Abstract
Molluscs fabricate shells of incredible diversity and complexity by localized secretions from the dorsal epithelium of the mantle. Although distantly related molluscs express remarkably different secreted gene products, it remains unclear if the evolution of shell structure and pattern is underpinned by the differential co-option of conserved genes or the integration of lineage-specific genes into the mantle regulatory program. To address this, we compare the mantle transcriptomes of 11 bivalves and gastropods of varying relatedness. We find that each species, including four Pinctada (pearl oyster) species that diverged within the last 20 Ma, expresses a unique mantle secretome. Lineage- or species-specific genes comprise a large proportion of each species’ mantle secretome. A majority of these secreted proteins have unique domain architectures that include repetitive, low complexity domains (RLCDs), which evolve rapidly, and have a proclivity to expand, contract and rearrange in the genome. There are also a large number of secretome genes expressed in the mantle that arose before the origin of gastropods and bivalves. Each species expresses a unique set of these more ancient genes consistent with their independent co-option into these mantle gene regulatory networks. From this analysis, we infer lineage-specific secretomes underlie shell diversity, and include both rapidly evolving RLCD-containing proteins, and the continual recruitment and loss of both ancient and recently evolved genes into the periphery of the regulatory network controlling gene expression in the mantle epithelium.
Keywords: mantle secretome, co-option, lineage-specific novelties, bivalve, gastropod, shell formation
Introduction
Mollusca is a morphologically diverse and speciose phylum with a long and rich history dating back to the Cambrian (Kocot et al. 2011; Smith et al. 2011; Vinther 2015). Their success can be partially attributed to the ability to build a strong shell (Marin et al. 2014). Shell synthesis occurs at the interface between mollusc and environment by specialized epithelial cells on the dorsal surface of the mantle, a highly muscularized and innervated organ that most likely arose early in mollusc evolution (reviewed by Furuhashi et al. 2009; Marin et al. 2012; Kocot et al. 2016). Secretions from the dorsal mantle epithelium include proteins, glycoproteins, lipids and polysaccharides. This macromolecular assemblage promotes the formation of calcium carbonate crystals and directs shell formation, ultimately regulating the architecture and pattern of the shell (Furuhashi et al. 2009; Marin et al. 2012; Kocot et al. 2016).
Molluscs are divided into two major clades, the Aculifera, which includes sclerite and shell plate-bearing neomeniomorphs, chaetodermomorphs and polyplacophorans, and the shelled and the speciose Conchifera, which includes gastropods, bivalves, scaphopods and cephalopods (Kocot et al. 2011; Smith et al. 2011; Vinther 2015). Conchiferan shells are layered and often covered by a thin, organic outer layer called the periostracum (Kocot et al. 2016). The underlying calcified layers, typically comprising aragonitic or calcite crystals, confer specific physical properties to the shell, and are classified based on their crystalline microstructures [i.e., prismatic, nacreous, crossed lamellar or homogeneous (Chateigner et al. 2000; Furuhashi et al. 2009)]. The shells and sclerites produced by different molluscan taxa exhibit noticeable differences in shape, color and ornamentation, however less obvious variation can also be observed at the deeper ultrastructural level (Chateigner et al. 2000; Furuhashi et al. 2009).
Perhaps unsurprisingly given the level of diversity seen in the shell, the mantle also displays significant variation, exhibiting different morphologies (e.g., mantle folds, grooves and/or tubules) in various species (Dix 1972; Jabbour-Zahab et al. 1992; Sud et al. 2002; McDougall et al. 2011; Budd et al. 2014). Within the mantle, distinct territories are responsible for the production of different layers of the shell (Jolly et al. 2004; Takeuchi and Endo 2005; Jackson et al. 2006; Marie et al. 2012). Gene expression is often restricted to a specific mantle territory, consistent with each shell layer having a different organic content (Jackson et al. 2006; Joubert et al. 2010; Kinoshita et al. 2011; Marie et al. 2012; Gao et al. 2015; Liao et al. 2015; Liu et al. 2015).
It might be expected that shell layers with similar microstructures (e.g., nacre) would be constructed from similar proteins in different mollusc species. However, comparison of genes expressed in the mantle that encode secreted proteins, the so-called “mantle secretome”, of the bivalve Pinctada maxima and the gastropod Haliotis asinina indicates that the inner nacreous layer of these two distantly related conchiferans are comprised of markedly different proteins (Jackson et al. 2010). These differences include a large number of novel proteins, such as the silk-like shematrins that are found only within pearl oyster species (McDougall et al. 2013). From this comparison, it has been inferred that the nacreous layers in these two species evolved independently (Jackson et al. 2010). Additional transcriptomic and proteomic studies of the mantle and shell have also revealed high levels of diversity in other molluscan species both within and between classes (reviewed by Kocot et al. 2016).
A number of common principles appear to underpin the differences observed in the limited number of mantle secretomes studied so far (Marin et al. 2012; Kocot et al. 2016). These include the presence of repetitive, low complexity domains (RLCDs) and the high degree of modularity and shuffling of functionally distinct domains (Shen et al. 1997; Jackson et al. 2010; McDougall et al. 2013). However, whether these principles are a common feature of mantle secretome evolution remains to be established.
Here, we investigate the expression and evolution of mantle secretomes for 11 adult bivalves and gastropods using a transcriptomics-based approach. We elected to use this approach instead of direct analysis of shell proteomes, which tend to be incomplete and do not capture secreted proteins necessary for biomineralization that are not incorporated into the shell (Joubert et al. 2010; Marie et al. 2010, 2012, 2013; Mann and Edsinger 2014; Mann and Jackson 2014; Gao et al. 2015; Liao et al. 2015; Liu et al. 2015). Given that the mantle secretome can change markedly during the lifespan of a mollusc (Jackson et al. 2007), our analyses were restricted to the mantles of mature adults. While these adults come from a range of marine habitats and have untraceable life histories, in all cases their mantle cells are expressing a terminally differentiated genetic program to create the final shell type. The analyses performed here include species that have diverged both recently [i.e., four Pinctada spp. that diverged within the last 20 Ma (Cunha et al. 2011)] and in the distant past (i.e., Cambrian period). This nested survey, which allows comparisons between molluscan classes, orders, families and species, reveals unexpectedly that genes encoding secreted proteins that evolved before the evolution of molluscs are continually being co-opted into the mantle gene regulatory network. This, coupled with lineage-specific gene family expansion and domain shuffling, has resulted in each lineage and species having a unique mantle secretome.
Results
The Evolutionary Birth of Secreted Mantle Proteins Ranges from the Origin of Cellular Life until the Present Day
Gene products expressed in the mantles of eight bivalves (Hyriopsis cumingii, Laternula elliptica, Crassostrea gigas, Mytilus edulis, Pinctada maxima, P. margaritifera, P. martensii and P. fucata) and three gastropods (Patella vulgata, Lottia gigantea and Haliotis rufescens) were classified in silico as being localized to the cytosol or to the membrane, or being destined for secretion (supplementary fig. S1, Supplementary Material online). For this study, we solely focused on the mantle genes encoding secreted proteins (the “mantle secretome”).
We first assessed if these mantle transcriptomes encode secreted proteins incorporated into the shell calcifying-matrix by investigating sequence similarity between our predicted mantle secretomes and previously published shell proteomes from C. gigas (Marie, Zanella-Cleon, et al. (2011); Zhang et al. 2012), P. fucata, P. margaritifera and P. maxima (Marie et al. 2012; Liu et al. 2015), and L. gigantea (Mann et al. 2012; Marie et al. 2013; Mann and Edsinger 2014). In all cases, there was a markedly greater number of secreted proteins predicted from our mantle transcriptomes than found in the shell proteomes; 5–50% of the secretomes derived from mantle transcriptomes had sequence similarity with proteins found in the shell of these species (supplementary table S1, Supplementary Material online). The consistent detection of more secreted proteins in the mantle transcriptomes compared with the shell proteomes suggests that some proteins secreted from the mantle are not incorporated into the shell.
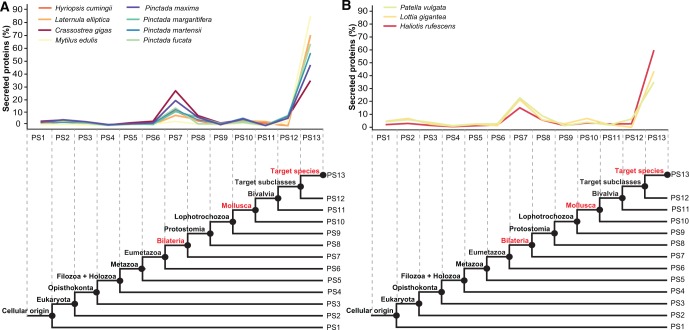
The ages of the genes encoding the mantle secretomes of the 11 molluscs were estimated using the phylostratigraphic approach (Domazet-Lošo et al. 2007). Thirteen phylogenetic ranks (phylostrata) were defined according to the NCBI Taxonomy database, with the first phylostratum (PS1) being at the origin of cellular life (i.e., the oldest genes), and the last phylostratum (PS13) being the lineage leading to each species under study (i.e., the youngest genes). For all species, the largest proportion of mantle secretome genes was classified into the youngest phytostratum (PS13) (fig. 1 and supplementary table S2, Supplementary Material online). In addition to these taxon-restricted genes, all mantle secretomes included a substantial number of genes that evolved along bilaterian (PS7) and mollusc (PS10) branches, before the origin of conchiferans (fig. 1 and supplementary fig. S2, Supplementary Material online).
Fig. 1.
Evolutionary origin of mantle genes encoding secreted proteins in different conchiferan taxa. Evolutionary origin of secreted mantle proteins for each bivalve (A) and gastropod (B) species with phylostrata (PS) depicted below. Phylostratum 12 corresponds to the subclass taxonomic rank for each bivalve (i.e., Palaeoheterodonta, Heterodonta and Pteriomorpha) and gastropod (i.e., Vetigastropoda and Patellogastropoda) species used in this study. Denoted in red are the three evolutionary periods associated with the emergence of most secreted proteins: the bilaterian stem (PS7), the mollusc stem (PS10), and taxon-restricted lineages (PS13).
Mantle Secretome Evolution Includes Extensive Gene Co-Option and Loss
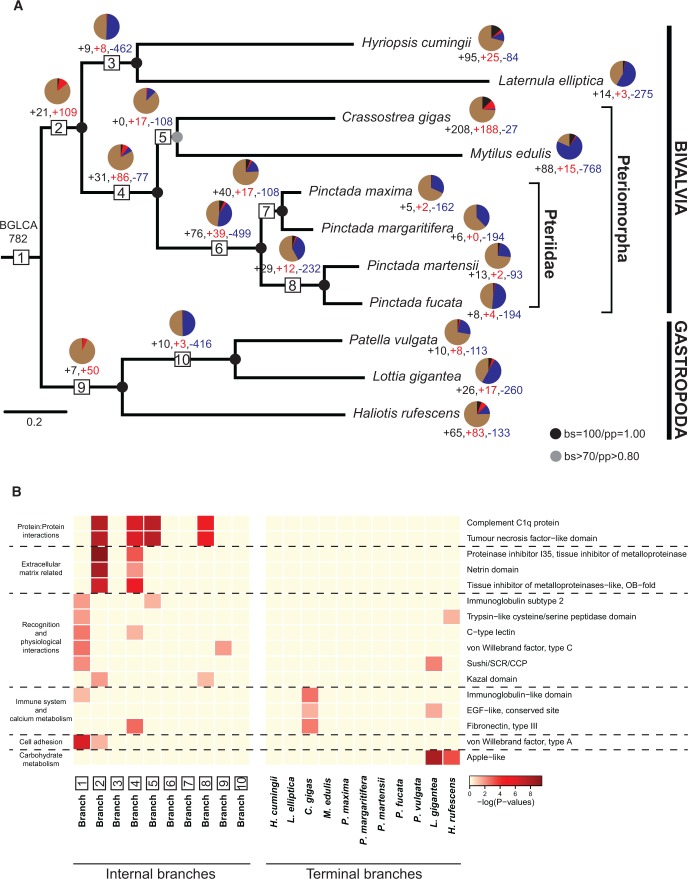
Using a phylogenetic framework, we investigated the distribution of expression of secretome genes in the mantles of these bivalves and gastropods over conchiferan evolution. First, a phylogenetic tree of the 11 species was constructed from a concatenated set of single-copy core genes (122 gene families). A single matrix of 13,604 aligned amino acids was curated and subjected to both Maximum Likelihood and Bayesian Inference analyses. Both phylogenetic approaches produced equivalent topologies, and all nodes but one were strongly supported by bootstrap percentages (BP = 100%) and posterior probabilities (PP = 1.0) (fig. 2A and supplementary fig. S3, Supplementary Material online), consistent with previous molluscan phylogenomic analyses (Kocot et al. 2011; Smith et al. 2011).
Fig. 2.
Evolutionary history of conchiferan mantle secretome gene family evolution. (A) Organismal tree (ML topology) showing the relationship of bivalves and gastropods. Black circles represent nodes with BS = 100 and PP = 1.00, and the gray circle represents the node with BS > 70% and PP > 0.80. Gene family acquisition was divided into lineage-specific gains (black) and independent co-options (red). Based on comparison of gastropod and bivalve mantle secretomes, the bivalve and gastropod last common ancestor (1; BGLCA) expressed 782 mantle secretome genes that are shared between at least one gastropod and one bivalve. From this ancestral condition, the LCA of the bivalves (BLCA) included in this study (2; BLCA) evolved 21 bivalve-specific mantle secretome gene families (black text and pie wedge) and co-opted 109 ancestral gene families into the mantle secretome (red text and pie wedge); the brown portion of the pie represents the 782 genes contributed from the BGLCA ancestor. The LCA of Hyriopsis cumingii and Laternula elliptica (3) gained 9 and 8 novel and co-opted genes into the mantle secretome, and lost 462 gene families (blue text and pie wedge) compared with the BLCA. All remaining (4–10) ancestral reconstructions, along with the evolution of species-specific secretome repertoires, follow the same interpretations. Gene family losses and gains are calculated based on the Dollo parsimony principle and do not take into account the independent co-option of the same gene family twice. (B) Enrichment of protein domains across conchiferan evolution (i.e., internal and terminal branches). Protein domains significantly enriched (P < 0.05, Fisher’s exact test) and present in newly gained secreted gene families from at least two branches are shown. The yellow-to-red scale, based on −log(P values), indicates the level of enrichment. InterPro protein domain descriptions of the over-represented secreted gene families are shown at the right. Broad functional categories representing each protein domain are shown to the left. For a comprehensive list of enriched protein domains across conchiferan evolution, see supplementary table S4, Supplementary Material online.
Second, 2,231 gene families encoding secreted proteins—“secretome gene families”—were identified from 19,134 protein coding sequences in these conchiferan mantle transcriptomes based on sequence similarity and clustering algorithms (Li et al. 2003). The presence and absence of these 2,231 gene families was assessed in the 11 species. Third, combining the phylogenetic analysis with the presence/absence gene family matrix, we reconstructed the evolution of bivalve and gastropod mantle secretomes (fig. 2A). The Dollo parsimony approach was applied to delineate the minimal gene set for the different ancestral branches (Farris 1977) and assumed that gene loss was irreversible and, thus, a gene family could not re-evolve (i.e., convergent evolution did not occur). In this study, gene loss means loss of expression in the mantle and not necessarily loss from the genome. It was assumed that the presence of orthologues in the mantle transcriptomes of two or more species supported the presence of this gene family in their last common ancestor; independent co-option events, while possible, were not considered.
From this analysis, 782 secretome gene families were determined to be expressed in the mantle of the bivalve and gastropod last common ancestor (BGLCA) (fig. 2A). These genes could have evolved any time before the divergence of bivalves and gastropods (i.e., phylostratigraphic levels PS1–PS10, fig. 1). Since diverging, both bivalve and gastropod lineages have undergone consistent and dramatic changes in the mantle secretome, which includes both (1) extensive loss of secretome genes expressed in the mantle, such as occurred along the branches leading to H. cumingii/L. elliptica, Pteriidae and Patellogastropoda clades (i.e., internal branches 3, 6 and 10, respectively, fig. 2A and supplementary table S3, Supplementary Material online), and (2) extensive gain of genes into mantle secretomes, such as occurred prior to the origin of bivalve, pteriomorph and pteriid last common ancestors (branches 2, 4, 6, respectively, fig. 2A and supplementary table S3, Supplementary Material online).
As gene gain is comprised of both co-option of older genes and by the incorporation of lineage-specific genes into the mantle regulatory architecture, we undertook phylostratigraphic profiling to separate these two categories. Although evolution of these two groups of mantle secretome genes occurred at different rates across bivalve and gastropod evolution, in general co-option of more ancient genes was more common than the expression of newer lineage-restricted genes (fig. 2A and supplementary table S3, Supplementary Material online). For instance 50 out of 57 new gene families expressed in the mantle of gastropods, after diverging from bivalves, were from co-option events; likewise 109 out of 130 bivalve mantle secretome families were from co-options (fig. 2A).
Analysis of the domain composition of pre-conchiferan proteins co-opted into the mantle secretome revealed that they are enriched in domains often associated with extracellular or cell surface proteins that facilitate protein–protein interactions, including immunoglobulin, EGF, Fibronectin, von Willebrand factor and C-type lectin domains (fig. 2B, supplementary fig. S4 and tables S4–S6,Supplementary Material online). These domain enrichments were typically restricted to a limited number of taxa, although there were cases where domain expansion appeared to continue over longer periods of evolution. For instance, complement C1q protein and tumour necrosis factor-like domains had continually expanded in the stems leading to bivalves, pteriomorphs and pteriids (fig. 2B).
To explore the relationship between gene origin/gene class and relative gene expression, we assessed the expression profiles of mantle secretome genes across phylostrata and gene classes (i.e., co-opted, lineage-specific and species-specific genes). We found that genes encoding secreted proteins have unique expression profiles across phylostrata (supplementary fig. S5, Supplementary Material online; no threshold for relative gene expression was used). We also found no correlation between gene age classes and expression levels (supplementary fig. S6, Supplementary Material online). Some species exhibited slightly higher expression levels for older co-opted genes (e.g., P. fucata, P. martensii and H. rufescens), while others showed high expression levels for lineage- and/or species-specific genes (e.g., C. gigas, H. cumingii and L. gigantea).
Additional Mantle Secretome Evolution Is Driven by the Gain of Novel Genes
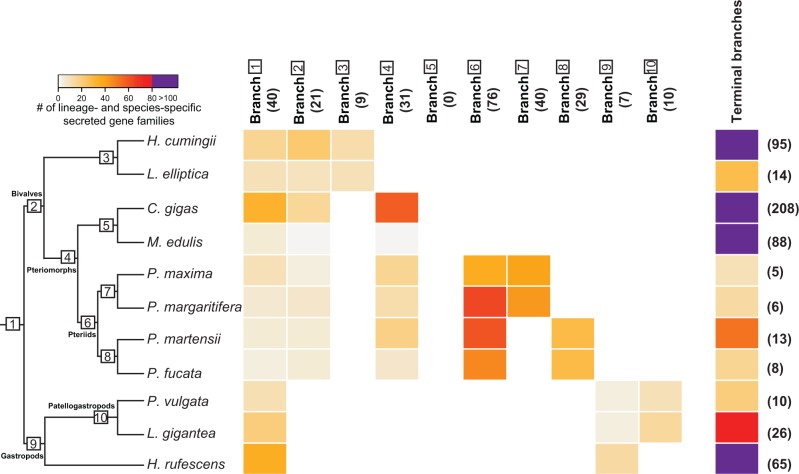
Mantle secretome gene families that evolved after the BGLCA comprised lineage- and species-specific gene families. The appearance of these gene families in the mantle transcriptomes varied markedly over bivalve and gastropod evolution as well as between species, with each lineage and species possessing a unique repertoire of novel genes (figs. 2A and 3). 21 of the 130 mantle secretome gene families (16%) specific to bivalves were deemed novelties (figs. 2A and 3, branch 2), while 7 of the 57 gene families (12%) restricted to gastropod mantle secretomes were innovations within this molluscan class (figs. 2A and 3, branch 9). The gain of new lineage-specific genes appears to continue throughout bivalve and gastropod evolution. For example, 26% of the newly gained secretome gene families in pteriomorphs (figs. 2A and 3, branch 4) were lineage-restricted.
Fig. 3.
Distribution and abundance of lineage- and species-specific secretome gene families across conchiferan evolution. Number of lineage- and species-specific secretome gene families depicted according to the color legend in the upper left. Black bold numbers in parenthesis depict the number of lineage-specific gene families at each evolutionary time point. These secreted gene families are grouped according to the phylogenetic tree shown in the left, where white squares with numbers indicate internal branches that correspond to figure 2A. Internal branches are indicated at the top of the heatmap, while terminal branches (i.e., species-specific secreted gene families) are indicated at the right. For the complete list of lineage- and species-specific secreted gene families, see supplementary table S7, Supplementary Material online.
The number of novel gene families also varied between species (fig. 3, terminal branches, supplementary table S7, Supplementary Material online). Analysis of Pinctada (pearl oyster) species, which have diverged over the last 20 My (Cunha et al. 2011), revealed a considerable diversity of lineage-specific novelties (66%) (figs. 2A and 3, branch 6). Some of these proteins (e.g., shematrins, KRMPs) are present in the organic calcifying-matrix of pearl oyster shells (Jackson et al. 2010; Kinoshita et al. 2011; McDougall et al. 2013), contributing to specific shell features and patterns (Funabara et al. 2014; Liang et al. 2015; Liu et al. 2015).
Many Lineage-Specific Mantle Secretome Genes Encode Repetitive, Low Complexity Domains
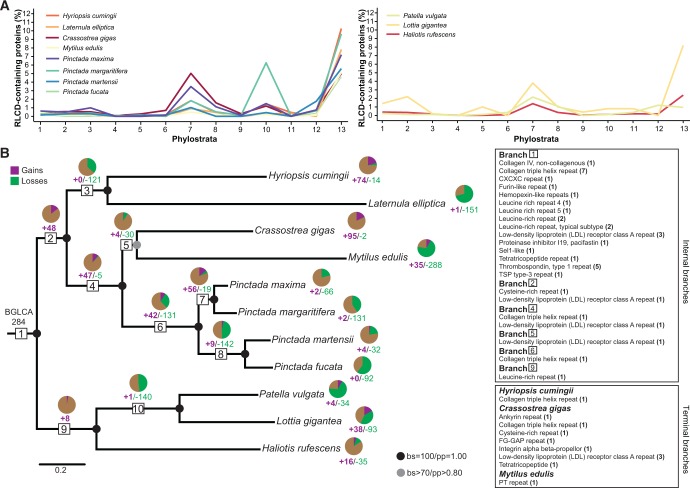
Repetitive, low complexity domains (RLCDs) are one of the most common features encoded by novel and lineage-restricted mantle secretome genes (McDougall et al. 2013, 2016). Using XSTREAM software (Newman and Cooper 2007), high abundances of RLCD-containing proteins were identified in both bivalves and gastropods (fig. 4A and supplementary table S1, Supplementary Material online). Most of these RLCD-containing proteins had no sequence similarity with other known proteins (supplementary table S8, Supplementary Material online). However, some RLCD-containing proteins appeared to have evolved before the divergence of gastropod and bivalve lineages. These proteins were largely paired with conserved domains associated with the ECM, including collagen domains, and leucine-rich and tetratricopeptide repeats (fig. 4B). These RLCD-containing secreted proteins were also expressed in particular conchiferan lineages, including some that were either bivalve- or gastropod-specific (supplementary table S8, Supplementary Material online).
Fig. 4.
Evolutionary dynamics of gain and loss of mantle secretome families with repetitive, low-complexity domains. (A) Phylostratigraphic maps of RLCD-containing proteins present in the secretomes of each bivalve (left panel) and gastropod (right panel) species. Phylostrata are labeled according to phylogenetic maps shown in figure 1. (B) Relationships among bivalve and gastropod lineages, depicting patterns of gains and losses of secreted gene families that contain RLCDs over conchiferan evolution. See figure 2A for a description on interpreting gene gain (purple) and loss (green). The pie charts display the proportion of secreted gene families that contain RLCD-containing proteins inherited from (brown), lost from (green) and gained since diverging from (purple) the previous node. The squares on the right represent RLCDs that show similarity with the InterPro sequence repeat database. Numbers in parenthesis indicate the number of secretome gene families that contain this specific RLCD. BGLCA: bivalve and gastropod last common ancestor.
Analysis of the evolutionary dynamics of RLCD-containing secretome families revealed that these families were lost more than gained (fig. 4B and supplementary table S3, Supplementary Material online). However, in the stems leading to Pteriomorpha, Pteriidae and P. maxima/P. margaritifera (fig. 4B, branches 4, 6 and 7, respectively), there were marked increases in the number of RLCD gene families. Independently acquired RLCD–containing families also appeared as a dominant part of the mantle secretome of H. cumingii, C. gigas, M. edulis, L. gigantea and H. rufescens (fig. 4B).
Conserved Secretome Families Have Undergone Independent Expansions and Domain Shuffling
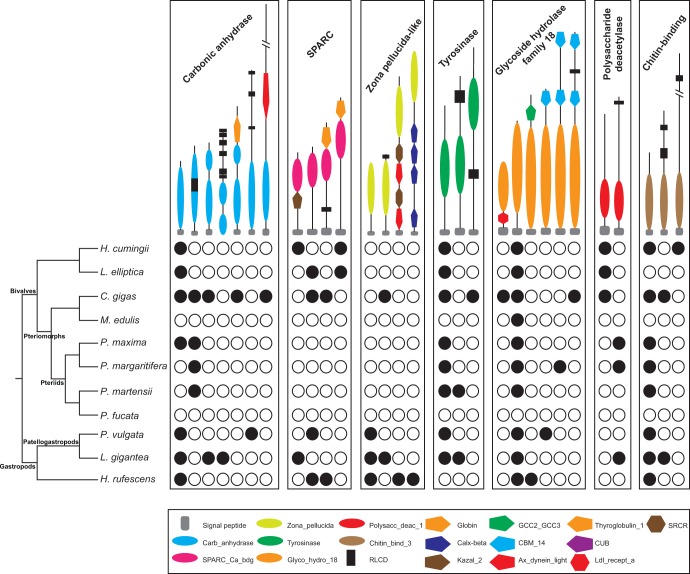
Amongst the secretome gene families expressed in the mantle was a suite of ancient, conserved families that appear to have gained further functionality through the acquisition of new domains. These novel domain architectures were often found restricted to a single species or clade (fig. 5). These gene families, which include variant forms of carbonic anhydrase, tyrosinase, SPARC and chitin-binding protein (fig. 5), often include extensive lineage-specific gene expansions and are highly expressed in the mantle (Zhang et al. 2012; Aguilera et al. 2014; Sleight et al. 2016). These also appear to have undergone multiple independent co-options into the mantle gene regulatory network (supplementary figs. S7–S11, Supplementary Material online).
Fig. 5.
Phylogenetic distribution of protein families with known domains associated with shell biomineralization. The domain architectures of secreted protein families are depicted at the top, and the key to protein domains at the bottom. A black and white dots indicate the presence and absence of a given protein domain architecture in the mantle secretome, respectively. Phylogenetic relationship among bivalve and gastropod species is shown to the left. RLCDs can vary between species and in most cases are not homologous. See supplementary figures S7–S11, Supplementary Material online for detailed Bayesian phylogenetic analysis for each mantle secreted gene family.
RLCDs were often part of these novel protein architectures. In the case of carbonic anhydrase, a RCLD inserted into the middle of the functional domain prior to pteriomorph cladogenesis has been maintained in multiple members of this lineage (fig. 5 and supplementary fig. S7, Supplementary Material online). There were also cases of different RCLDs becoming part of the same gene family independently, as occurred in gastropod and pteriomorph bivalve carbonic anhydrases, which each possess different RCLDs (fig. 5 and supplementary fig. S7, Supplementary Material online).
Some Protein Domains Are Repeatedly Co-Opted into Mantle Secretomes
Functional convergences, detected by the presence of particular protein domains, can occur independent of the level of the gene and may be missed using orthology inference. Phylostratigraphic analysis of domains present within proteins encoded by mantle secretome genes (supplementary table S9, Supplementary Material online) revealed a suite of known functional domains that originated before conchiferan cladogenesis (fig. 6). Together, these domains spanned a range of known and putative functionalities, including hydrolysis, protein phosphorylation, oxidation–reduction processes, extracellular interactions, immunity, and calcium and carbohydrate metabolism (fig. 6).
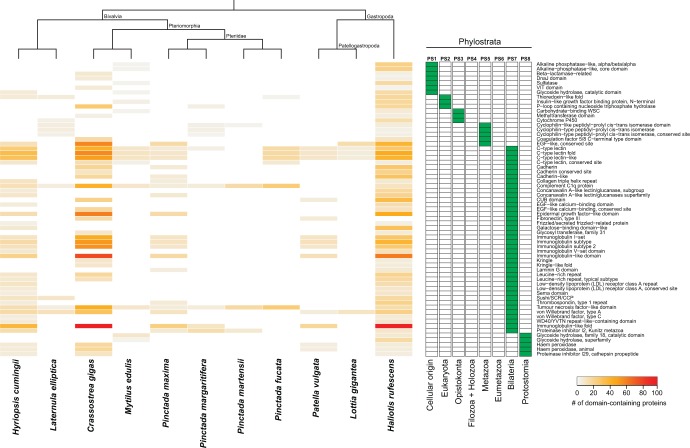
Fig. 6.
Distribution and abundance of enriched protein domains expressed in mantle secretomes. The heatmap depicts the number of domain-containing proteins expressed in the mantle secretome of a given species. Enriched protein domains (rows) are clustered according to their evolutionary origin (green boxes), and species (columns) are grouped according to their phylogenetic relationship. The eight phylostrata correspond to those in figure 1. Only enriched protein domains present in at least two species are shown. For a comprehensive list of enriched protein domains across phylostrata and species, see supplementary table S9, Supplementary Material online.
Each species expressed a unique combination of these conserved domains in their mantles, with some species having a far greater representation (e.g., H. rufescens) than other species (e.g., L. gigantea) (fig. 6 and supplementary table S9, Supplementary Material online). The differential enrichment of these various domains in the mantle transcriptomes in particular subsets of species was despite the high likelihood that these domains were encoded in the genomes of all these conchiferans.
Discussion
Mantle Secretomes are Encoded by Genes with Disparate Origins
Proteins secreted from the dorsal epithelium of the mantle contribute to both the shell matrix and the templating of the shell architecture (Kocot et al. 2016). Determination of the age of mantle-expressed genes by phylostratigraphy (fig. 1) indicates that the mantle secretomes of these 11 species have similar age profiles. The largest proportion of genes are in the youngest age category and appear to be species- or lower rank-specific, lending support to the premise that mantle secretomes consist of many unique and rapidly evolving genes (Jackson et al. 2006, 2010). However, these secretomes also have a wide range of gene families that originated before the evolution of conchiferans. For instance, there is a marked enrichment in genes estimated to have originated before bilaterian and molluscan cladogenesis. As these gene families originated before the evolution of the conchiferan mantle, they would have been co-opted into their role in shell formation.
This more ancient class of secretome proteins are enriched in domains that have known hydrolytic activity, and are involved in protein–protein interactions that occur on the cell surface or in the extracellular matrix, including EGF, immunoglobulin, laminin, fibronectin, cadherin and C-type lectin domains. Other domains detected in this analysis include those present in shell proteomes, such as EF-hand, C-type lectin, EGF, von Willebrand factor, Low-density lipoprotein, sushi, and Kunitz proteinase inhibitor domains (Marie et al. 2010, 2012, 2013; Marie, Trinkler, et al. 2011; Marie, Zanella-Cleon, et al. 2011; Mann and Edsinger 2014; Mann and Jackson 2014). Additionally, several of these domains are present in echinoderm skeletons, including thrombospondin, semaphorin and leucine-rich repeat domains (Mann et al. 2008, 2010). The domains shared in the matrix of biomineralized structures in molluscs and sea urchins are likely playing a common role in the biomineralization process in these disparate bilaterians.
Most of the mantle secretome genes that evolved after the divergence of bivalves and gastropods originated along specific lineages, with most taxa possessing a high number of gene families that appear to be species-specific (fig. 3, terminal branches), except for Pinctada spp., L. elliptica and P. vulgata. In the case of Pinctada spp., this may be because of the short divergence times between the four species included in this study (Cunha et al. 2011). It is likely that lineage-restricted gene families have been evolving at an equal rate over the course of bivalve and gastropod evolution, with newly emerged genes either being regularly lost from the mantle regulatory network, or evolving at rates sufficient to prevent detection of orthology. Thus, this apparent enrichment of novel genes along the terminal branches (i.e., species-specific genes) probably reflects the sampling of extant mantle transcriptomes.
Lineage-specific secreted gene families that evolved in early bivalves (21 genes) or gastropods (7 genes) and that have been maintained in at least two species in each class are likely to be playing an important function in shell formation. The same can be said for other lineage-specific innovations that have been conserved. For instance, 76 new gene families evolved in the stem leading to the pearl oysters (Pinctada spp.) and have been maintained since the divergence of P. maxima + P. margaritifera and P. fucata + P. martensii lineages some 20 Ma (Cunha et al. 2011).
Many of these taxon-specific secretome gene products are known shell matrix proteins, including lustrin A, sometsuke, basic protein N23, N16, shematrin, and KRMP. They often possess RLCDs and tend to be amongst the most highly expressed genes in the mantle (Jackson et al. 2010; Marie et al. 2010, 2012, 2013; Kinoshita et al. 2011; Mann and Jackson 2014). Together, these observations are consistent with these taxon-restricted genes playing essential roles in the structure, function and/or patterning of shells of diverse conchiferans.
RLCD-containing proteins also appear to have diverse origins. In some cases, RLCDs are present in evolutionarily ancient proteins, such as carbonic anhydrase, while in other cases they comprise evolutionarily young proteins that contain no additional domains, such as the shematrins and KRMPs in pearl oysters (McDougall et al. 2013). Regardless of overall domain architecture, RLCD domains appear to have a proclivity to expand and contract, and are likely to be intrinsically unstable (Evans 2012). For these reasons they have been proposed to be a driving factor in the evolvability of the molluscan shell (McDougall et al. 2013).
Mantle Secretomes are Complex and Diverse
Given the mantle tissue is considered a molluscan synapomorphy (Kocot et al. 2016), it is reasonable to infer that shell diversity is explained by an essential and conserved set of structural proteins that are secreted and guide shell fabrication—a shell biomineralization “toolkit”. From a nested comparison of conchiferan mantle secretomes, which includes sister Pinctada species that diverged ∼20 Ma (Cunha et al. 2011), other pteriomorphs, more distantly related bivalves, and gastropods, we find no compelling evidence for such a toolkit. Instead we find that these mantle secretomes are typified by their complexity and disparate molecular composition.
Despite broad-scale differences, there are a number of commonalities in the bivalve and gastropod mantle secretomes that appear to have evolved convergently. First, these secretomes are complex and are encoded by hundreds of genes. Second, despite the lack of clear orthology, many of these gene products are comprised of a suite of functionally similar domains, including those involved in hydrolysis, and protein interactions in the extracellular matrix or on the cell surface. The consistent independent co-option of genes encoding these domains into the mantle regulatory network suggests that these classes of domains are critical to shell fabrication. Consistent with the premise that there are extensive convergences in the domains expressed in the mantle secretome is the presence of species-specific RLCD repertoires. Although these rapidly evolving domains vary markedly within and between species, they have structural commonalities, including having biased amino acid composition (glycine- or alanine-rich), and being highly repetitive, modular and intrinsically disordered (Evans 2012; McDougall et al. 2013, 2016).
The Periphery of the Mantle Gene Regulatory Network Is Highly Evolvable
Shell formation is the terminal morphogenetic process of the dorsal mantle epithelium, and is downstream of the developmental processes that specify, determine and pattern these cells (reviewed by Jackson and Degnan 2016). The extensive lineage- and species-specific gene novelties and co-option of ancient genes into the mantle secretome indicates that entering and exiting the regulatory program underlying shell fabrication occurs continuously through evolution. This is consistent with these genes being on the periphery of a gene regulatory network that controls terminal outputs leading to shell fabrication. Terminal outputs of differentiated cells are typically under the control of a small number of transcription factors (Davidson 2010). Leaving or falling under the controls of this relatively simple regulatory regime typically requires less changes in the cis-regulatory architecture of the target gene (i.e., mantle secretome genes). This allows genes to enter and leave this network at a relatively high rate, which is observed by the dynamic changes in the mantle secretomes of the bivalves and gastropods investigated here. For example, expression levels of orthologous tyrosinase genes differ between the sister species P. maxima and P. margaritifera, indicating that the regulation of these genes has changed since these species diverged (Aguilera et al. 2014).
Conclusion
The genes encoding proteins secreted from the mantle vary markedly between species of gastropods and bivalves. The evolution of gene regulation appears to be the major driver underpinning these differences, with genes of different ages and composition being continuously co-opted into and lost from this regulatory network. Being at the terminus of the mantle gene regulatory network, relatively small changes in the cis-regulatory architecture of these genes allows for inclusion in and exclusion from the mantle secretome. While these regulatory mechanisms can account for the broad-scale differences in the composition of the mantle secretomes in these conchiferan species, this process is not the only contributor to shell diversity. In addition, many of the secretome coding sequences are rapidly evolving. This appears to be primarily driven by the high prevalence of RLCD-containing proteins in the mantle secretome. These simple, modular and intrinsically disordered sequences evolve rapidly and have a tendency to expand and contract in size over short evolutionary periods (McDougall et al. 2013). The continual recruitment and deletion of genes from the regulatory architecture controlling expression of mantle secretomes, and the rapid evolution of the protein domains comprising these secretomes, together provide a molecular explanation for the evolution and diversity of the molluscan shell.
Materials and Methods
Conchiferan Mantle Transcriptome Data Collection
All mantle transcriptomes were generated from adult animals, allowing for direct comparisons. Most mantle transcriptomes were made by pooling RNA from different individuals, except for H. cumingii, C. gigas, P. vulgata and L. gigantea (supplementary table S10, Supplementary Material online). Mantle transcriptomes were sequenced using different technologies including Sanger, 454 pyrosequencing and Illumina (Clark et al. 2010; Joubert et al. 2010; Kinoshita et al. 2011; de Wit and Palumbi 2012; Zhang et al. 2012; Bai et al. 2013; Freer et al. 2014), and were downloaded from publicly available databases (supplementary table S10, Supplementary Material online). Raw 454 reads from P. martensii whole adult mantle tissue were kindly provided by Dr. Yaohua Shi (Hainan University, China) (Shi et al. 2013). Assembled contigs from P. vulgata whole adult mantle tissue were kindly provided by Dr. Sebastian Shimeld (University of Oxford, UK) (Werner et al. 2013).
Total RNA was extracted from whole adult mantle of six adult P. maxima provided by Clipper Pearls/Autore Pearling, Broome, Western Australia, Australia. These were pooled in equal amounts to generate a mixed sample for library preparation and sequencing using a 454 FLX Plus sequencer. P. maxima 454 mantle reads were submitted to NCBI SRA database (accession number NCBI-SRA: SRR4020114).
De Novo Assemblies and Prediction of Mantle Secreted Proteins
Raw 454 sequencing reads from P. fucata, which correspond to mantle edge and mantle pallial from adult specimens (Kinoshita et al. 2011), were pooled and then processed as whole mantle tissue for further comparative analysis. 454 and Illumina raw reads were filtered and trimmed using the NGS QC Toolkit v2.2.3 with default settings (Patel and Jain 2012). For H. cumingii, L. elliptica, M. edulis, P. maxima, P. margaritifera, P. martensii and P. fucata, filtered-454 sequences were de novo assembled using MIRA v.3.4.0 (Chevreux et al. 2004), with the following parameters: job = denovo,est,accurate,454 -fastq COMMON_SETTINGS -noclipping -notraceinfo -GE:not = 4 -AS:sep = yes:urd = no 454_SETTINGS -AS:mrl = 200 -AS:mrpc = 1 -OUT:sssip = yes. For C. gigas and H. rufescens, filtered-Illumina reads were de novo assembled using Trinity software (v2013-02-25) (Grabherr et al. 2011) with default settings and a minimum transcript length of 200 nucleotides. For L. gigantea, Sanger-sequenced ESTs were assembled using CAP3 software (v12/21/07) (Huang and Madan 1999) with a sequence identity of 95%. To remove redundant sequences from each mantle transcriptome assembly, clustering was performed using CAP3 software (v12/21/07) (Huang and Madan 1999) requiring at least 95% sequence identity and a maximum unmatched overhang of 30 nucleotides. Resulting contigs and singletons from each species were translated into the six possible open reading frames. The longest open reading frames (ORFs) beginning with a methionine residue were selected using a custom ruby script. ORFs shorter than 50 amino acids were removed from further analysis. Statistics from the assemblies and sequence analysis are presented in supplementary tables S11–S12, Supplementary Material online.
Predicted ORFs were searched for signal peptides using a local installation of SignalP v4.1 (Petersen et al. 2011), as per Jackson et al. 2010, with the following parameters: -s best –t euk –u 0.45 -U 0.50. Proteins predicted as nonsecretory were classified as cytosolic proteins. Signal peptide-positive proteins were additionally screened for transmembrane domains using the THMHH v2.0 webserver (Krogh et al. 2001), or for mitochondrial, Golgi, or lysosomal targeting using the TargetP v1.1 webserver (Emanuelsson et al. 2007). Positive proteins were classified as transmembrane or cytosolic, respectively.
To evaluate whether secreted proteins predicted from transcriptome data have evidence of being involved in molluscan shell formation, we performed BLASTP v2.2.28+ (Camacho et al. 2009) comparisons between mantle secretomes and previously published molluscan shell proteomes, using an e-value cut-off of 10−6. Molluscan shell proteomes include 267 proteins from C. gigas reported by Marie, Zanella-Cleon, et al. (2011) and Zhang et al. (2012), 75 proteins from P. fucata reported by Liu et al. (2015), 78 proteins from P. margaritifera reported by Marie et al. (2012), 42 proteins from P. maxima reported by Marie et al. (2012), and 827 proteins from L. gigantea reported by Mann et al. (2012), Marie et al. (2013) and Mann and Edsinger (2014).
Phylostratigraphic Analysis
Gene age estimations were based on the phylostratigraphic approach, as described previously (Domazet-Lošo et al. 2007), using a consensus phylogenetic tree from the NCBI Taxonomy database. Using the BLASTP v2.2.28+ (e-value cut-off of 10−3) (Camacho et al. 2009), secreted protein sequences from each conchiferan species were compared with a custom nonredundant protein database comprised of 15,637,497 protein sequences from 1,848 species across the three domains of life (supplementary table S13, Supplementary Material online). In addition, TBLASTN searches v2.2.28+ (e-value cut-off of 10−15) (Camacho et al. 2009) were performed against EST sequences of Bivalvia, Gastropoda and other molluscan classes (supplementary table S13, Supplementary Material online), as complete annotated genomes are still limited or lacking for these internodes. Using the obtained BLAST outputs, we mapped the secreted proteins onto the consensus phylogenetic map of each species. If no BLAST hit was reported, the corresponding protein was assigned to the newest phylostratum (PS13). Otherwise, we used the most phylogenetically distant BLAST match to assign the evolutionary origin to a gene.
Estimation of Relative Gene Expression
Expression levels of genes encoding secreted proteins were assessed for all studied species, except P. vulgata. First, high-quality reads from each species were mapped to their respective mantle transcriptome using BWA v0.7.12-r1039 (Li and Durbin 2009), with default parameters. Then, eXpress v1.5.1 software (Roberts and Pachter 2013) was used to calculate the expression levels for each gene in the transcriptome. Expression levels were normalized by sequencing depth and converted into TPM (Transcripts Per Million) using eXpress (Roberts and Pachter 2013).
To determine whether genes classified as co-opted, lineage-specific and species-specific, as well as the gene age of secreted proteins, are correlated with relative gene expression levels, all genes encoding secreted proteins were classified as quartiles of levels of expression. No cut-off, in terms of TPM, was applied for these analyses.
Detection of Orthologous Secreted Proteins and Inference of the Organismal Tree
To group sequences into secreted gene families, a similarity search was performed (all-against-all BLASTP; e-value cut-off of 10−5) using the predicted secreted proteins from all 11 mantle transcriptomes. Gene families were constructed using OrthoMCL v2.0.9 (Li et al. 2003). Different inflation values were evaluated to identify the optimal parameters that generated the maximum number of OrthoMCL clusters (i.e., secreted gene families). From this analysis, an inflation value of 2.7 was selected (supplementary fig. S12, Supplementary Material online).
Secreted gene families with sequences from at least 6 of the 11 taxa were kept for further organismal tree inference. For every secreted gene family, a multiple alignment was performed with MAFFT v5 (Katoh et al. 2005) using the default alignment strategy, and trimmed with Gblocks v0.91b (Castresama 2000) to select conserved regions. Any orthologous groups shorter than 50 amino acids in length after trimming were discarded for further analysis. To screen secreted gene families for evidence of paralogy, splice variants or assembly errors, parsimonious trees (1,000 bootstrap replicates) were inferred with MEGA v5.2.2 (Tamura et al. 2011). All but one of the sequences from the same taxon were excluded from the orthologous group if they were monophyletic with a bootstrap support of >80% (i.e., paralogues). Secreted gene families that still had taxa with multiple sequences were visually inspected and excluded if orthology was unable to be determined. Remaining alignments were concatenated into a supermatrix with ScaFoS v1.2.5 (Roure et al. 2007).
Phylogenomic analyses were conducted using Maximum Likelihood (ML) in RAxML v8.0.2 (Stamatakis 2014) and Bayesian Inference (BI) in Phylobayes MPI-version 1.5a (Lartillot et al. 2013). Leaf stability and taxonomic instability were calculated for all taxa using the RogueNaRok algorithm (Aberer et al. 2013) (supplementary table S14, Supplementary Material online). ML analysis was performed using the PROTGAMMAWAGF substitution model and the topological support was assessed with 100 replicates of nonparametric bootstrapping. BI analysis was performed using the CAT-GTR substitution model (Lartillot and Philippe 2004). Three independent Markov Chain Monte Carlo (MCMC) chains were run for 15,000 cycles each, with the first 20% discarded as burn-in. Stationary state was assessed from acceptable values of “maxdiff: 0.0625” for the construction of the consensus tree. A 50% majority-rule consensus tree was computed from the combined remaining trees from three independent runs.
Patterns of Gene Family Gain-and-Loss in Conchiferans
The most parsimonious evolutionary scenario for the gain and loss of mantle secreted gene families within branches of the organismal tree was inferred using the DOLLOP program from the PHYLIP package v3.695 (Felsenstein 2005). The DOLLOP program is based on the Dollo’s parsimony law, which assumes that genes arise once on the evolutionary tree and can be lost independently in different evolutionary lineages (Farris 1977). This means that once a gene family is predicted to be lost in one or more lineages, based on its expression in the mantle, it can no longer be regained during evolution.
To determine whether gene gain was due to the birth of novel genes or the co-option of pre-existing genes into a mantle-specific role, we estimated the emergence of each secreted gene family using phylostratigraphic profiling, as is described earlier. From this analysis, we were able to classify ancient and lineage-specific gene families, and doing so, infer co-option events during conchiferan evolution.
Functional Annotation of Gene Families and Enrichment Analyses
Secreted gene families were functionally annotated using Blast2GO v1.1.4 with a BLASTP e-value filter of 10−6, an annotation cut-off value of 45, and GO (Gene Ontology) weight of 5 (Conesa et al. 2005). InterPro domain searches were performed using the built-in feature of Blast2GO, and protein domain enrichment was conducted using the FatiGO software on the interactive online platform Babelomics (Al-Shahrour et al. 2007; Medina et al. 2010). Proteins that mapped to a particular GO category were explicitly included into all parental categories. GO annotations per secreted gene family were obtained by listing the GO labels for all the proteins within that particular family. Fisher’s exact tests were employed to estimate whether secreted gene families from a given evolutionary time point (i.e., internal and terminal branches in the species tree) were enriched in specific GO categories or InterPro domains when compared against the background dataset (i.e., all secreted gene families) (Al-Shahrour et al. 2007). P values were adjusted by False Discovery Rate (FDR) (Benjamin and Hochberg 1995), and an adjusted P value of <0.05 was chosen as the significant threshold. Heatmaps were built using the heatmap.2 function in the gplots R package (R Development Core Team 2014).
Identification of RLCD-Containing Proteins
Secreted gene families were screened for the presence of RLCDs by using XSTREAM v1.73 software (Newman and Cooper 2007) with the following parameters: minimum character identity: 0.7; minimum TR domain length: 10 amino acids; minimum period: 1; and maximum gaps: 3. Shematrin and KRMP proteins, two well-known RLCD-containing proteins in pearl oysters (McDougall et al. 2013), were used as positive controls. Sequences that were found to contain RLCDs were additionally interrogated for the presence of sequence repeats in the InterPro database (Quevillon et al. 2005).
Secreted Gene Family Phylogenetic and Domain Structure Analyses
To reconstruct the evolutionary history of known shell-forming gene families and gene families containing domains known to be important for biomineralization, targeted searches were performed on the mantle secretome datasets. This search was also extended to molluscan shell proteomes (Marie et al. 2010, 2012, 2013; Mann et al. 2012; Zhang et al. 2012; Mann and Jackson 2014), the NCBI nonredundant database, and genomes of eukaryotic biomineralizing taxa (e.g., calcareous sponge, coccolithophorid, coral, chicken, human, mouse and sea urchin).
HMMER v3.1b2 searches (Finn et al. 2014) were performed using default parameters, with an inclusive E-value of 0.05, and the following PFAM domains were used as HMM profiles: carbonic anhydrase (PF00194); SPARC (PF10591); zona pellucida-like domain (PF00100); tyrosinase (PF00264); glycoside hydrolase family 18 (PF00704); polysaccharide deacetylase (PF01522); and chitin-binding domain (PF03067). The retrieved protein sequences from the eukaryote biomineralizing taxa were searched for a signal peptide as described earlier, and only secreted proteins were kept for further analysis. Domain architecture for each protein was determined using the PFAM database (Finn et al. 2014), and representations of domain architectures were undertaken using the MyDomain tool (Hulo et al. 2008). Phylogenetic analyses were performed as described elsewhere (Aguilera et al. 2013), and all phylogenetic trees were visualized and edited using FigTree v1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/; last accessed 30 December 2016). All alignments are available upon request.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
All authors conceived and designed the study, and wrote the article. F.A. carried out all the experimental and computational analyses. C.M. contributed with P. maxima sample collection.
Supplementary Material
Acknowledgments
We thank Patrick Moase and Clipper Pearls/Autore for kindly providing P. maxima specimens to undertake mantle transcriptome using 454 pyrosequencing; Dr Sebastian Shimeld (University of Oxford, UK) for providing mantle transcriptome assemblies from P. vulgata; and Dr Yaohua Shi (Hainan University, China) for providing raw 454 mantle data from P. martensii. F.A. was supported by a Becas Chile scholarship from CONICYT, Chile. This study was supported by funding from the Australian Research Council to B.M.D.
References
- Aberer AJ, Krompass D, Stamatakis A.. 2013. Pruning rouge taxa improves phylogenetic accuracy: an efficient algorithm and webservice. Syst Biol. 62:162–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera F, McDougall C, Degnan BM.. 2014. Evolution of the tyrosinase gene family in bivalve molluscs: independent expansion of the mantle gene repertoire. Acta Biomater. 10:3855–3865. [DOI] [PubMed] [Google Scholar]
- Aguilera F, McDougall C, Degnan BM.. 2013. Origin, evolution and classification of type-3 copper proteins: lineage-specific gene expansions and losses across the Metazoa. BMC Evol Biol. 13:96.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shahrour F, Minguez P, Tárraga J, Medina I, Alloza E, Montaner D, Dopazo J.. 2007. FatiGO+: a functional profiling tool for genomic data. Integration of functional annotation, regulatory motifs and interaction data with microarray experiments. Nucleic Acids Res. 35:W91–W96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Z, Zheng H, Lin J, Wang G, Li J.. 2013. Comparative analysis of the transcriptome in tissues secreting purple and white nacre in the pearl mussel Hyriopsis cumingii. PLoS One 8:e53617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 57:289–300. [Google Scholar]
- Budd A, McDougall C, Green K, Degnan BM.. 2014. Control of shell pigmentation by secretory tubules in the abalone mantle. Front Zool. 11:62. [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL.. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresama J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Chateigner D, Hedegaard C, Wenk H-R.. 2000. Mollusc shell microstructures and crystallographic textures. J Struct Geol. 22:1723–1735. [Google Scholar]
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WEG, Wetter T, Suhai S.. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14:1147–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MS, Thorne MAS, Vieira FA, Cardoso JCR, Power DM, Peck LS.. 2010. Insights into shell deposition in the Antarctic bivalve Laternula elliptica: gene discovery in the mantle transcriptome using 454 pyrosequencing. BMC Genomics 11:362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Götz S, Garcia-Gómez JM, Terol J, Talón M, Robles M.. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. [DOI] [PubMed] [Google Scholar]
- Cunha RL, Blanc F, Bonhomme F, Arnaud-Haond S.. 2011. Evolutionary patterns in pearl oysters of the genus Pinctada (Bivalvia: Pteriidae). Mar Biotechnol. 13:181–192. [DOI] [PubMed] [Google Scholar]
- Davidson EH. 2010. Emerging properties of animal gene regulatory networks. Nature 468:911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit P, Palumbi SR.. 2012. Transcriptome-wide polymorphisms of red abalone (Haliotis rufescens) reveal patterns of gene flow and local adaptation. Mol Ecol. 22:2884–2897. [DOI] [PubMed] [Google Scholar]
- Dix TG. 1972. Histochemistry of mantle and pearl sac secretory cells in Pinctada maxima (Lamellibranchia). Australian J Zool. 20:359–368. [Google Scholar]
- Domazet-Lošo T, Brajković J, Tautz D.. 2007. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 23:533–539. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H.. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2:953–971. [DOI] [PubMed] [Google Scholar]
- Evans JS. 2012. Aragonite-associated biomineralization proteins are disordered and contain interactive motifs. Bioinformatics 28:3182–3185. [DOI] [PubMed] [Google Scholar]
- Farris JS. 1977. Phylogenetic analysis under Dollo’s law. Syst Zool. 26:77–88. [Google Scholar]
- Felsenstein J. 2005. PHYLIP (Phylogeny Inference Package) version 3.6. Seattle: Department of Genome Sciences, University of Washington. [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. 2014. Pfam: the protein families database. Nucleic Acids Res. 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freer A, Bridgett S, Jiang J, Cusack M.. 2014. Biomineral proteins from Mytilus edulis mantle tissue transcriptome. Mar Biotechnol. 16:34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabara D, Ohmori F, Kinoshita S, Koyama H, Mizutani S, Ota A, Osakabe Y, Nagai K, Maeyama K, Okamoto K, et al. 2014. Novel genes participating in the formation of prismatic and nacreous layers in the pearl oyster as revealed by their tissue distribution and RNA interference knockdown. PLoS One 9:e84706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi T, Schwarzinger C, Miksik I, Smrz M, Beran A.. 2009. Molluscan shell evolution with review of shell calcification hypothesis. Comp Biochem Physiol B Biochem Mol Biol. 154:351–371. [DOI] [PubMed] [Google Scholar]
- Gao P, Liao Z, Wang X-x, Bao L-f, Fan M-h, Li X-m, Wu C-w, Xia S-w.. 2015. Layer-by-layer proteomic analysis of Mytilus galloprovincialis shell. PLoS One 10:e0137487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A.. 1999. CAP3: A DNA sequence assembly program. Genome Res. 9:868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJ.. 2008. The 20 years of PROSITE. Nucleic Acid Res. 36:D245–D249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbour-Zahab R, Chagot D, Blanc F, Grizel H.. 1992. Mantle histology, histochemistry and ultrastructure of the pearl oyster Pinctada margaritifera (L.). Aquat Living Resour. 5:287–298. [Google Scholar]
- Jackson DJ, Degnan BM.. 2016. The importance of evo-devo to an integrated understanding of molluscan biomineralisation. J Struct Biol. 196:67–74. [DOI] [PubMed] [Google Scholar]
- Jackson DJ, McDougall C, Green K, Simpson F, Worheide G, Degnan BM.. 2006. A rapidly evolving secretome builds and patterns a sea shell. BMC Biol. 4:40.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DJ, McDougall C, Woodcroft B, Moase P, Rose RA, Kube M, Reinhardt R, Rokhsar DS, Montagnani C, Joubert C, et al. 2010. Parallel evolution of nacre building gene sets in molluscs. Mol Biol Evol. 27:591–608. [DOI] [PubMed] [Google Scholar]
- Jackson DJ, Wörheide G, Degnan BM.. 2007. Dynamic expression of ancient and novel molluscan shell genes during ecological transitions. BMC Evol Biol. 7:160.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Berland S, Milet C, Borzeix S, Lopez E, Doumenc D.. 2004. Zonal localization of shell matrix proteins in mantle of Haliotis tuberculata (Mollusca, Gastropoda). Mar Biotechnol. 6:541–551. [DOI] [PubMed] [Google Scholar]
- Joubert C, Piquemal D, Marie B, Manchon L, Pierrat F, Zanella-Cleon I, Cochnnec-Laureau N, Gueguen Y, Montagnani C.. 2010. Transcriptome and proteome analysis of Pinctada margaritifera calcifying mantle and shell: focus on biomineralization. BMC Genomics 11:613.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K-i, Toh H, Miyata T.. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita S, Wang N, Inoue H, Maeyama K, Okamoto K, Nagai K, Kondo H, Hirono I, Asakawa S, Watabe S.. 2011. Deep sequencing of ESTs from nacreous and prismatic layer producing tissues and a screen for novel shell formation-related genes in the pearl oyster. PLoS One 6:e21238.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocot KM, Aguilera F, McDougall C, Jackson DJ, Degnan BM.. 2016. Sea shell diversity and rapidly evolving secretomes: insights into the evolution of biomineralization. Front Zool. 13:23.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocot KM, Cannon JT, Todt C, Citarella MR, Kohn AB, Meyer A, Santos SR, Schander C, Moroz LL, Lieb B, et al. 2011. Phylogenomics reveals deep molluscan relationships. Nature 477:452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL.. 2001. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol. 305:567–580. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H.. 2004. A bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 21:1095–1109. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J.. 2013. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 62:611–615. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ Jr, Roos DS.. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Xu G, Xie J, Lee I, Xiang L, Wang H, Zhang G, Xie L, Zhang R.. 2015. Dual roles of the lysine-rich matrix protein (KRMP)-3 in shell formation of pearl oyster, Pinctada fucata. PLoS One 10:e0131868.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Z, Bao L-f, Fan M-h, Gao P, Wang X-x, Qin C-l, Li X-m.. 2015. In-depth proteomic analysis of nacre, prism, and myostracum of Mytilus shell. J Proteomics 122:26–40. [DOI] [PubMed] [Google Scholar]
- Liu C, Li S, Kong J, Liu Y, Wang T, Xie L, Zhang R.. 2015. In-depth proteomic analysis of shell matrix proteins of Pinctada fucata. Sci Rep. 5:17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Edsinger E.. 2014. The Lottia gigantea shell matrix proteome: re-analysis including MaxQuant iBAQ quantitation and phosphoproteome analysis. Prot Sci. 12:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Edsinger-Gonzales E, Mann M.. 2012. In-depth proteomic analysis of a mollusc shell: acid-soluble and acid-insoluble matrix of the limpet Lottia gigantea. Prot Sci. 10:28.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Jackson DJ.. 2014. Characterization of the pigmented shell-forming proteome of the common grove snail Cepaea nemoralis. BMC Genomics 15:249.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Poustka AJ, Mann M.. 2008. The sea urchin (Strongylocentrotus purpuratus) test and spine proteomes. Prot Sci. 6:22.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Wilt FH, Poustka AJ.. 2010. Proteomic analysis of sea urchin (Strongylocentrotus purpuratus) spicule matrix. Prot Sci. 8:33.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie B, Jackson DJ, Ramos-Silva P, Zanella-Cleon I, Guichard N, Marin F.. 2013. The shell-forming proteome of Lottia gigantea reveals both deep conservations and lineage-specific novelties. Febs J. 280:214–232. [DOI] [PubMed] [Google Scholar]
- Marie B, Joubert C, Tayalé A, Zanella-Cléon I, Belliard C, Piquemal D, Cochennec-Laureau N, Marin F, Gueguen Y, Montagnani C.. 2012. Different secretory repertoires control the biomineralization processes of prism and nacre deposition of the pearl oyster shell. Proc Natl Acad Sci U S A. 109:20986–20991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie B, Marie A, Jackson DJ, Dubost L, Degnan BM, Milet C, Marin F.. 2010. Proteomic analysis of the organic matrix of the abalone Haliotis asinina calcified shell. Prot Sci. 8:54.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie B, Trinkler N, Zanella-Cleon I, Guichard N, Becchi M, Paillard C, Marin F.. 2011. Proteomic identification of novel proteins from the calcifying shell matrix of the Manila clam Venerupis philippinarum. Mar Biotechnol. 13:955–962. [DOI] [PubMed] [Google Scholar]
- Marie B, Zanella-Cleon I, Guichard N, Becchi M, Marin F.. 2011. Novel proteins from the calcifying shell matrix of the Pacific oyster Crassostrea gigas. Mar Biotechnol. 13:1159–1168. [DOI] [PubMed] [Google Scholar]
- Marin F, Le Roy N, Marie B.. 2012. The formation and mineralization of mollusk shell. Front Biosci. 4:1099–1125. [DOI] [PubMed] [Google Scholar]
- Marin F, Le Roy N, Marie B, Ramos-Silva P, Bundeleva I, Guichard N, Immel F.. 2014. Metazoan calcium carbonate biomineralizations: macroevolutionary trends – challenges for the coming decade. Bull Soc Geol France 185:217–232. [Google Scholar]
- McDougall C, Woodcroft BJ, Degnan BM.. 2016. The widespread prevalence and functional significance of silk-like structural proteins in metazoan biological materials. PLoS One 11:e0159128.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougall C, Aguilera F, Degnan BM.. 2013. Rapid evolution of pearl oyster shell matrix proteins with repetitive, low-complexity domains. J R Soc Interface 10:20130041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougall C, Green K, Jackson DJ, Degnan BM.. 2011. Ultrastructure of the mantle of the gastropod Haliotis asinina and mechanisms of shell regionalization. Cells Tissues Organs 194:103–107. [DOI] [PubMed] [Google Scholar]
- Medina I, Carbonell J, Pulido L, Madeira SC, Goetz S, Conesa A, Tárraga J, Pascual-Montano A, Nogales-Cadenas R, Santoyo J, et al. 2010. Babelomics: an integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic Acids Res. 38:W210–W213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AM, Cooper JB.. 2007. XSTREAM: a practical algorithm for identification and architecture modeling of tandem repeats in protein sequences. BMC Bioinformatics 8:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RK, Jain M.. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7:e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H.. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. [DOI] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R.. 2005. InterProScan: protein domains identifier. Nucleic Acids Res. 33:W116–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2014. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Roberts A, Pachter L.. 2013. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods 10:71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roure B, Rodriguez-Ezpeleta N, Philippe H.. 2007. SCaFoS: a tool for selection, concatenation and fusion of sequences for phylogenomics. BMC Evol Biol. 7(Suppl 1):S2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Belcher AM, Hansma PK, Stucky GD, Morse DE.. 1997. Molecular cloning and characterization of lustrin A, a matrix protein from shell and pearl nacre of Haliotis rufescens. J Biol Chem. 272:32472–32481. [DOI] [PubMed] [Google Scholar]
- Shi Y, Yu C, Gu Z, Zhan X, Wang Y, Wang A.. 2013. Characterization of the pearl oyster (Pinctada martensii) mantle transcriptome unravels biomineralization genes. Mar Biotechnol. 15:175–187. [DOI] [PubMed] [Google Scholar]
- Sleight VA, Thorne MAS, Peck LS, Arivalagan J, Berland S, Marie A, Clark MS.. 2016. Characterisation of the mantle transcriptome and biomineralisation genes in the blunt-gaper clam, Mya truncata. Mar Genomics 27:47–55. [DOI] [PubMed] [Google Scholar]
- Smith SA, Wilson NG, Goetz FE, Feehery C, Andrade SC, Rouse GW, Giribet G, Dunn CW.. 2011. Resolving the evolutionary relationships of molluscs with phylogenomics tools. Nature 480:364–367. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sud D, Poncet J-M, Saïhi A, Lebel J-M, Doumenc D, Boucaud-Camou E.. 2002. A cytological study of the mantle edge of Haliotis tuberculata L. (Mollusca, Gastropoda) in relation to shell structure. J Shellfish Res. 21:201–210. [Google Scholar]
- Takeuchi T, Endo K.. 2005. Biphasic and dually coordinated expression of the genes encoding major shell matrix proteins in the pearl oyster Pinctada fucata. Mar Biotechnol. 8:52–61. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S.. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinther J. 2015. The origins of molluscs. Palaeontology 58:19–34. [Google Scholar]
- Werner GDA, Gemmell P, Grosser S, Hamer R, Shimeld SM.. 2013. Analysis of a deep transcriptome from the mantle tissue of Patella vulgata Linnaeus (Mollusca: Gastropoda: Patellidae) reveals candidate biomineralising genes. Mar Biotechnol. 15:230–243. [DOI] [PubMed] [Google Scholar]
- Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, Yang P, Zhang L, Wang X, Qi H, et al. 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490:49–54. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.