA were then used in PCR reactions to qualitatively evaluate the degree of KD, while substituting genomic DNA or water for template cDNA as controls (supplementary fig. S4, Supplementary Material online). To facilitate comparisons of expression levels, equal amounts of KD and control cDNA were used in each reaction.
Fertility Assays
We measured male fertility by mating single pairs of KD or control males and wild-type, Canton S females. Each sex was 3–5 days old at mating. Males were removed immediately after the mating, and females were allowed to lay eggs for the next four days and then discarded. Progeny were counted by counting the number of pupal cases on the sides of the vials once all progeny had reached the pupal stage.
Sperm Motility
We evaluated sperm motility qualitatively in saturn KD and control males by performing testis squashes. Testes from 3- to 5-day old males were dissected into cold testis buffer (White-Cooper 2004), gently torn open with dissecting forceps, and squashed under the weight of a cover slip. Sperm motility was observed under phase contrast with a Leica SP5 microscope and images were taken using the LASAF program. An average of three images were taken every 6.45 s and were made into movies using the xyt setting in LASAF. In this qualitative assay, motility was evaluated by eye.
Sperm Production, Sperm Transfer, and Sperm Localization
To produce KD males in which mature sperm had GFP-labeled nuclei, we crossed UAS-RNAi lines (KK 109920 for goddard, GD 41108 for saturn) and the AttP control line to Bam-GAL4, UAS-Dicer2 females that also carried Mst35Bb-GFP (Manier et al. 2010). This latter construct allowed the nuclei of late-stage spermatids and mature sperm to be visualized by fluorescence confocal microscopy on the Leica SP5 microscope. Images were captured using the LASAF program, using a Z-stack height of 1–1.5 μm and 1,024 × 1,024 pixel resolution. Z-stack images were formatted with ImageJ, where they were compressed into a flattened image consisting of the maximum intensity of all images in the stack. The sperm cells on the flattened image were counted using the Cell Counter feature on ImageJ, utilizing color-coded tallying to prevent under and double counting.
To evaluate sperm production by saturn KD males, we dissected testes from 3- to 5-day-old KD and control males that had been aged in single-sex vials since their collection on day 0. We then counted the number of sperm in each seminal vesicle (SV). Because a few SVs were damaged during dissection, we counted on each slide the number of sperm per intact SV and report the average number of sperm per SV in each male. To evaluate sperm transfer to females, 3- to 5-day-old KD and control males were single-pair mated to Canton S females. Matings were observed, and females were flash frozen in liquid nitrogen 30 min ASM. Female reproductive tracts were then dissected, and we counted all sperm observed in the bursa, as well as any in the oviduct and the sperm storage organs. To evaluate the entry of sperm into storage and their persistence there, we performed similar single-pair mating experiments but instead froze females at 60 or 120 min ASM. Female reproductive tracts were dissected, and all sperm present in the SR (the primary sperm storage organ in D. melanogaster at this time point; Manier et al. 2010) were counted. Counting was done by hand in ImageJ as described above.
To compare sperm production and sperm in females at each time point between mates of saturn KD and control males, we performed two-sample Welch’s t-tests with unequal variances. The results were equivalent when evaluated with nonparametric Mann–Whitney U tests.
Identification of Orthologs and Paralogs and Sequence Alignment
We identified orthologs of the goddard and saturn proteins from the sets of annotated proteins from the original 12 sequenced Drosophila species (Clark et al. 2007) by using precomputed gene group annotations (Attrill et al. 2016) and BLASTP searches. To identify unannotated orthologs in these species, and orthologs in additional species for which protein annotations are not yet available on FlyBase, we used a combination of genome-wide tblastn searches and LASTZ- and Exonerate-based analyses of the syntenic regions (Slater and Birney 2005; Harris 2007). Since both saturn and goddard reside within a large intron of a conserved gene (nuclear fallout and omega, respectively), these genes were used to identify syntenic regions.
Orthologous and paralogous protein sequences were aligned with the T-COFFEE algorithm (Notredame et al. 2000). Alignments were visualized in MEGA 6 (Tamura et al. 2013) and refined by eye. DNA alignments of protein-coding sequences were obtained by back-translating the protein alignments using PAL2NAL (Suyama et al. 2006). Phylogenetic trees were constructed with RAxML version 8 using the Gtrcat model of evolution (Stamatakis 2014). To resolve the timing of the saturn gene duplication event, we also constructed a protein tree using the orthologs and paralogs found in the original set of sequenced Drosophila species. For this tree, we used only the N-terminal portion of the saturn protein (amino acids 1–105 in D. melanogaster), since the C-terminal portion could not be confidently aligned. We obtained equivalent results when analyzing the data with the programs Muscle version 6 (Edgar 2004) and PHYLIP version 3.695 (Felsentein 2005).
Bioinformatic Analysis of Saturn and Goddard Proteins
We used the IUPred short algorithm to predict intrinsic disorder in the D. melanogaster protein sequences based on the frequency of disorder-promoting amino acids (Dosztanyi et al. 2005). As a measurement of folding potential, we employed Seq-HCA, which analyzes clusters of hydrophobic amino acids (Faure and Callebaut 2013). We compared predicted hydrophobic clusters in saturn and goddard with a database of hydrophobic clusters from well-established globular proteins to assess possible differences in folding patterns, as previously described (Bitard-Feildel et al. 2015). We also calculated the hexamer score for goddard and saturn using CPAT (Wang et al. 2013). Hexamer score was developed to distinguish coding from noncoding sequences (Fickett and Tung 1992). Accordingly, a difference in hexamer score between saturn and goddard and other protein coding genes would support the evolution of these genes from noncoding sequence. To make such comparisons, we also calculated the hexamer score, disorder content, and HCA for all D. melanogaster protein-coding genes as annotated in FlyBase (release 6.12), using only the longest isoform of each gene. We used the JPred4 web server for secondary structure prediction (Drozdetskiy et al. 2015) and TMHMM version 2.0c (Krogh et al. 2001) to predict potential transmembrane domains.
Tests for Adaptive Evolution
We used the back-translated cDNA alignments and phylogenetic trees to test for adaptive evolution using the codeml program in the PAML package (Yang 2007). For each gene, we tested for selection in two groups: The melanogaster group, which contained 16 species for goddard and 15 species for saturn (the latter due to difficulty in aligning the D. kikkawai ortholog), and a nested set of species, the melanogaster subgroup, which contained six species. Within each group, we estimated the whole-gene dN/dS ratio (ω) using model M0 and tested for specific sites under selection using the PAML sites tests that compared model M8, which allows a class of positively selected sites, to null models M7 and M8a, which do not allow for positive selection (Yang et al. 2000; Swanson et al. 2003). If Model M8 was a significantly better fit to the data than the null models, Bayes Empirical Bayes (BEB) analysis was used to infer sites likely to be under positive selection. Finally, when testing saturn, we could confidently align the full-length protein in only the melanogaster subgroup, due to rapid divergence and indel variation in the C terminus. Therefore, we also partitioned the sequence alignment after amino acid position 105 in D. melanogaster and tested the N-terminus only in both the melanogaster subgroup and group, and the C-terminus only in just the melanogaster subgroup. The melanogaster subgroup species were D. melanogaster, D. simulans, D. sechellia, D. mauritiana, D. yakuba, and D. erecta. The melanogaster group species included those listed above, as well as D. rhopaloa, D. elegans, D. ficusphila, D. biarmipes, D. suzukii, D. eugracilis, D. takahashii, D. kikkawai, D. bipectinata, and D. ananassae.
Gene Expression
We tested for conservation of gene expression patterns of the identified orthologs and paralogs in a variety of species using previously described methods (Findlay et al. 2014; Sirot et al. 2014). We tested for expression of both genes in D. ananassae and D. ficusphila, as divergent members of the melanogaster group. Additionally, we tested goddard expression in D. pseudoobscura, and saturn expression in D. virilis and D. mojavensis. RNA was isolated, and cDNA synthesized, from whole males and whole females as described above.
To determine whether saturn paralogs in D. virilis and D. mojavensis showed conservation of testis-specific expression, we dissected testes from eight to nine males of each species and used the testes-lacking carcasses for RNA isolation and RT-PCR as above. We ensured that other dissected male reproductive organs (e.g., accessory glands, ejaculatory bulbs, and ducts) were included with the carcasses. We used the same samples to test the expression of the D. virilis and D. mojavensis ORFs found in the goddard syntenic region.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation (CAREER award #1652013 to G.D.F.), the National Institutes of Health (F32-GM097789 to G.D.F., R01-HD038921 to M.F.W.), the Human Frontiers Science Program (RGP0006/2013 to E.B.B.), and internal funding from the College of the Holy Cross to A.M.G. and G.D.F. We thank Meaghan McGeary, Rob Bellin, Karen Ober, and Willie Swanson for technical assistance; Jainy Thomas for additional bioinformatic analysis; Harmit Malik, John Belote, the Vienna Drosophila RNAi Center, the Bloomington Stock Center and the Drosophila Species Stock Center for fly lines; and, Purva Rumde, Michelle Mondoux, Dustin Rubinstein, Chip Aquadro, Cedric Feschotte, Barbara Wakimoto, and members of the Findlay Lab for valuable discussions and feedback.
References
- Abrusan G. 2013. Integration of new genes into cellular networks, and their structural maturation. Genetics 195:1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attrill H, Falls K, Goodman JL, Millburn GH, Antonazzo G, Rey AJ, Marygold SJ, FlyBase C.. 2016. FlyBase: establishing a Gene Group resource for Drosophila melanogaster. Nucleic Acids Res. 44:D786–D792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila FW, Wolfner MF.. 2009. Acp36DE is required for uterine conformational changes in mated Drosophila females. Proc Natl Acad Sci U S A. 106:15796–15800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Lindfors HA, Thompson ME, Holloway AK.. 2006. Recently evolved genes identified from Drosophila yakuba and Drosophila erecta accessory gland expressed sequence tags. Genetics 172:1675–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitard-Feildel T, Heberlein M, Bornberg-Bauer E, Callebaut I.. 2015. Detection of orphan domains in Drosophila using "hydrophobic cluster analysis". Biochimie 119:244–253. [DOI] [PubMed] [Google Scholar]
- Bono JM, Matzkin LM, Kelleher ES, Markow TA.. 2011. Postmating transcriptional changes in reproductive tracts of con- and heterospecifically mated Drosophila mojavensis females. Proc Natl Acad Sci U S A. 108:7878–7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JB, Boley N, Eisman R, May GE, Stoiber MH, Duff MO, Booth BW, Wen JY, Park S, Suzuki AM, et al. 2014. Diversity and dynamics of the Drosophila transcriptome. Nature 512:393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, et al. 2012. Proto-genes and de novo gene birth. Nature 487:370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SD, Krinsky BH, Long MY.. 2013. New genes as drivers of phenotypic evolution. Nat Rev Genet. 14:645–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SD, Zhang YE, Long MY.. 2010. New genes in Drosophila quickly become essential. Science 330:1682–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z-X, Sturgill D, Qu J, Jiang H, Park S, Boley N, Suzuki AM, Fletcher AR, Plachetzki DC, FitzGerald PC, et al. 2014. Comparative validation of the D. melanogaster modENCODE transcriptome annotation. Genome Res. 24:1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JAT.. 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 39:715–720. [DOI] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, et al. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450:203–218. [DOI] [PubMed] [Google Scholar]
- Cui X, Lv Y, Chen ML, Nikoloski Z, Twell D, Zhang DB.. 2015. Young genes out of the male: an insight from evolutionary age analysis of the pollen transcriptome. Mol Plant 8:935–945. [DOI] [PubMed] [Google Scholar]
- Ding Y, Zhao L, Yang SA, Jiang Y, Chen YA, Zhao RP, Zhang Y, Zhang GJ, Dong Y, Yu HJ, et al. 2010. A young Drosophila duplicate gene plays essential roles in spermatogenesis by regulating several Y- linked male fertility genes. PLoS Genet. 6:e1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Loso T, Tautz D.. 2003. An evolutionary analysis of orphan genes in Drosophila. Genome Res. 13:2213–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorus S, Freeman ZN, Parker ER, Heath BD, Karr TL.. 2008. Recent origins of sperm genes in Drosophila. Mol Biol Evol. 25:2157–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosztanyi Z, Csizmok V, Tompa P, Simon I.. 2005. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21:3433–3434. [DOI] [PubMed] [Google Scholar]
- Drozdetskiy A, Cole C, Procter J, Barton GJ.. 2015. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 43:W389–W394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure G, Callebaut I.. 2013. Comprehensive repertoire of foldable regions within whole genomes. Plos Computational Biology 9:e1003280.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsentein J. 2005. PHYLIP - Phylogenetic Inference Package, version 3.6. Version 3.6. Seattle: Department of Genome Sciences, University of Washington. [Google Scholar]
- Fickett JW, Tung CS.. 1992. Assessment of protein coding measures. Nucleic Acids Res. 20:6441–6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GD, MacCoss MJ, Swanson WJ.. 2009. Proteomic discovery of previously unannotated, rapidly evolving seminal fluid genes in Drosophila. Genome Res. 19:886–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GD, Sitnik JL, Wang W, Aquadro CF, Clark NL, Wolfner MF.. 2014. Evolutionary rate covariation identifies new members of a protein network required for Drosophila melanogaster female post-mating responses. PLoS Genet. 10:e1004108.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GD, Yi X, MacCoss MJ, Swanson WJ.. 2008. Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. PLoS Biol. 6:1417–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EW, Fedele G, Giorgini F, Kyriacou CP.. 2014. A Drosophila RNAi collection is subject to dominant phenotypic effects. Nat Methods 11:222–223. [DOI] [PubMed] [Google Scholar]
- Guerzoni D, McLysaght A.. 2011. De novo origins of human genes. PLoS Genet. 7:e1002381.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS. 2007. Improved pairwise alignment of genomic DNA. State College (PA: ): ProQuest. [Google Scholar]
- Heinen T, Staubach F, Haming D, Tautz D.. 2009. Emergence of a new gene from an intergenic region. Curr Biol. 19:1527–1531. [DOI] [PubMed] [Google Scholar]
- Kaessmann H. 2010. Origins, evolution, and phenotypic impact of new genes. Genome Res. 20:1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser VB, Bachtrog D.. 2010. Evolution of sex chromosomes in insects. Annu Rev Genet. 44:91–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn RC, Clark NL, Nguyen ED, Swanson WJ.. 2008. Adaptive evolution in rodent seminal vesicle secretion proteins. Mol Biol Evol. 25:2301–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher ES, Watts TD, LaFlamme BA, Haynes PA, Markow TA.. 2009. Proteomic analysis of Drosophila mojavensis male accessory glands suggests novel classes of seminal fluid proteins. Insect Biochem Mol Biol. 39:366–371. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE.. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protocols 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW.. 2006. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 23:1891–1901. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305:567–580. [DOI] [PubMed] [Google Scholar]
- Levine MT, Jones CD, Kern AD, Lindfors HA, Begun DJ.. 2006. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc Natl Acad Sci U S A. 103:9935–9939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loppin B, Lepetit D, Dorus S, Couble P, Karr TL.. 2005. Origin and neofunctionalization of a Drosophila paternal effect gene essential for zygote viability. Curr Biol. 15:87–93. [DOI] [PubMed] [Google Scholar]
- Manier MK, Belote JM, Berben KS, Novikov D, Stuart WT, Pitnick S.. 2010. Resolving mechanisms of competitive fertilization success in Drosophila melanogaster. Science 328:354–357. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu SN, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res. 43:D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow TA, O'Grady PM.. 2005. Evolutionary genetics of reproductive behavior in Drosophila: connecting the dots. Annu Rev Genet. 39:263–291. [DOI] [PubMed] [Google Scholar]
- McLysaght A, Guerzoni D.. 2015. New genes from non-coding sequence: the role of de novo protein-coding genes in eukaryotic evolutionary innovation. Philosophical Transactions of the Royal Society B-Biological Sciences 370:20140332.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght A, Hurst LD.. 2016. Open questions in the study of de novo genes: what, how and why. Nat Rev Genet. 17:567–578. [DOI] [PubMed] [Google Scholar]
- Meslin C, Mugnier S, Callebaut I, Laurin M, Pascal G, Poupon A, Goudet G, Monget P.. 2012. Evolution of genes involved in gamete interaction: evidence for positive Selection, duplications and losses in vertebrates. PLoS One 7:e44548.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metta M, Schlotterer C.. 2008. Male-biased genes are overrepresented among novel Drosophila pseudoobscura sex-biased genes. BMC Evol Biol. 8:182.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyers BA, Zhang JZ.. 2015. Phylostratigraphic bias creates spurious patterns of genome evolution. Mol Biol Evol. 32:258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neme R, Tautz D.. 2016. Fast turnover of genome transcription across evolutionary time exposes entire non-coding DNA to de novo gene emergence. Elife 5:e09977.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neme R, Tautz D.. 2013. Phylogenetic patterns of emergence of new genes support a model of frequent de novo evolution. BMC Genomics 14:117.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J.. 2000. T-coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 302:205–217. [DOI] [PubMed] [Google Scholar]
- Nurminsky DI, Nurminskaya MV, De Aguiar D, Hartl DL.. 1998. Selective sweep of a newly evolved sperm-specific gene in Drosophila. Nature 396:572–575. [DOI] [PubMed] [Google Scholar]
- Obbard DJ, Maclennan J, Kim KW, Rambaut A, O'Grady PM, Jiggins FM.. 2012. Estimating divergence dates and substitution rates in the Drosophila phylogeny. Mol Biol Evol. 29:3459–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri N, Kosiol C, Schlotterer C.. 2014. The life cycle of Drosophila orphan genes. Elife 3:e01311.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanicolaou A, Schetelig MF, Arensburger P, Atkinson PW, Benoit JB, Bourtzis K, Castanera P, Cavanaugh JP, Chao H, Childers C, et al. 2016. The whole genome sequence of the Mediterranean fruit fly, Ceratitis capitata (Wiedemann), reveals insights into the biology and adaptive evolution of a highly invasive pest species. Genome Biol. 17:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitnick S, Markow T, Spicer GS.. 1999. Evolution of multiple kinds of female sperm-storage organs in Drosophila. Evolution 53:1804–1822. [DOI] [PubMed] [Google Scholar]
- Raja SJ, Renkawitz-Pohl R.. 2005. Replacement by Drosophila melanogaster protamines and Mst77F of histones during chromatin condensation in late spermatids and role of sesame in the removal of these proteins from the male pronucleus. Mol Cell Biol. 25:6165–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramm SA, Scharer L, Ehmcke J, Wistuba J.. 2014. Sperm competition and the evolution of spermatogenesis. Mol Hum Reprod. 20:1169–1179. [DOI] [PubMed] [Google Scholar]
- Rathke C, Baarends WM, Jayaramaiah-Raja S, Bartkuhn M, Renkawitz R, Renkawitz-Pohl R.. 2007. Transition from a nucleosome-based to a protamine-based chromatin configuration during spermiogenesis in Drosophila. J Cell Sci. 120:1689–1700. [DOI] [PubMed] [Google Scholar]
- Reinhardt JA, Wanjiru BM, Brant AT, Saelao P, Begun DJ, Jones CD.. 2013. De novo ORFs in Drosophila are important to organismal fitness and evolved rapidly from previously non-coding sequences. PLoS Genet. 9:e1003860.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartain CV, Cui J, Meisel RP, Wolfner MF.. 2011. The poly(A) polymerase GLD2 is required for spermatogenesis in Drosophila melanogaster. Development 138:1619–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudan P, Hauck K, Soller M, Choffat Y, Ottiger M, Sporri M, Ding ZB, Hess D, Gehrig PM, Klauser S, et al. 2002. Ductus ejaculatorius peptide 99B (DUP99B), a novel Drosophila melanogaster sex-peptide pheromone. Eur J Biochem. 269:989–997. [DOI] [PubMed] [Google Scholar]
- Scharer L, Da Lage JL, Joly D.. 2008. Evolution of testicular architecture in the Drosophilidae: a role for sperm length. BMC Evol Biol. 8:143.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlotterer C. 2015. Genes from scratch - the evolutionary fate of de novo genes. Trends Genet. 31:215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira AB, Trontin C, Cortijo S, Barau J, Del Bem LEV, Loudet O, Colot V, Vincentz M.. 2013. Extensive natural epigenetic variation at a de novo originated gene. PLoS Genet. 9:e1003437.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirot LK, Findlay GD, Sitnik JL, Frasheri D, Avila FW, Wolfner MF.. 2014. Molecular characterization and evolution of a gene family encoding both female- and male-specific reproductive proteins in Drosophila. Mol Biol Evol. 31:1554–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater GS, Birney E.. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinform. 6:31.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snook RR, Karr TL.. 1998. Only long sperm are fertilization-competent in six sperm-heteromorphic Drosophila species. Curr Biol. 8:291–294. [DOI] [PubMed] [Google Scholar]
- Sorourian M, Kunte MM, Domingues S, Gallach M, Ozdil F, Rio J, Betran E.. 2014. Relocation facilitates the acquisition of short cis-regulatory regions that drive the expression of retrogenes during spermatogenesis in Drosophila. Mol Biol Evol. 31:2170–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P.. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34:W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Nielsen R, Yang QF.. 2003. Pervasive adaptive evolution in mammalian fertilization proteins. Mol Biol Evol. 20:18–20. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telonis-Scott M, Kopp A, Wayne ML, Nuzhdin SV, McIntyre LM.. 2009. Sex-specific splicing in Drosophila: widespread occurrence, tissue specificity and evolutionary conservation. Genetics 181:421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacquier VD, Swanson WJ, Lee YH.. 1997. Positive Darwinian selection on two homologous fertilization proteins: what is the selective pressure driving their divergence? J Mol Evol. 44:S15–S22. [DOI] [PubMed] [Google Scholar]
- Vibranovski MD, Lopes HF, Karr TL, Long MY.. 2009. Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet. 5:e1000731.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers JHA, Manning SA, Kulkarni A, Harvey KF.. 2016. A Drosophila RNAi library modulates Hippo pathway-dependent tissue growth. Nat Commun. 7:10368.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff BJ, Begun DJ.. 2007. Adaptive evolution of recently duplicated accessory gland protein genes in desert Drosophila. Genetics 177:1023–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff BJ, Begun DJ.. 2005. Comparative genomics of accessory gland protein genes in Drosophila melanogaster and D. pseudoobscura. Mol Biol Evol. 22:818–832. [DOI] [PubMed] [Google Scholar]
- Wakimoto BT, Lindsley DL, Herrera C.. 2004. Toward a comprehensive genetic analysis of male fertility in Drosophila melanogaster. Genetics 167:207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Park HJ, Dasari S, Wang SQ, Kocher JP, Li W.. 2013. CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 41:e74.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasbrough ER, Dorus S, Hester S, Howard-Murkin J, Lilley K, Wilkin E, Polpitiya A, Petritis K, Karr TL.. 2010. The Drosophila melanogaster sperm proteome-II (DmSP-II). J Proteomics 73:2171–2185. [DOI] [PubMed] [Google Scholar]
- White-Cooper H. 2004. Spermatogenesis: analysis of meiosis and morphogenesis In: Henderson DS, editor. Methods in molecular biology. Clifton (NJ: ): Springer; p. 45–75. [DOI] [PubMed] [Google Scholar]
- White-Cooper H. 2012. Tissue, cell type and stage-specific ectopic gene expression and RNAi induction in the Drosophila testis. Spermatogenesis 2:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilburn DB, Swanson WJ.. 2016. From molecules to mating: rapid evolution and biochemical studies of reproductive proteins. J Proteomics 135:12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissler L, Gadau J, Simola DF, Helmkampf M, Bornberg-Bauer E.. 2013. Mechanisms and dynamics of orphan gene emergence in insect genomes. Genome Biol Evol. 5:439–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DD, Irwin DM, Zhang YP.. 2011. De novo origin of human protein-coding genes. PLoS Genet. 7:e1002379.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang ZH, Nielsen R, Goldman N, Pedersen AMK.. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda GK, Schubiger G, Wakimoto BT.. 1995. Genetic characterization of MS(3) K81, a paternal effect gene of Drosophila melanogaster. Genetics 140:219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh SD, Do T, Chan C, Cordova A, Carranza F, Yamamoto EA, Abbassi M, Gandasetiawan KA, Librado P, Damia E, et al. 2012. Functional evidence that a recently evolved Drosophila sperm-specific gene boosts sperm competition. Proc Natl Acad Sci U S A. 109:2043–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE, Vibranovski MD, Krinsky BH, Long MY.. 2010. Age-dependent chromosomal distribution of male-biased genes in Drosophila. Genome Res. 20:1526–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Saelao P, Jones CD, Begun DJ.. 2014. Origin and spread of de novo genes in Drosophila melanogaster populations. Science 343:769–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Zhang GJ, Zhang Y, Xu SY, Zhao RP, Zhan ZB, Li X, Ding Y, Yang SA, Wang W.. 2008. On the origin of new genes in Drosophila. Genome Res. 18:1446–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.