Abstract
Lipids are essential components of the brain. Here, we conducted a comprehensive mass spectrometry-based analysis of lipidome composition in the prefrontal cortex of 40 humans, 40 chimpanzees, and 40 rhesus monkeys over postnatal development and adulthood. Of the 11,772 quantified lipid peaks, 7,589 change significantly along the lifespan. More than 60% of these changes occur prior to adulthood, with less than a quarter associated with myelination progression. Evolutionarily, 36% of the age-dependent lipids exhibit concentration profiles distinct to one of the three species; 488 (18%) of them were unique to humans. In both humans and chimpanzees, the greatest extent of species-specific differences occurs in early development. Human-specific lipidome differences, however, persist over most of the lifespan and reach their peak from 20 to 35 years of age, when compared with chimpanzee-specific ones.
Keywords: lipidome, brain, development, evolution
Introduction
Human cognitive abilities are unique, suggesting the evolution of novel molecular features in brain organization and function. Presence of such uniquely human features has been investigated at different levels of phenotype, from single cell genomics and epigenetic DNA and chromatin modifications to construction of cerebral connectome maps (Zeng et al. 2012; Li et al. 2013; Darmanis et al. 2015; Vermunt et al. 2016). At present, however, we still lack a comprehensive picture of the molecular mechanisms underlying human cognition. Distinctive cognitive abilities have emerged on the human evolutionary lineage within the past 6–8 million years, after the separation of the human and the chimpanzee/bonobo ancestors. Given the long generation time of humans and human ancestors, however, only a limited number of adaptive changes might be expected to have occurred during this short time interval, leading to striking cerebral enlargement, modifications of connectivity and changes in neurodevelopment (Somel et al. 2013).
Evolutionarily, small changes in developmental regulation can result in large phenotypic adaptations (Carroll 2000). The human brain might fit this expectation, as human cognitive abilities form and mature during ontogeny. Supporting this notion, previous investigations of synaptic gene expression changes in the human, chimpanzee, and macaque pre-frontal cortex (PFC) during postnatal development reveled an extension of the maturation period in humans (Liu et al. 2012). Similarly, neocortical myelination is prolonged in humans (Miller et al. 2012), suggesting possible mechanisms underlying the development of human cognitive uniqueness.
One of the levels of the human brain phenotype that is yet to be fully investigated is the lipidome. Lipids constitute the majority of the brain’s dry weight and are known to be important for neural function as structural and signaling molecules (Wenk 2005; Piomelli et al. 2007). Disruptions of the brain lipid metabolism have been linked to disorders including Alzheimer’s disease, Parkinson’s disease, depression and anxiety (Wenk 2005; Abbott et al. 2015; Muller et al. 2015). Furthermore, lipid concentration analysis conducted in three brain regions of adult humans, chimpanzees, and macaques revealed an acceleration of the brain lipidome composition changes on the human evolutionary lineage, especially in the neocortex (Bozek et al. 2015). At the same time, as indicated by biochemical analysis of the lipid classes’ abundance, it has been shown that the human brain lipidome composition is not static, but changes during development and aging (Rouser and Yamamoto 1968).
We conducted a systematic investigation of lipid concentration changes during postnatal PFC development and identified lipidome features unique to humans by measuring concentrations of >11,000 hydrophobic compounds in samples from humans, chimpanzees, and rhesus monkeys.
Results
Data Description
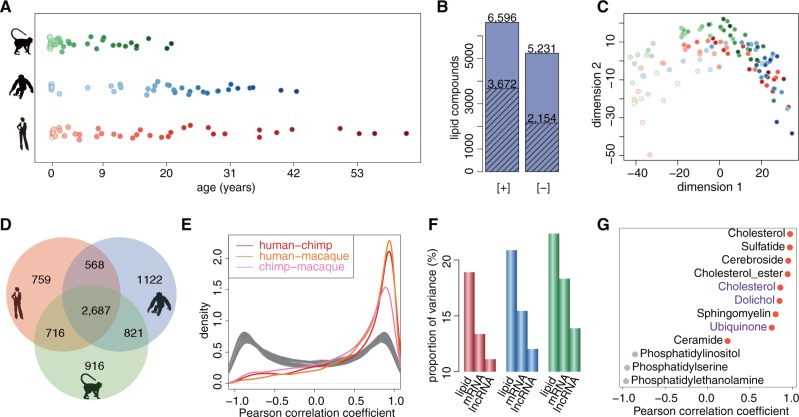
We measured concentrations of hydrophobic compounds in samples of the superior frontal gyrus of the dorsolateral PFC from 40 humans, 40 chimpanzees, and 40 rhesus monkeys of different ages (supplementary table S1, Supplementary Material online). The ages of individuals ranged from 2 days to 61 years in humans, from newborns to 42 years in chimpanzees, and from 14 weeks post-conception to 21 years in macaques, with a higher number of samples representing early development ( fig. 1A). The lipidome composition of all samples was measured using liquid chromatography coupled with mass spectrometry at positive ([+] LC–MS) and negative ([-] LC–MS) ionization modes. This resulted in 6,596 and 5,231 distinct MS features representing hydrophobic compounds (lipids) with molecular weights below 2,000 Da detected in at least 95% of all samples in positive and negative modes, respectively. Among them, 3,672 and 2,154 compounds were computationally annotated using probabilistic matching to the lipid compound database (fig. 1B;supplementary data S1, Supplementary Material online).
Fig. 1.
Lipidome variation. (A) Samples used for LC–MS measurements. Each point represents an individual (red—humans, blue—chimpanzees, green—macaques). Points filled with lighter shades of colors indicate younger ages. (B) Number of lipid peaks detected by LC–MS procedure in positive ([+]) or negative ([-]) ionization modes. Shaded bars indicate number of detected lipid peaks that were annotated based on probabilistic matching to the LIPID MAPS database and HMDB. (C) Multidimensional scaling (MDS) based on concentrations of all 11,772 quantified lipid peaks. Each point represents an individual (red—humans, blue—chimpanzees, green—macaques). Points filled with lighter shades of colors indicate younger ages. (D) Venn diagram showing the number of age-dependent lipid peaks detected in each species (red—humans, blue—chimpanzees, green—macaques). (E) The distributions of Pearson correlation coefficients between concentration profiles of the 7,589 age-dependent lipid peaks calculated in pairwise comparisons between species. Gray curves indicate distributions of Pearson correlation coefficients expected by chance, obtained by randomly matching the lipids between species 100 times. (F) Proportion of total variance explained by age for quantified lipids, mRNAs and lncRNAs in each species (red—humans, blue—chimpanzees, green—macaques). (G) Pearson correlation coefficients calculated for 12 lipid classes based on concentration measurements in human samples and concentrations reported for human (black) and rat (purple) brains in previous studies. Red points indicate positive correlation.
The PFC lipidome measurements were accompanied by transcriptome measurements conducted in a subset of PFC samples from 38 humans, 39 chimpanzees, and 40 rhesus monkeys using RNA sequencing (RNA-seq) on the Illumina platform (He et al. 2014) (supplementary table S1, Supplementary Material online). We quantified expression levels of 13,777 mRNAs and 420 long non-coding RNAs (lncRNA) detected in at least 95% of all samples.
In addition to the developmental data, we identified lipidome markers of myelination by measuring lipid concentrations in white matter samples dissected from corpus callosum and gray matter samples from the PFC of five adult humans (supplementary table S1, Supplementary Material online). Furthermore, we identified lipidome changes induced by postmortem delay (PMD) by examining lipidome measurements conducted in additional PFC samples of three rhesus monkeys (supplementary table S1, Supplementary Material online).
The analysis of lipidome composition in macaque PFC samples collected immediately and with PMD of 4–6 h revealed substantial concentration changes for 55 (0.5%) of the 11,827 lipids detected in the developmental dataset, which is similar to the proportion (1.5%) reported previously (Bozek et al. 2015). These compounds were excluded from further analyses.
The PFC Lipidome Undergoes Substantial Age-Dependent Change
Multidimensional scaling (MDS) based on concentrations of all 11,772 quantified MS features revealed clear effect of age in all three species (supplementary data S2, Supplementary Material online; fig. 1C). Consistent with this observation, 7,589 lipid peaks (64%) showed significant age-dependent concentration change in humans, chimpanzees, or macaques (F-test, permutations P < 0.001) (fig. 1D). The majority of these changes correlated positively and strongly among species (fig. 1E). Notably, in all three species, age-dependent lipidome changes exceeded changes in mRNA and lncRNA expression, in both the proportion of total variance explained by age and the percentage of significant age-dependent changes (fig. 1F;supplementary fig. S1, Supplementary Material online).
Lipid concentration changes identified in our data were compared with results of previous studies at the lipid class level: nine lipid classes were reported to change concentration over the lifespan in the human brain and three in the rat brain (Rouser and Yamamoto 1968; Zhang et al. 1996). Lipid concentration changes observed in our data positively correlated with previous observations for 9 of these 12 lipid classes, an agreement that exceeded by chance expectations (permutations, P = 0.04) (fig. 1G). Similarly, in comparison to GC–MS measurements conducted in human PFC across lifespan (Fu et al. 2011), 7 out of 8 lipids detected in both datasets correlated positively and significantly (supplementary table S2, Supplementary Material online).
Furthermore, age-dependent lipid concentration changes could be linked to the expression changes of corresponding enzymes. Of the 13,777 mRNAs expressed in human PFC over development, 667 were associated with 2,064 age-dependent lipids based on the human metabolome database (HMDB) annotation. The expression of these transcripts over the lifespan showed a stronger positive correlation with concentrations of their associated compounds than expected by chance (supplementary fig. S2, Supplementary Material online).
Accurate Lipid Age Markers
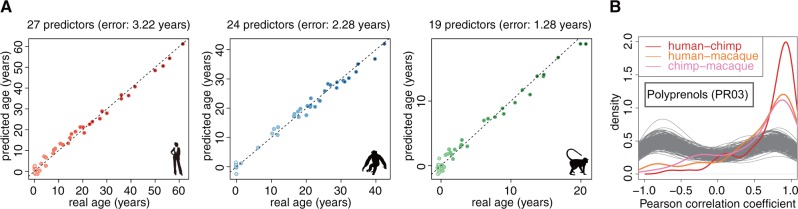
We next tested whether lipid concentrations could be used as predictors of individuals’ age in any of the three species. We identified 27 age predictor lipids in humans, 24 in chimpanzees, and 19 in rhesus monkeys using support vector regression (SVR) (supplementary table S3, Supplementary Material online). Strikingly, the error range of these predictions was under three and a half years for humans, under two and a half years for chimpanzees and under one and a half years for macaques (fig. 2A). To the best of our knowledge, this is one of the most accurate age predictions based on molecular or phenotypic markers obtained for humans (Horvath 2013; Chen et al. 2015).
Fig. 2.
Lipids as accurate age markers. (A) Real ages and predicted ages of samples in each species (red—humans, blue—chimpanzees, green—macaques). The predicted ages were obtained by 10-fold cross-validation based on the age predictors selected using SVR in each species. The numbers above each panel indicate the number of age predictors and the prediction error range calculated as two times the average of the absolute differences between real ages and predicted ages across all the age points. Points filled with lighter shades indicate younger ages. (B) The distributions of Pearson correlation coefficients between concentration profiles of the 65 age predictor lipid peaks calculated in pairwise comparisons between species. Gray curves indicate distributions of Pearson correlation coefficients expected by chance, obtained by randomly matching the lipids between species 100 times. The insert shows the lipid class enriched in the 65 age predictor lipids.
Despite only partial overlap of age predictors among species (supplementary fig. S3, Supplementary Material online), their concentration changes with age were well correlated. Taken together, these age predictors were enriched in the lipid class polyprenols (PR03) containing the known age markers dolichols (hypergeometric test, Bonferroni corrected P < 0.05) (fig. 2B).
Analysis of Lipidome Differences between Infant and Adult PFC
Changes in brain lipidome composition were not distributed uniformly over the lifespan. In human PFC, the yearly amplitude of lipidome changes of the 7,589 age-dependent lipids was ∼5 times greater during the first year of life compared with any year between 20 and 60 years of age (fig. 3A). Consequently, in the human PFC, >61% of all lipid concentration changes took place during the first 20 years of life. The same trend was observed for chimpanzees and macaques, when corrected for difference in the maximal lifespan of the species (supplementary fig. S4, Supplementary Material online).
Fig. 3.
Characteristics of age-dependent lipidome changes. (A) Yearly amplitude of lipidome changes calculated as the ratio of concentration changes occurring within a one-year interval to the concentration changes occurring across the lifespan in human brains based on concentrations of the 7,589 age-dependent lipids. The points represent the mean change amplitude. The area shows the variance of the change amplitude estimate (±1 SD). The lighter shades of colors indicate younger ages. The dark red numbers show the percentage of concentration changes occurring during the first 20 years of life and during 20–60 years interval. (B) Amplitude of cumulative concentration changes in adult brains relative to infant brains based on lipids of the corresponding lipid class in humans. The data are shown for the top 10 most abundant lipid classes jointly constituting 70% of the human brain lipidome and the remaining lipids (others). The numbers next to bars show percentage of the human brain lipidome contributed by the lipid class. (C) The proportion of white matter in the human samples estimated using quadratic programming and simulated annealing. The circles filled with lighter shades of color indicate younger ages. The slope and P-value were calculated using the linear regression model fitted to the white matter proportions.
On the basis of concentrations of 4,284 putatively annotated lipid peaks quantified in human PFC, the top 10 most abundant lipid classes cumulatively constitute 70% of the brain lipidome (fig. 3B). Between infant (0–1 years of human age) and adult (25–50 years of human age) brains, concentrations of lipids within these classes change by 7–14% (fig. 3B). Taking together the concentrations of all 7,589 age-dependent lipids in human PFC, the lipidome changes by 15% between infants and adults, with the largest change (20%) observed for the polyprenols (PR03) containing known age markers, dolichols. Similar proportions were observed for chimpanzee and macaque brains after correction for differences in the species’ maximal lifespan (supplementary fig. S5, Supplementary Material online). Overall, we detect 377 lipid peaks with large concentration changes between infant and adult brains (fold-change > 2, t-test, Benjamini–Hochberg (BH) corrected P < 0.05) in at least one of the three species. More than 84% of these changes correlated well between species (Pearson correlation, r > 0.6, P < 0.05), with 104 lipids showing high concentration in infant brains and 210—in adults of all three species (supplementary fig. S6 and table S4, Supplementary Material online).
Association between PFC Lipidome Composition and Myelination
Myelination is one of the major age-dependent processes potentially affecting brain lipidome composition. To assess the association between the age-dependent lipidome changes and myelination progression, we defined lipid myelination markers by comparing lipidome composition of cortical gray matter and white matter dissected from corpus callosum in five adult humans (supplementary table S1, Supplementary Material online). Among 1,900 lipid peaks detected in both developmental and white/gray matter datasets (supplementary data S3, Supplementary Material online), 15 had significantly and substantially higher concentrations in white matter (t-test, BH corrected P < 0.05, fold-change > 2; supplementary fig. S7, Supplementary Material online). Consistent with myelination progress with age, these myelination markers were enriched among lipids showing higher concentration in adult PFC of all three species (hypergeometric test, P = 0.06; supplementary fig. S8, Supplementary Material online). Notably, cerebrosides, which are known myelin markers (Jurevics et al. 2001), were similarly enriched in the adult brain (hypergeometric test, P = 0.0001; supplementary fig. S8, Supplementary Material online).
To determine the extent of age-dependent lipid concentration changes associated with myelination, we estimated the proportion of white matter in the developmental samples based on the relative concentrations of lipids in gray and white matter. The white matter proportion showed increased trend in all three species, mainly remaining under 20% (fig. 3C;supplementary fig. S9, Supplementary Material online), consistent with the sample dissection procedure preferentially targeting cortical gray matter. The white matter increase with age correlated either positively or negatively with concentration changes of ∼15% of 7,589 age-dependent lipids (Pearson correlation, > 0.6, P < 0.05), suggesting that only a minority of the detected age-dependent lipidome changes is associated with myelination.
Evolution of the PFC Lipidome Composition
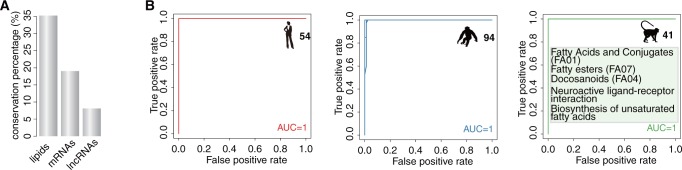
Among the three species, lipid abundance profiles show approximately 2-fold greater conservation compared with mRNA, and approximately 4-fold greater conservation compared with lncRNA (F-test, P < 0.01, FDR < 5%; fig. 4A).
Fig. 4.
PFC lipidome changes across species. (A) Percentage of age-dependent concentration changes conserved among species for lipids, mRNAs, and lncRNAs. (B) Receiver operating characteristic (ROC) curves obtained by 10 × 10 cross-validation based on optimal parameters from which the lipid predictors distinguishing one species from the other two derived using logistic regression with elastic net penalty. The numbers next to the species silhouette figures show the number of lipid predictors. The area under the receiver operating characteristic curve (AUC) was calculated as the average AUC across the 10 × 10 cross-validation. The inserts show lipid classes and KEGG pathways enriched in the lipid predictors.
Still, there are significant and apparent differences in the lipidome composition among species. Between humans and chimpanzees, 1,824 lipids showed significant concentration profile differences. These 1,824 lipids were enriched in specific lipid classes and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (hypergeometric test, Bonferroni corrected P < 0.05, supplementary table S5, Supplementary Material online). Furthermore, enzymes associated with these 1,824 lipids based on HMDB annotation tended to have more gene expression differences between humans and chimpanzees than the enzymes associated with the other detected age-dependent lipids (one-sided Wilcoxon test, P = 0.07, supplementary fig. S10, Supplementary Material online).
Consequently, each of the species’ identity can be identified with 100% accuracy based on the concentration levels of relatively small lipid groups using logistic regression with elastic net penalty and 10 × 10 cross-validation: 54 lipid peaks distinguish humans from the other two species, 94 lipid peaks distinguish chimpanzees, and 41 distinguish rhesus monkeys (fig. 4B;supplementary table S6, Supplementary Material online).
Previous work conducted in adult individuals indicated greater lipidome divergence on the human evolutionary lineage compared with the chimpanzee lineage (Bozek et al. 2015). Of the 251 lipid peaks showing significant concentration differences between adult humans and chimpanzees in developmental data (t-test, BH corrected P < 0.05), 85 could match lipid peaks quantified in the previous study (supplementary table S7, Supplementary Material online). The majority of these lipid peaks showed consistent concentration difference between species, as well as an approximate 2-fold excess of lipids showing human-specific concentration profiles compared with those showing chimpanzee-specific concentration profiles (fig. 5A).
Fig. 5.
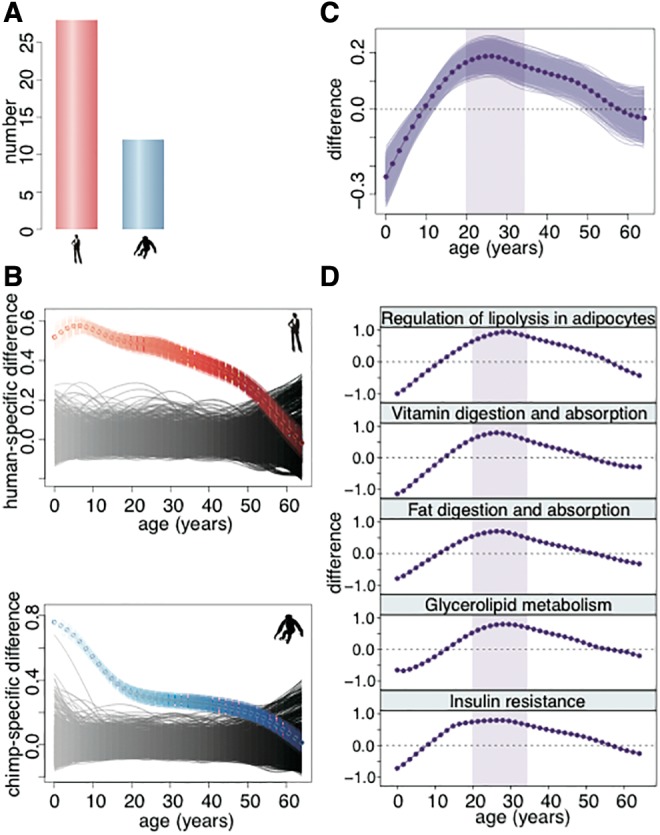
Human-specific lipidome features. (A) Numbers of lipids showing humans-specific (red) and chimpanzee-specific (blue) concentration profiles among the lipids showing significant difference between adult humans and chimpanzees in the current data and detected in the published dataset (Bozek et al. 2015). (B) The colored curves show the average human-specific difference and chimpanzee-specific difference along the lifespan calculated based on lipids showing human-specific (top, red) or chimpanzee-specific (bottom, blue) concentration profiles. The colored areas show the variation of the species-specific difference estimates calculated by bootstrapping the lipids showing human-specific or chimpanzee-specific concentration profiles 1,000 times. The black areas show the average species-specific difference expected by chance calculated based on 1,000 permutations of the species labels. (C) The dark purple curve shows difference between human-specific and chimpanzee-specific difference curves shown in panel B. The purple area shows the variation of the difference estimates calculated by bootstrapping the lipids showing human-specific and chimpanzee-specific concentration profiles 1,000 times. The shaded area shows the age interval with the greatest excess of human-specific difference. (D) The difference between human-specific and chimpanzee-specific difference curves calculated as in panel B based on lipids located within the pathways enriched in lipids showing human-specific profiles. The shaded area shows the 20–35 years age interval, as in panel C.
Notably, the human-specific and chimpanzee-specific lipidome differences, defined as the difference between humans and chimpanzees based on lipids showing human-specific concentration profiles or chimpanzee-specific ones, were not distributed uniformly across the lifespan: on each lineage the greatest extent of species-specific concentration differences took place during early postnatal development (supplementary data S4, Supplementary Material online; fig. 5B). Unexpectedly, the greatest excess of the human-specific concentration differences over chimpanzee-specific ones did not occur early childhood, but fell within the early adulthood period: from 20 to 35 years of human age (fig. 5C).
Lipids showing human-specific concentration profiles cluster in specific KEGG pathways: regulation of lipolysis in adipocytes, vitamin digestion and absorption, fat digestion and absorption, glycerolipid metabolism, and insulin resistance (hypergeometric test, Bonferroni corrected P < 0.05; supplementary table S8, Supplementary Material online). All these pathways show the largest excess of human-specific lipid concentration differences during early adulthood (fig. 5D). Notably, even though pathways identified using all 1,824 lipids showing significant concentration differences between humans and chimpanzees did not overlap with pathways identified using lipids showing human-specific concentration profiles only, they also showed greater changes on the human evolutionary lineage (human-specificity ratio > 1, supplementary table S5, Supplementary Material online).
Discussion
Previous biochemical studies demonstrating age-dependent concentration changes for several major lipid classes in brains of humans and rats showed that the lipidome is developmentally dynamic (Rouser and Yamamoto 1968; Zhang et al. 1996). Our study, employing LC–MS-based methods to examine concentrations of 11,772 hydrophobic compounds in PFC of humans, chimpanzees and macaques over postnatal lifespan, confirms and extends previous observations. It provides a more comprehensive view of lipidome changes with age and identifies its uniquely human features.
We show that lipid concentrations change substantially across lifespan in all three species, exceeding changes in mRNA and even lncRNA abundance. At the same time, lipid concentration profiles are twice as conserved among species compared with mRNA profiles and four times as conserved compared with lncRNA profiles. This indicates that extensive age-dependent lipidome changes taking place during postnatal development are not caused by lack of constraint, but are dictated by their functional roles in the three species.
Similar to mRNA and lncRNA (supplementary fig. S11, Supplementary Material online), the greatest extent of lipid concentration changes takes place during the first several years of life, potentially accompanying changes in the connectivity of the developing neocortex. Between 20 and 60 years of life, the rate of the lipidome changes in humans remains nearly constant and approximately six-times lower than the rate of change during the first year of life. Approximately 15% of the age-dependent lipidome changes could be related to the progression of axonal myelination, however, leaving the majority of the changes unexplained.
The substantial lipidome changes over lifespan allow for high accuracy of age predictions (±1.6 years for humans) based on concentrations of 27 lipids. This accuracy exceeds age prediction based on facial morphology, and is comparable to results obtained using DNA methylation levels (Horvath 2013; Chen et al. 2015). This suggests that lipids in other tissues, such as blood, might also be accurate indicators of age.
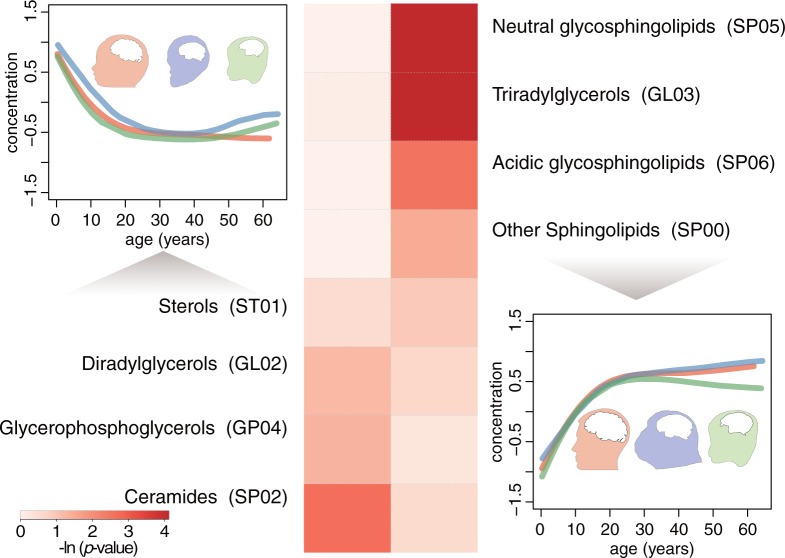
Cumulatively, the lipid concentrations change by ∼15% between the infant and adult human PFC. Lipids showing higher concentrations in the adult brains cluster in specific lipid classes, including: neutral glycosphingolipids (SP05), triradylglycerols (GL03) and acidic glycosphingolipids (SP06) (fig. 6). Glycosphingolipids together with other membrane components are involved in glycosynapse formation (Hakomori Si 2002). Glycosynapses are involved in cell adhesion and signaling and may form during axonal myelination (Boggs et al. 2004; Boggs 2014) reported to proceed in the human PFC up until the end of third decade (Miller et al. 2012). Lipids showing higher concentrations in the infant brains cluster in lipid classes including ceramides (SP02) (fig. 6), which are involved in cellular signaling including regulation of differentiation and apoptosis (Hannun and Obeid 2008).
Fig. 6.
Lipid classes preferentially present in the infant and adult PFC. The curves show the mean concentration profiles of lipids preferentially present in the infant (left) or adult (right) brains. The colors represent species (red—humans, blue—chimpanzees, green—macaques). Heatmap shows the hypergeometric test P-values of the lipid classes’ enrichment in the infant (left) and adult (right) brains.
Evolutionarily, 1,824 lipids show significant concentration profile differences between humans and chimpanzees and 3,324 between humans and macaques. The greatest species-specific concentration differences occur within the first 10 years of human age. Thus, the lipidome of humans and chimpanzees at one year of human age are approximately two-times more distinct from one another than at 40 years of human age (after correction of chimpanzee age to the difference in gestation time and maximal lifespan). The ratio of human-specific differences over the chimpanzee-specific ones, however, reaches the maximum not during early age, but in the early adulthood period: between 20 and 35 years of human age. Notably, all five functional pathways enriched in lipids showing human-specific concentration profiles exhibit greater human-specific difference over chimpanzee-specific one during the same period. This implies that these lipid concentration changes may reflect functional rearrangements of the PFC lipidome unique to humans.
Taken together, we show that the PFC lipidome composition changes substantially during human, chimpanzee, and macaque postnatal development. We also describe a number of uniquely human lipidome features. Although these observations might imply important roles of lipids in human brain development and evolution, they also highlight the need for further in-depth studies that include a greater range of brain structures to examine brain lipidome functionality.
Materials and Methods
Samples
Human samples were obtained from the NICHD Brain and Tissue Bank for Developmental Disorders and Maryland Population Research Center at the University of Maryland, USA and the Chinese Brain Bank Center (CBBC), Wuhan, China. Written consent for the use of human tissues for research was obtained from all donors or their next of kin. All subjects were defined as healthy controls by pathologists or neuropathologists at the corresponding brain bank. All subjects suffered sudden death with no prolonged agonal state. According to the protocol of the CBBC, use of human autopsy tissue is considered non-human subject research and is IRB exempt under NIH guidelines. Chimpanzee samples were obtained from the Alamogordo Primate Facility, NM, USA, Max Planck Institute for Evolutionary Anthropology in Leipzig, Germany, and Southwest National Primate Research Center in the Texas Biomedical Research Institute, USA. Rhesus monkey samples were obtained from the Suzhou Experimental Animal Center, China. All non-human primates used in this study suffered sudden deaths for reasons other than their participation in this study and without any relation to the tissue used. PFC dissections were made from the frontal part of the superior frontal gyrus. Special care was taken to preferentially dissect gray matter for all samples.
White/Gray Matter Samples
To identify lipidome markers of myelination, we collected five adult human samples, dissecting the corpus callosum and the pure subcortical gray matter from PFC.
Postmortem Samples
To identify lipids affected by PMD in primates, we collected three rhesus monkey samples dissected 4–6 h after death. Other macaque samples used in this study had PMDs lower than 20 min.
MS Sample Preparation and Measurements
Lipids were extracted from the frozen brain tissue powder by methyl-tert-butyl-ether (MTBE): methanol (3:1, v/v) (Khrameeva et al. 2014). In brief, ∼25 mg of frozen powdered tissue was re-suspended in 1ml MTBE:MeOH (3:1) solution containing the internal standard (1 µg/mL of 1,2-diheptadecanoyl-sn-glycero-3-phosphocholine (PC 34:0)). The samples were incubated for 10 min at 4 °C on an orbital shaker, before subjecting them to ultrasonication for 10 min in an ice-cooled bath-type sonicator. The insoluble tissue material (including proteins) was pelleted by a centrifugation step (5 min; 14,000 × g) and the supernatant was transferred to a fresh 2 ml Eppendorf tube. To separate the organic from the aqueous phase, 650 ml of a H2O:methanol mixture (3:1, v/v) was added to the supernatant, mixed by vortexing and centrifuged (5 min; 14,000 × g). Five hundred microliters of the upper-lipid-phase (MTBE) was transferred to a fresh 1.5-ml Eppendorf tube, concentrated in a speed vacuum and the pellet was re-suspended in 100 µl of an acetonitrile:isopropanol mixture (7:3, v/v) before liquid chromatography–mass spectrometry analysis. For the LC–MS analysis two times 3 µl of the re-suspended lipid extract were injected onto the ultra performance liquid chromatography C8-reversed phase column (BEH C8, Waters), connected to an Orbitrap Exactive mass spectrometer and analyzed once in positive and once in negative ion mode (Hummel et al. 2011). Data alignment and pre-processing was performed using the QI software (Version 2.2, www.nonlinear.com).
Normalization
The concentration values of all identified peaks were log2-transformed and quantile normalized across all developmental samples. Before log2 transformation, the concentration values were increased by one in order to avoid infinite values.
Identification of Lipids Affected by PMD
We identified lipids affected by PMD by comparing the concentrations from three rhesus monkey samples collected with substantial PMD with those from the other macaque samples without substantial PMD. For each lipid, we defined the difference between the actual concentration value and the corresponding interpolated value from the spline curve fitted to the concentrations from the short PMD macaques with four degrees of freedom as residual. Lipids with the difference between the actual value and the interpolated value falling outside of the 1.96 SD of the residuals for all the three macaque samples with substantial PMD, were considered to be affected by PMD and excluded from further analysis.
RNA-Seq Read Alignment and Quantification
Reads sequenced in each sample were aligned to a consensus reference genome constructed by whole-genome alignment of humans (hg19), chimpanzees (panTro4) and rhesus monkeys (rheMac3) as described in He et al. (2014), using “STAR” mapper with default parameter settings. On the basis of reads aligned to a unique genomic location and GENCODE v19 annotation, expression level of each gene was quantified as reads per kilobase per million mapped reads (RPKM). mRNAs/lncRNAs with expression level detected in at least 95% of all samples and with maximal expression level across samples >1 were kept for further analysis.
Age Scale
For ages of all samples, we took gestation time into account by adding 280 days, 220 days and 165 days to the ages of humans, chimpanzees and rhesus monkeys, respectively. We then normalized individuals’ age according to the maximal lifespan differences among species by multiplying chimpanzee ages by 1.5 and macaque ages by 3, based on the following maximal lifespan estimates: 105 years for humans, 70 years for chimpanzees, and 35 years for macaques.
Age-Dependent Change Detection
In order to detect age-dependent lipid concentration or gene expression changes across the lifespan, we conducted age-test, constructing polynomial models with age as a regressor and concentration or expression levels as the regressand, described in (Somel et al. 2009). For each lipid/gene, the best polynomial regression model was chosen based on families of polynomial regression models and “adjusted r2” criteria and the significance of the chosen model was estimated using F-test. Each species was tested independently. The permutation for the age-test was performed by permuting ages across samples 1,000 times. The P-value for the permutation was estimated as the frequency of cases when the number of lipids/genes passing a given age-test P-value in a permutation was equal or greater than the one obtained in the actual age-test. The age-dependent lipids or genes were defined as those with age-test P-value <0.01, corresponding to permutation P-value <0.001.
Missing Concentration Value Imputation
For age-dependent lipids, the missing concentrations at given age points were estimated as the interpolated values from the spline curve fitted to the concentrations from remaining age points with four degrees of freedom. For lipids showing no significant age-dependent concentration changes, the missing concentrations at given age points were estimated as the average concentration from remaining age points.
Lipid Concentration Profile Correlation among Species
To estimate the correlation of age-dependent lipid concentration changes between each pair of the three species (humans, chimpanzees and rhesus monkeys) for a given lipid group, Pearson correlation coefficient was calculated between two vectors of 40 values uniformly interpolated from spline curves fitted to concentration data points of each species with four degrees of freedom for each lipid within the lipid group (He et al. 2014). The distribution of correlation coefficients expected by chance was estimated by matching lipids randomly between species 100 times.
Proportion of Concentration/Expression Variance Explained by Age Factor
To estimate the proportion of concentration/expression variance explained by age factor for lipids/genes, we used the best polynomial regression model applied to ages and concentrations/expressions obtained by the abovementioned age-test. The proportion was then defined as one minus the ratio of residual sum of squares from the regression model and residual sum of squares from the null model (variance multiplied by n - 1, where n is the sample size). The mean proportion across all detected lipids/genes was then determined.
Peak Annotation
The mass spectrometry peaks were annotated to known compounds based on the LIPID MAPS database and HMDB (Fahy et al. 2009; Wishart et al. 2013). For each lipid class we defined a list of possible adducts in positive and negative ionization modes (supplementary table S9, Supplementary Material online). The m/z values obtained by allowing adducts were then searched against the LC–MS measured m/z values with a mass tolerance of 10 ppm for the annotation of lipid [+] and lipid [-] datasets. For each annotated compound, lipid class and pathway were assigned to it based on LIPID MAPS database and KEGG. The enrichment of lipid classes and KEGG pathways in a given compound group was tested based on hypergeometric test followed by Bonferroni correction for multiple testing.
Comparison of Lipid Concentration Changes with Previous Studies
To compare the lipid concentration changes observed in our data with that previously reported, we searched the literature for lipid classes described to change concentration over the lifespan, which resulted in nine such lipid classes in human brains and three in rat brains. For each lipid class, the Pearson correlation coefficient was then calculated between the mean concentration profile based on our human data and that from previous studies, leading to 9 of the 12 lipid classes showing a positive correlation. To estimate the significance of this observation, we randomly sub-sampled the same number of compounds as in a given lipid class and calculated the correlation between the mean concentration profile of the sampled compounds and previously described profile for this lipid class, 1,000 times. P-value was then defined as the frequency of cases when the number of lipid classes showing a positive correlation in the sub-sampling was = or >9.
To compare the age-dependent concentration changes between our LC–MS measurements and published GC–MS measurements at the individual compound level, we identified common lipid compounds in both datasets based on the annotations, yielding eight lipids. For each of the eight lipids, Pearson correlation coefficients were calculated using the splined concentration profiles fit to LC–MS and the GC–MS measurements with four degrees of freedom.
Enzyme Expression
To test the connection between the age-dependent lipid concentration changes and the expression changes of corresponding enzymes, the enzymes corresponding to a lipid compound were determined based on HMDB annotation. For each lipid–enzyme pair, Pearson correlation coefficient was then calculated between the concentration profile of the lipid and the expression profile of the enzyme. The significance of the relationship was estimated by conducting 1,000 permutations of the enzyme-compound assignments. P-value was then defined as the frequency of cases when the median correlation coefficient across all lipid–enzyme pairs in a permutation was equal or greater than the actual median correlation coefficient.
To calculate the expression differences between humans and chimpanzees for enzymes associated with lipids, the associated enzyme was defined as the one showing the best correlation with the lipid concentration profile among all enzymes linked to the lipid using HMDB annotation. The gene expression differences between humans and chimpanzees were calculated by averaging the absolute expression differences between the two species calculated at 40 age points uniformly interpolated from the spline curves fitted to the actual expression data across matched age range.
Age Marker Identification
To identify lipids that could be used as predictors of individuals’ ages in each species, we trained regression models based on ages of samples and lipid concentrations at each age point using SVR combined with recursive feature elimination (RFE) (Guyon et al. 2002). Briefly, iteratively, the ranking for features (lipids) was based on the weight magnitude in the linear SVR and the feature with the smallest ranking was removed. The model was then retrained based on the remaining features and the model accuracy was estimated based on 10-fold cross-validation. The process stopped when one feature was left. Then, among all iterations we identified the one resulting in the subset of lipids with the best prediction accuracy. The identified set of age predictor lipids was stable based on 100 repetitions of the feature identification procedure.
Yearly Amplitude of Lipid Concentration/Gene Expression Change Calculation
To calculate the yearly amplitude of lipid concentration/gene expression change in each species, we uniformly interpolated 15 values for each one-year interval from spline curves fitted to lipid concentration/gene expression data points along the lifespan with four degrees of freedom. The sum of the absolute successive differences of the 15 interpolated values was then defined as the difference within a one-year interval. The ratio of the difference within a one-year interval to sum of differences across lifespan was defined as the yearly amplitude of change.
Lipidome Differences between Infant and Adult Brains
To search for large concentration changes between infant (0–1 years of human age) and adult (25–50 years of human age) brains in each species, t-test was conducted between concentrations in infant samples and concentrations in adult samples for the 7,589 age-dependent lipids. Lipids with two-sided t-test P-value after Benjamini Hochberg (BH) correction <0.05 and with fold-change >2 were defined to show large concentration changes between infant and adult brains.
Concentration Abundance of Lipid Class in Human PFC Lipidome
For each lipid class, the ratio of the total concentration of lipids within this lipid class to the total concentration of all annotated lipids based on LIPID MAPS database was calculated for each human sample to represent the abundance contribution of this lipid class in the human PFC lipidome. Concentrations of lipids annotated with multiple lipid classes were evenly proportioned among these lipid classes. The top 10 lipid classes were defined as those with the highest average abundance across all human samples.
Extent of Lipid Concentration Changes between Infant and Adult PFC
To estimate the extent of concentration changes between infant and adult brains for a given lipid class or lipid group, we calculated the absolute difference between the mean concentration within infant brains (0–1 years of human age) and the mean concentration within adult brains (25–50 years of human age) for each lipid. The sum of the calculated difference across the lipid class or lipid group was defined as d. The sum of the mean concentration within infant brains across the lipid class or lipid group was defined as t. The ratio of d to t was considered as the extent of concentration changes between infant and adult brains.
Myelination Marker Identification
We identified myelination markers based on lipid concentrations measured in white matter dissected from corpus callosum and cortical gray matter in five adult humans. Concentration values of all lipid peaks were log2-transformed. Before log2 transformation, the concentration values were increased by one to avoid infinite values. Lipid peaks with concentrations from the five white matter samples or the five gray matter samples larger than the fifth percentile of all peak concentrations were kept for following analysis. In order to link this dataset to the developmental dataset, lipid peaks with unique correspondence to each other between the two datasets under a mass tolerance of 5 ppm and RT difference of 0.3 min were matched, resulting in 1,900 lipid peaks detected in both datasets. We used t-tests to compare concentrations from white matter samples with those from gray matter samples for the 1,900 lipids. Lipids with two-sided t-test P-value after BH correction <0.05 and with fold-change calculated between white matter and gray matter >2 were defined as myelination markers. For known myelination marker cerebrosides, we collected lipid peaks corresponding to them based on LIPID MAPS database and HMDB. The enrichment of myelination markers in a given lipid group was conducted using hypergeometric test.
White Matter Proportion Estimation
We estimated the proportion of white matter in the developmental samples based on concentrations of age-dependent lipids detected in both the developmental dataset and the gray/white matter dataset, using quadratic programming and simulated annealing (SA). The mean concentration across the five white matter samples and the mean concentration across the five gray matter samples for each lipid were used for white matter proportion estimation. Before estimation, the concentrations were standardized across lipids for both the gray/white matter dataset and the developmental dataset.
Quadratic programming was conducted based on the following criteria using R package “quadprog”:
where represents the white matter proportion, represents the gray matter proportion, is the lipid concentration in white matter, is the lipid concentration in gray matter, and is the lipid concentration in the developmental sample.
For SA, the goal function , with parameter representation the same as above. The initial temperature was set to be 100,000 and for each temperature value the procedures were repeated 10 times. The final state was chosen as the one with the temperature below 0.001.
To estimate the proportion of age-dependent lipid concentration changes associated with myelination progression, for each age-dependent lipid we calculated the Pearson correlation coefficients between the concentrations of the lipid and the white matter proportions in developmental samples from humans, chimpanzees, and macaques, or the three species, respectively. The age-dependent lipids with absolute Pearson correlation coefficient >0.6 and with correlation test P-value <0.05 were considered to be associated with myelination progression.
Lipidome/Transcriptome Differences among Species
We used the analysis of covariance (ANCOVA) with linear, quadratic, and cubic models to identify lipids/genes showing differential concentration/expression profile between a pair of species based on all age-dependent lipids/genes. Briefly, as described in Somel et al. (2009), based on the polynomial regression model obtained using the above age-test, we tested if a regression model with species-specific parameters was significantly better than the model with common parameters for both species. The null model (with no species-specific parameters) and alternative models were compared using F-test. For each pair of species, the differential concentration/expression profile test was conducted twice with either species as a reference for obtaining the age-test model. To preserve the age structure in the data, the permutations were performed by dividing the age-range into 10 sections and randomly permuting the species assignments across samples within each section 1,000 times. Lipids/genes showing significant differential concentration/expression profile between two species were defined as those with F-test P-value <0.01 under both two tests, corresponding to FDR <5%.
Lipids/genes with species-specific (human-specific, chimpanzee-specific, macaque-specific) concentration profiles were defined as those showing significant differential concentration/expression profile between this species and the other two species, but showing no significant differential concentration/expression profile between the other two species. Lipids/genes with conserved concentration/expression profile across human, chimpanzee and rhesus monkeys were defined as those showing no significant differential concentration/expression profile between any pair of the three species.
Identification of Species Predictor Lipids
Binary classifiers, which distinguish one species from the remaining species, were constructed based on age-dependent lipids, using logistic regression with elastic net penalty within R package “glmnet.” The parameter α was set to 0.5. For each 10-fold cross-validation run, λ parameter was set to the one that yielded the cross-validated error within one standard error of the minimum. Species predictor lipids were defined as the union of selected lipids under each run given parameters α and λ. The accuracy of the classifier was estimated as the average of the area under the receiver operating characteristic curve (AUC) across the 10 × 10 cross-validation run.
Lipidome Difference between Adult Humans and Chimpanzees
To identify lipids showing significant concentration differences between adult humans and chimpanzees (25–50 years of human age) in developmental data, we performed t-tests between concentrations from adult humans and those from adult chimpanzees. Lipids with two-sided t-test P-value after BH correction <0.05 were considered to show significant difference between adults of the two species. To determine how many such lipids could match previous study (Bozek et al. 2015), we connected lipids annotated to the same known compound in our developmental dataset and the previously reported dataset. Among the matched lipids, the number of lipids showing human-specific concentration profiles and those showing chimpanzee-specific concentration profiles (defined above) was determined.
Human-Specific and Chimpanzee-Specific Lipid Concentration Differences
We calculated the human-specific and chimpanzee-specific concentration difference as the difference between humans and chimpanzees along the lifespan for lipids showing human-specific concentration profiles or those showing chimpanzee-specific concentration profiles (defined in the part “lipidome/transcriptome differences across species”). Before the species-specific difference estimation, the lipid concentrations were standardized across human and chimpanzee samples. For each species, we then interpolated concentration values at 40 equally distributed points along the age range. Interpolation was performed using cubic spline regression with four degrees of freedom. The difference between humans and chimpanzees at each of the 40 time points was then defined as the average of the absolute difference between the concentration value interpolated from humans and that from chimpanzees at the given time point across the lipids showing human-specific profiles or those showing chimpanzee-specific profiles. The permutations were conducted by dividing the age-range into 10 sections and randomly permuting the species assignments within samples in each section to preserve the age-structure in the data 1,000 times.
The species-specific difference was normalized by subtracting the mean species-specific difference of the 1,000 permutation distribution at each age point. The excess of human-specific differences over chimpanzee-specific ones was determined as the difference between the two measurement vectors.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
P.K. and P.G. conceived and designed the experiments; J.S., Y.L., and P.G. performed experiments; S.P., C.C.S., P.R.H., J.J.E., L.W., and P.G. contributed reagents/materials/samples; Q.L., K.B., C.X., and Y.G. analyzed the data; S.P., C.C.S., P.R.H., P.K., P.G., and Q.L. wrote the paper.
Supplementary Material
Acknowledgments
We thank the NICHD Brain and Tissue Bank for Developmental Disorders, the Maryland Population Research Center and the Chinese Brain Bank Center, and Dr H.R.Z. and Dr J.D. for providing the human samples; the Alamogordo Primate Facility, Max Planck Institute for Evolutionary Anthropology, Southwest National Primate Research Center and National Chimpanzee Brain Resource (NINDS grant NS092988) and Dr R.M. and Dr W.S. for providing the chimpanzee samples; Suzhou Drug Safety Evaluation and Research Center and C.L., H.C. and X.Z. for providing the macaque samples. We thank Dr G.L.B. for his helpful comments on the manuscript. This work was supported by Strategic Priority Research Program of the Chinese Academy of Sciences (grant number XDB13010200); the National Natural Science Foundation of China (grant number 31420103920); the National Natural Science Foundation of China (grant number 91331203); the National One Thousand Foreign Experts Plan (grant number WQ20123100078); the Bureau of International Cooperation, Chinese Academy of Sciences (grant number GJHZ201313); and the Russian Science Foundation (grant number 16-14-00220).
References
- Abbott SK, Jenner AM, Spiro AS, Batterham M, Halliday GM, Garner B.. 2015. Fatty acid composition of the anterior cingulate cortex indicates a high susceptibility to lipid peroxidation in Parkinson's disease. J Parkinsons Dis 5:175–185. [DOI] [PubMed] [Google Scholar]
- Boggs JM. 2014. Role of galactosylceramide and sulfatide in oligodendrocytes and CNS myelin: formation of a glycosynapse. Adv Neurobiol 9:263–291. [DOI] [PubMed] [Google Scholar]
- Boggs JM, Wang H, Gao W, Arvanitis DN, Gong Y, Min W.. 2004. A glycosynapse in myelin? Glycoconj J 21:97–110. [DOI] [PubMed] [Google Scholar]
- Bozek K, Wei Y, Yan Z, Liu X, Xiong J, Sugimoto M, Tomita M, Paabo S, Sherwood CC, Hof PR, et al. 2015. Organization and evolution of brain lipidome revealed by large-scale analysis of human, chimpanzee, macaque, and mouse tissues. Neuron 85:695–702. [DOI] [PubMed] [Google Scholar]
- Carroll SB. 2000. Endless forms: the evolution of gene regulation and morphological diversity. Cell 101:577–580. [DOI] [PubMed] [Google Scholar]
- Chen WY, Qian W, Wu G, Chen WZ, Xian B, Chen XW, Cao YQ, Green CD, Zhao FH, Tang K, et al. 2015. Three-dimensional human facial morphologies as robust aging markers. Cell Res 25:574–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Hayden Gephart MG, Barres BA, Quake SR.. 2015. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A 112:7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CR, Shimizu T, Spener F, van Meer G, Wakelam MJ, Dennis EA.. 2009. Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res 50(Suppl):S9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Giavalisco P, Liu X, Catchpole G, Fu N, Ning ZB, Guo S, Yan Z, Somel M, Paabo S, et al. 2011. Rapid metabolic evolution in human prefrontal cortex. Proc Natl Acad Sci U S A 108:6181–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon I, Weston J, Barnhill S, Vapnik V.. 2002. Gene selection for cancer classification using support vector machines. Mach Learn 46:389–422. [Google Scholar]
- Hakomori Si SI. 2002. The glycosynapse. Proc Natl Acad Sci U S A 99:225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannun YA, Obeid LM.. 2008. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol 9:139–150. [DOI] [PubMed] [Google Scholar]
- He Z, Bammann H, Han D, Xie G, Khaitovich P.. 2014. Conserved expression of lincRNA during human and macaque prefrontal cortex development and maturation. RNA 20:1103–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. 2013. DNA methylation age of human tissues and cell types. Genome Biol 14:R115.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel J, Segu S, Li Y, Irgang S, Jueppner J, Giavalisco P.. 2011. Ultra performance liquid chromatography and high resolution mass spectrometry for the analysis of plant lipids. Front Plant Sci 2:54.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurevics H, Hostettler J, Muse ED, Sammond DW, Matsushima GK, Toews AD, Morell P.. 2001. Cerebroside synthesis as a measure of the rate of remyelination following cuprizone-induced demyelination in brain. J Neurochem 77:1067–1076. [DOI] [PubMed] [Google Scholar]
- Khrameeva EE, Bozek K, He L, Yan Z, Jiang X, Wei Y, Tang K, Gelfand MS, Prufer K, Kelso J, et al. 2014. Neanderthal ancestry drives evolution of lipid catabolism in contemporary Europeans. Nat Commun 5:3584.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Hu X, Preuss TM, Glasser MF, Damen FW, Qiu Y, Rilling J.. 2013. Mapping putative hubs in human, chimpanzee and rhesus macaque connectomes via diffusion tractography. Neuroimage 80:462–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Somel M, Tang L, Yan Z, Jiang X, Guo S, Yuan Y, He L, Oleksiak A, Zhang Y, et al. 2012. Extension of cortical synaptic development distinguishes humans from chimpanzees and macaques. Genome Res 22:611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DJ, Duka T, Stimpson CD, Schapiro SJ, Baze WB, McArthur MJ, Fobbs AJ, Sousa AMM, Sestan N, Wildman DE, et al. 2012. Prolonged myelination in human neocortical evolution. Proc Natl Acad Sci U S A 109:16480–16485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller CP, Reichel M, Muhle C, Rhein C, Gulbins E, Kornhuber J.. 2015. Brain membrane lipids in major depression and anxiety disorders. Biochim Biophys Acta 1851:1052–1065. [DOI] [PubMed] [Google Scholar]
- Piomelli D, Astarita G, Rapaka R.. 2007. A neuroscientist's guide to lipidomics. Nat Rev Neurosci 8:743–754. [DOI] [PubMed] [Google Scholar]
- Rouser G, Yamamoto A.. 1968. Curvilinear regression course of human brain lipid composition changes with age. Lipids 3:284–287. [DOI] [PubMed] [Google Scholar]
- Somel M, Franz H, Yan Z, Lorenc A, Guo S, Giger T, Kelso J, Nickel B, Dannemann M, Bahn S, et al. 2009. Transcriptional neoteny in the human brain. Proc Natl Acad Sci U S A 106:5743–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Liu X, Khaitovich P.. 2013. Human brain evolution: transcripts, metabolites and their regulators. Nat Rev Neurosci 14:112–127. [DOI] [PubMed] [Google Scholar]
- Vermunt MW, Tan SC, Castelijns B, Geeven G, Reinink P, de Bruijn E, Kondova I, Persengiev S, Netherlands Brain B, Bontrop R, et al. 2016. Epigenomic annotation of gene regulatory alterations during evolution of the primate brain. Nat Neurosci 19:494–503. [DOI] [PubMed] [Google Scholar]
- Wenk MR. 2005. The emerging field of lipidomics. Nat Rev Drug Discov 4:594–610. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, et al. 2013. HMDB 3.0 – the human metabolome database in 2013. Nucleic Acids Res 41:D801–D807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Konopka G, Hunt BG, Preuss TM, Geschwind D, Yi SV.. 2012. Divergent whole-genome methylation maps of human and chimpanzee brains reveal epigenetic basis of human regulatory evolution. Am J Hum Genet 91:455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Appelkvist EL, Kristensson K, Dallner G.. 1996. The lipid compositions of different regions of rat brain during development and aging. Neurobiol Aging 17:869–875. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.