Abstract
WUS and WOX5, which are expressed, respectively, in the organizing center (OC) and the quiescent center (QC), are essential for shoot/root apical stem-cell maintenance in flowering plants. However, little is known about how these stem-cell factors evolved their functions in flowering plants. Here, we show that the WUS/WOX5 proteins acquired two distinct capabilities by a two-step functional innovation process in the course of plant evolution. The first-step is the apical stem-cell maintenance activity of WUS/WOX5, which originated in the common ancestor of ferns and seed plants, as evidenced by the interspecies complementation experiments, showing that ectopic expression of fern Ceratopteris richardii WUS-like (CrWUL) surrounding OC/QC, or exclusive OC-/QC-expressed gymnosperms/angiosperms WUS/WOX5 in Arabidopsis wus-1 and wox5-1 mutants, could rescue their phenotypes. The second-step is the intercellular mobility that emerged in the common ancestor of seed plants after divergence from the ferns. Evidence for this includes confocal imaging of GFP fusion proteins, showing that WUS/WOX5 from seed plants, rather than from the fern CrWUL, can migrate into cells adjacent to the OC/QC. Evolutionary analysis showed that the WUS-like gene was duplicated into two copies prior to the divergence of gymnosperms/angiosperms. Then the two gene copies (WUS and WOX5) have undergone similar levels of purifying selection, which is consistent with their conserved functions in angiosperm shoot/root stem-cell maintenance and floral organ formation. Our results highlight the critical roles and the essential prerequisites that the two-step functional innovation of these genes performs and represents in the origin of flowering plants.
Keywords: experimental evolution, plant molecular evolution, developmental biology, plant stem cells.
Introduction
The development of flowering plants depends on the activity of stem cells situated in specific niches (Dinneny and Benfey 2008; Heidstra and Sabatini 2014; Bennett et al. 2014). In higher plants, stem-cell niches most prominently occur in the shoot apical meristem (SAM) and the root apical meristem (RAM) (Scheres 2001; Weigel and Jürgens 2002; Busch et al. 2010). SAM activity controls the development of the aerial parts of the plant body, whereas RAM activity controls the development of the belowground parts. These plant stem-cell niches are maintained by two small groups of the so-called organizing cells, which occur at a site known as the organizing center (OC) in the SAM and a site known as the quiescent center (QC) in the RAM. The OC and QC provide microenvironments that function in determining the fate of surrounding stem cells (Heyman et al. 2013; Gaillochet et al. 2015).
WUSCHEL (WUS) and WUSCHEL-related homeobox (WOX) genes encode a large family (WOX gene family) of homeodomain (HD)-containing transcription factors that are known to regulate the formation and development of plant tissues; these genes are found in plant lineages, but not in other organisms such as bacteria or animals. (van der Graaff et al. 2009). Phylogenetic analyses have divided the WOX gene family into three major lineages: the WUS lineage, which contains WUS/WOX5 genes that are mainly present in seed plants (i.e., gymnosperms and angiosperms), the WOX9 lineage (mainly present in vascular plants), and the WOX13 lineage (present in all major plant lineages, including green algae and non-vascular mosses). With the exception of the HD that is common to all WOX family members, other unique motifs are shared only within one of the three lineages of WOX proteins (Deveaux et al. 2008; Nardmann et al. 2009; van der Graaff et al. 2009; Nardmann and Werr 2012; Lian et al. 2014).
In flowering plants, within the SAM, WUS is exclusively expressed in the OC; it controls shoot stem cell development. WOX5 is specifically expressed in the QC within the RAM, where it regulates root stem cell homeostasis (Scheres 2005; Forzani et al. 2014; Zhou et al. 2015). Additionally, WUS is known to be essential for the formation of functional floral organs (Sarkar et al. 2007). The flowering plants are the most diverse group of land plants; with about 350,000 species, they comprise about 90% of the plant kingdom. The ancestors of flowering plants emerged in the Triassic Period sometime between 202 and 245 million years ago (Ma). They diversified extensively during the Low Cretaceous, replacing the previously-dominant conifers (Bond and Scott 2010). The floral organs are the defining characteristics of the flowering plants, which increase the successful ratio of fertilization and facilitate the flowering plants rapid propagation after their divergence from the nonflowering plants. However, little is known concerning how flowering plants emerged with flower organs during plant evolution.
The members of the WUS/WOX5 family (WUS lineage) contain the WUS motif and the ERF-associated amphiphilic repression (EAR) motif (Nardmann et al. 2009; van der Graaff et al. 2009; Nardmann and Werr 2012), in addition to the invariably conserved, characteristic HD. The WUS motif is involved in transcriptional repression via cooperation with the EAR motif. Recent work has established that the WUS motif can recruit TPL/TPR corepressors to regulate the genes that control cell differentiation (Ikeda et al. 2009; Lin et al. 2013; Zhang et al. 2014; Pi et al. 2015). The stem-cell factor WUS establishes the shoot apical stem-cell niche through a CLAVATA3 (CLV3)−WUS feedback loop (Mayer et al. 1998; Brand et al. 2000; Schoof et al. 2000; Yadav et al. 2011; Perilli et al. 2012). The cell-to-cell movement of the WUS proteins is essential for this feedback loop (Yadav et al. 2011). Likewise, in the RAM, WOX5 establishes the root stem-cell niche through a feedback circuit involving auxin-related response factors (Sabatini et al. 1999; Blilou et al. 2005; Ding and Friml 2010; Yang et al. 2015). REPRESSOR OF WUSCHEL1 (ROW1) maintains both RAM and SAM development by confining the expression of WUS to the OC, and by confining WOX5 expression to the QC (Han et al. 2008; Han and Zhu 2009; Zhang et al. 2015; Kong et al. 2015). A recent study showed that HAIRY MERISTEM controls the development of the shoot and root stem-cell niches by interacting with, respectively, WUS and WOX5 (Zhou et al. 2015).
A previous study showed that the occurrence of WUS and WOX5 as separate genes was an evolutionary innovation of angiosperms, as only a single copy of WUS/WOX5 was identified in gymnosperms (Nardmann et al. 2009). However, both the separate WUS and WOX5 genes were recently identified in the gymnosperm Picea abies (Hedman et al. 2013). Interestingly, WOX5 and WUS have been shown to be functionally interchangeable in Arabidopsis shoot and root stem cell maintenance (Sarkar et al. 2007).
Despite the importance of WUS/WOX5 in plant apical stem-cell homeostasis and flower morphogenesis, little is known about how these conserved stem-cell factors evolved these important functions in flowering plants. Here, we expressed various ancestral WUS/WOX5 from extant plant species in the Arabidopsis WUS or WOX5 knockout mutants with the goal of studying the function of these stem-cell factors and elucidating their underlying evolutionary processes during the course of plant evolution. Our results reveal that a two-step functional innovation of WUS/WOX5 endowed these stem-cell factors with two distinct capabilities: apical stem-cell maintenance activity and intercellular mobility. These innovations enable WUS/WOX5 to noncell-autonomously regulate shoot and root stem-cell niches of the flowering plant and to control floral organ formation. This evolutionary innovation also seems to have been a critical prerequisite step that facilitated the emergence of functional floral organs in the origin of flowering plants.
Results
Survey of the WUS and WOX5 Genes in Plant Kingdom
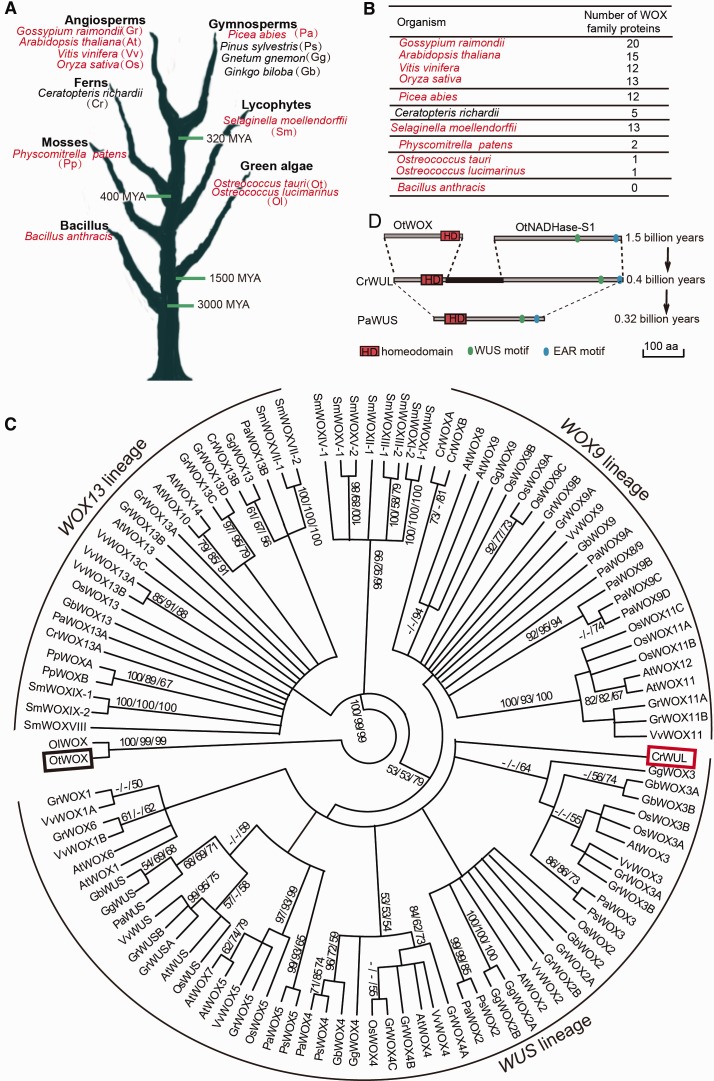
To obtain a broad view of the evolution of the WOX gene family, we surveyed the sequences of WOX family members in the plant kingdom. We used the full-length protein sequence of Arabidopsis thaliana WUS (AtWUS) as a query (Nardmann et al. 2009) to search against the available genome sequences from the bacterium Bacillus anthracis and 13 species from the plant kingdom that represented unicellular green algae, mosses, lycophytes, ferns, gymnosperms, and angiosperms (fig. 1A). We found that the number of WOX family genes increased substantially with the emergence of the vascular plant lineages (i.e., lycophytes, ferns, gymnosperms, and angiosperms) (WOX number >10), in contrast to the nonvascular plant lineages (i.e., mosses and green algae) (WOX number ≤ 2) (fig. 1B). Note that only five WOX genes were identified in the fern Ceratopteris richardii, which likely reflects the incomplete status of the genome sequence of this species.
Fig. 1.
Evolution and origin of the WUS/WOX5 genes. (A) Summary of the investigated plant species, from green algae to angiosperms, according to evolutionary history. Red indicates species whose genomes have been fully sequenced. (B) Number of WOX proteins in various organisms. The distribution of the WOX members showed that the number of the WOX genes is substantially increased in the vascular plants (i.e., lycophytes, ferns, gymnosperms, and angiosperms) as compared with the nonvascular plants (i.e., mosses and green algae). (C) Phylogenetic analysis of the WOX family in the plant kingdom. The tree was divided into three lineages: the WUS lineage, the WOX9 lineage, and the WOX13 lineage. The black box highlights the single WOX identified in the green alga O. tauri. The red box highlights the single copy of a WUS-like gene, belonging to the WUS lineage, in the fern Ceratopteris richardi (CrWUL). A phylogenetic analysis was performed using the NJ/MP/ML method with full-length protein sequences. Bootstrap values are shown on the branch points. Ot, Ostreococcus lucimarinus; Ot, Ostreococcus tauri; Pp, Physcomitrella patens; Sm, Selaginella moellendorffii; Cr, Ceratopteris richardii; Gb, Ginkgo biloba; Gg, Gnetum gnemon; Ps, Pinus sylvestris; Pa, Picea abies; Os, Oryza sativa; Vv, Vitis vinifera; At, Arabidopsis thaliana; Gr, Gossypium raimondii. (D) Alignment of CrWUL with OtWOX and OtNADHase-S1 indicates that the three conserved domains common to WUS/WOX5 might have originated in fern via gene fusion. The PaWUS protein from the gymnosperm P. abies is substantially smaller than the fern CrWUL. Scale bars: 100 amino acids (aa).
The deduced amino acid sequences of the WOX genes were aligned, and a phylogenetic tree was constructed (fig. 1C). The tree is similar to that of a previous report (Nardmann et al. 2009; Nardmann and Werr 2012) and can be divided into three major lineages according to the features of the conserved domain: the WOX13, the WOX9, and the WUS lineages (fig. 1C). The WOX13 lineage proteins are present in all major plant lineages, including the green algae and mosses. In addition to the conserved HD, the WOX13 proteins contain the WOX13 OG domain (supplementary fig. S1, Supplementary Material online). The WOX9 lineage is present in vascular plants; these proteins possess motifs that proteins of the other WOX lineages lack, such as the LQxG WOX8 motif (supplementary fig. S1, Supplementary Material online). The WUS lineage is mainly present in the seed plants. WUS lineage proteins are characterized by their WUS motifs (supplementary fig. S1, Supplementary Material online). Only one WOX protein, belonging to the WOX13 lineage, was identified in the green alga Ostreococcus tauri (Deveaux et al. 2008). However, the fern C. richardii (Cr) was found to have one or two WOX genes from each of the three lineages (fig. 1C). Notably, one WOX family member in C. richardii was grouped in the WUS lineage, because, in addition to its HD, this C. richardii WUS-like (CrWUL) protein also contains a WUS motif common to the WUS lineage and an EAR motif specific to the WUS/WOX5 proteins (Nardmann and Werr 2012) (supplementary fig. S1, Supplementary Material online). Rather than a single copy of WUS-like gene was found in the fern C. richardii, both the WUS and WOX5 genes are present in seed plants, including the gymnosperm P. abies and other angiosperm species (fig. 1C) (Hedman et al. 2013). This is consistent with the timing of the first whole-genome duplication event that was postulated to have occurred at the base of the seed plant lineage approximately 320 Ma (Jiao et al. 2011).
Although no WUS/WOX5 member was found in the green alga Ostreococcus tauri, we identified an nicotinamide adenine dinucleotide reduced dehydrogenase subunit 1 protein (OtNADHase-S1) that contains both WUS and EAR motifs, but no HD, that exists in Ostreococcus tauri (OtWOX) (fig. 1D and supplementary fig. S2, Supplementary Material online). Therefore, the fern C. richardii WUS-like (CrWUL) predecessor may have originated through a gene-fusion event that generated a protein containing all three conserved domains of WUS/WOX5. Moreover, the third, functionally essential HD is apparently missing in all plant species that originated prior to the fern lineage in which we theorize an important gene-fusion event occurred (supplementary fig. S3, Supplementary Material online). Notably, the fern CrWUL has a much longer protein sequence (591aa) than the WUS/WOX5 protein sequences of seed plants such as the gymnosperm P. abies WUS (PaWUS, 285aa) and PaWOX5 (207aa) (supplementary fig. S4, Supplementary Material online), among others (supplementary fig. S1, Supplementary Material online).
The Apical Stem-Cell Maintenance Functions of WUS and WOX5 in Shoots
All WOX genes in seed plants are known to have diverged from a single common ancestral WOX gene predecessor. With the goal of exploring which of the Arabidopsis thaliana WOX genes that evolved from the single green-algae–like ancestor function in maintaining shoot apical stem-cell niches (similar to WUS genes function), we analyzed all of the members of the WOX family with genetic complementation experiments using the Arabidopsis wus-1 mutant, which carries a loss-of-function mutation in WUS. In accordance with previous reports, we found that, in contrast to wild-type Arabidopsis, wus-1 mutant plants have a defective shoot meristem that terminates prematurely in an aberrant flat structure (fig. 2A and B, mid-upper panels) and fails to develop into a normal inflorescence (fig. 2A and B, upper panels) (Laux et al. 1996; Sarkar et al. 2007). Furthermore, the green fluorescent protein (GFP) of enhancer trap line J2341, which showed specific expression in the SAM of shoots (Kim et al. 2005) and in the distal meristem of roots in wild-type Arabidopsis (Ding and Friml 2010), was absent from wus-1 mutant background (fig. 2A and B, mid-lower panels), confirming the aborted development of the shoot meristem in the wus-1 mutant. Additionally, in contrast to the wild-type, wus-1 mutants showed severe defects in floral organ development, with only a single stamen and no gynoecium (fig. 2A and B , lower panels). Genetic complementation experiments showed that the expression of AtWOX5 and AtWUS driven by the native WUS promoter rescued both the premature termination of the shoot meristem and the floral organ developmental defects in Arabidopsis wus-1 (supplementary fig. S5a and b, Supplementary Material online). Other WOX members of WUS lineage, including AtWOX1, AtWOX2, or AtWOX3, when driven by Arabidopsis WUS promoter, could also partially rescue the aborted SAM with an indeterminate inflorescence meristem (supplementary fig. S5c–e, Supplementary Material online). Notably, the expression of the WUS lineage AtWOX4 failed to rescue the Arabidopsis wus-1 phenotype (supplementary fig. S5f, Supplementary Material online). AtWOX9 (WOX9 lineage) and AtWOX13 (WOX13 lineage) were also unable to rescue wus-1 shoot meristem defects (supplementary fig. S5g and h, Supplementary Material online). These results indicate that the ability to maintain the Arabidopsis shoot stem-cell niche is not a general property of all the AtWOX family members that ultimately diverged from the green-algae–like ancestor.
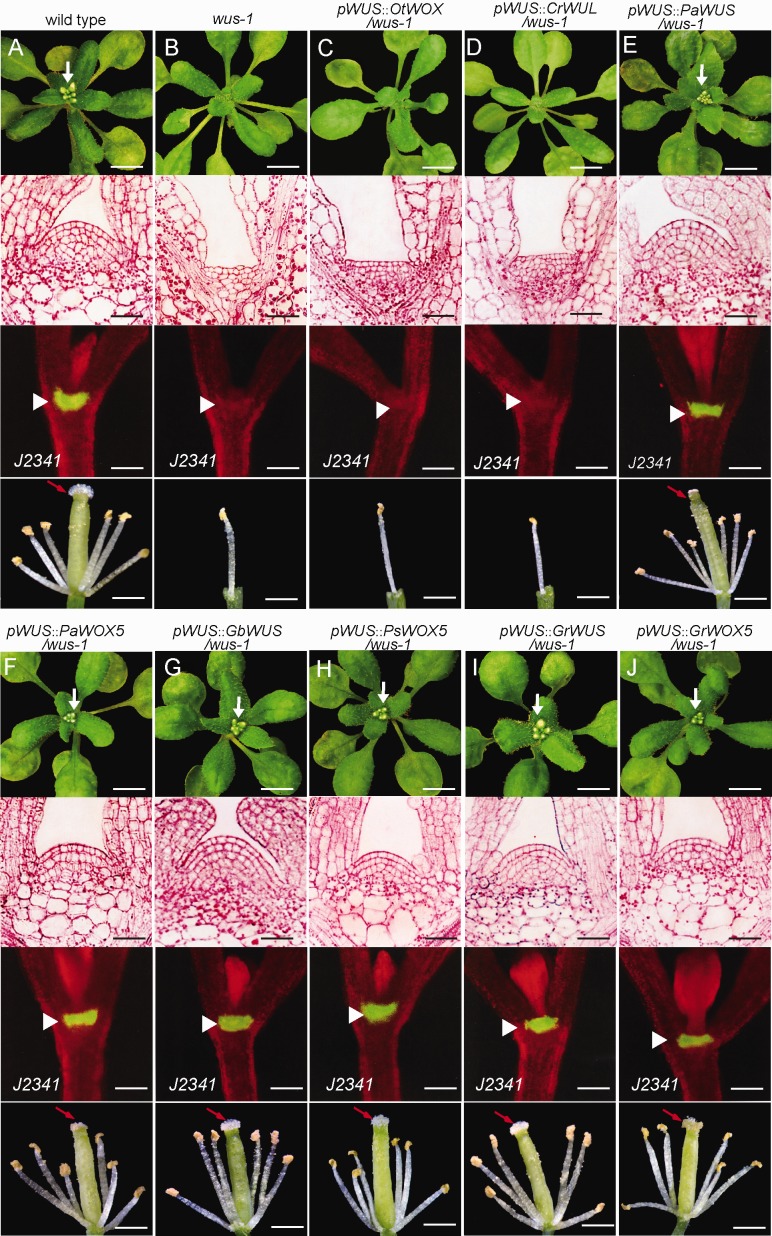
Fig. 2.
Evolutionary analysis of WUS/WOX5 function in the maintenance of the Arabidopsis shoot apical stem-cell niche and in floral organ development. (A, B) In contrast to the wild-type shoot (A), the wus-1 mutant (B) had defective shoot meristem developent (upper panel) that terminated prematurely in an aberrant flat structure (mid-upper panel). The GFP of enhancer trap line J2341, which showed specific expression in the SAM of wild-type Arabidopsis shoots, was absent from the wus-1 mutant (mid-lower panel). Flowers with four sepals and four petals removed (lower panel). Wild-type flowers contain six stamens and central gynoecium (A), while wus-1 flowers had only one central stamen present and lacked central gynoecium (B). (C–J) Interspecies complementation experiments with WUS/WOX5 orthologues from the green alga lineage to the angiosperm lineage expressed in Arabidopsis wus-1 mutants showed that OtWOX (green alga) and CrWUL (fern) are unable to maintain the shoot stem cell-niche and regulate floral organ morphogenesis (C, D), while the WUS/WOX5 orthologues from seed plants can rescue the defective wus-1 shoot meristem niche and floral organ phenotypes. White arrows indicate the shoot apex. Red arrows indicate the gynoecium. Scale bars: 1 cm (upper panel), 20 μm (mid-upper panel), 100 μm (mid-lower panel), and 1 mm (lower panel).
We next explored the functional origin of WUS and WOX5 proteins that maintain the Arabidopsis shoot apical stem-cell niche and floral organ development during plant evolution. Interspecies genetic complementation experiments revealed that expression of OtWOX (green alga), which lacks the WUS and EAR motifs, in Arabidopsis wus-1 was not able to rescue the phenotype with defective shoot meristem in aberrant flat structure and floral organ containing a single stamen and no gynoecium (fig. 2C). Strikingly, despite containing both WUS and EAR motifs, expression of the fern CrWUL protein in the Arabidopsis wus-1 mutant, using the WUS promoter, still failed to complement its defects in SAM maintenance or flower organ formation (fig. 2D). However, the expression of gymnosperm proteins, including PaWUS/PaWOX5, WUS from G. biloba (GbWUS), and WOX5 from P. sylvestris (PsWOX5), in Arabidopsis wus-1 mutant rescued Arabidopsis defects in both shoot apical stem-cell maintenance and flower organ formation (fig. 2E−H). Expression of GrWUS and GrWOX5 from the angiosperm Gossypium raimondii rescued the Arabidopsis wus-1 phenotypes, suggesting that GrWUS/GrWOX5 and AtWUS/AtWOX5 are functionally equivalent in determining shoot apical stem-cell fate and in flower morphogenesis (fig. 2I and J). These results imply that the functional WUS/WOX5 predecessor, maintaining the Arabidopsis SAM and floral organ development, might have originated in the common ancestor of the gymnosperms/angiosperms after its divergence from the fern lineage. These successful interspecies functional complementation experiments in which WUS/WOX5 proteins from various seed plants expressed in Arabidopsis wus-1 shoots reveals that the shoot apical stem-cell maintenance function of these proteins has been evolutionarily conserved since the separation of angiosperms and gymnosperms.
The Apical Stem-Cell Maintenance Functions of WUS and WOX5 in Roots
We performed a similar functional analysis of WOX5 in the maintenance of root stem-cell homeostasis using wox5-1, a loss-of-function mutant for AtWOX5. In contrast to wild-type Arabidopsis, wox5-1 mutants have a defective root distal meristem that seems to have undergone premature differentiation and is characterized by the accumulation of starch granules (fig. 3A and B,upper panel). Besides being absent from wus-1 mutants, the GFP of enhance-trap line J2341 showed specific expression in wild-type root distal stem cells (DSCs) and was also absent from wox5-1 mutant roots (fig. 3A and B, lower panel), confirming the aborted development of DSCs in wox5-1 roots. AtWOX5 and AtWUS, as driven by WOX5 promoter, were able to complement the DSC defects in the wox5-1 mutant (supplementary fig. S6a and b, Supplementary Material online). No other WOX members from any of the three lineages had the ability to maintain the Arabidopsis root stem-cell niche (supplementary fig. S6c–h, Supplementary Material online).
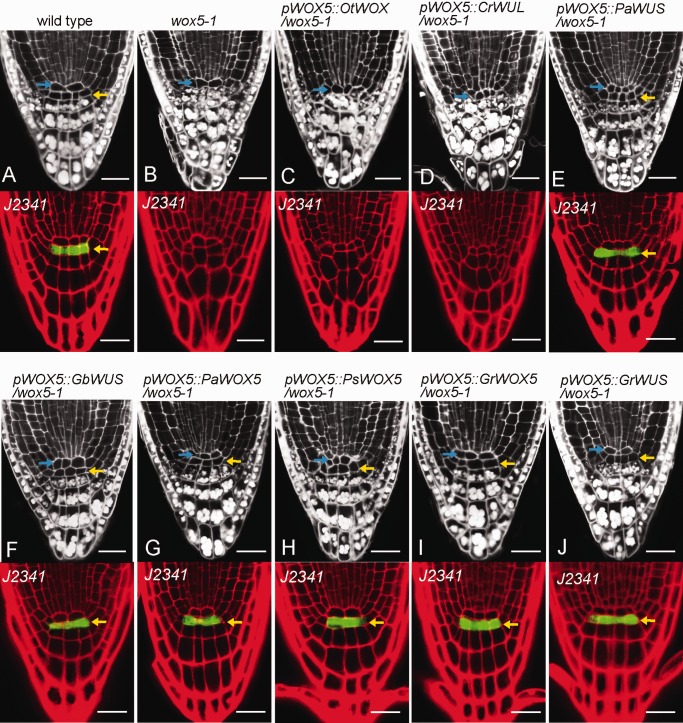
Fig. 3.
Evolutionary analysis of WOX5/WUS function in the maintenance of the Arabidopsis root stem-cell niche. (A) Wild-type root showing a normal root stem-cell niche. In the upper panel, undifferentiated DSCs (yellow arrowheads) below the QC (blue arrowheads) are characterized by the absence of starch, whereas white starch granules stained by the mPS-PI method are visible in differentiated columella cells below the DSCs. (B) The wox5-1 mutant failed to maintain root stem cells, lacked DSCs, and showed premature differentiation, as visualized by the accumulation of starch granules in the cell tier below the QC (upper panel). In contrast to the single tier of GFP signals in the wild-type root DSCs, the DSC marker J2341 in wox5-1 roots showed no GFP expression (lower panel). (C–J) Interspecies complementation with WUS/WOX5 orthologues from the green alga lineage to the angiosperm lineage expressed in Arabidopsis wox5-1 mutants showed that OtWOX (green alga) and CrWUL (fern) are unable to maintain the Arabidopsis root stem cell niche, whereas WUS/WOX5 orthologues from seed plants are able to rescue the defective wox5-1 root meristem. Scale bars, 20 µm.
Similar to the results found in the shoot meristem, OtWOX (green alga), driven by Arabidopsis WOX5 promoter, failed to rescue the Arabidopsis wox5-1 mutant (fig. 3C). The fern CrWUL, which contains the conserved HD and the WUS/EAR motifs as in the seed plant WUS/WOX5 proteins (supplementary fig. S3, Supplementary Material online), was not able to complement the Arabidopsis wox5-1 defects when also driven by native WOX5 promoter (fig. 3D). Both WUS and WOX5 from seed plants, including those from both gymnosperms and angiosperms, were able to replace the function of AtWOX5 in maintaining the Arabidopsis root distal meristem stem cell niche (fig. 3E–J), implying that functional WUS/WOX5 molecular in flowering plant root stem-cell niche maintenance might originate in the recent common ancestor of gymnosperm/angiosperm after the divergence from fern lineage, and the function has been highly conserved during evolution course after the gymnosperms/angiosperms split.
Intercellular Mobility and Biological Activity of WUS in Shoots
Even though both the seed plant WUS and the fern CrWUL contain the three conserved domains of WUS/WOX5 subclade, we want to know why the seed plant WUS proteins, but not the fern CrWUL, were able to maintain the shoot apical stem-cell niche and control flower formation in Arabidopsis. Given that the fern CrWUL is much longer than the seed plant WUS proteins (supplementary fig. S1, Supplementary Material online), we began by investigating how this change of the molecular mass might have influenced WUS/WOX5 function. Using a Arabidopsis transcriptional reporter pWUS::ER-GFP construct, we found that WUS is specifically expressed in the OC driven by WUS promoter, which is localized to the L3 and deeper layers of the SAM (Yadav et al. 2013); it is not expressed in the L1 or L2 layers (fig. 4A). To observe the distribution and mobility of the WUS proteins in the Arabidopsis shoot meristem, we further generated various chimeric WUS-GFP proteins using three WUS protein-coding sequences from fern and seed plants. These fusion WUS–GFP proteins were driven from the native Arabidopsis WUS promoter. In both pWUS::AtWUS-GFP and pWUS::PaWUS-GFP transgenic lines, we observed bright fluorescence signals in OC cells, indicating WUS expression. We also observed relatively weak fluorescence signals in adjacent cells of the L1 and L2 layers, indicating the mobility of these fusion proteins (fig. 4B). By contrast, pWUS::CrWUL-GFP signals were strictly restricted to the OC, with no detectable signals in the L1 and L2 layers (fig. 4B). Genetic complementation experiments also showed that the mobile seed plant AtWUS-GFP and PaWUS-GFP fusion proteins, but not the immobile fern CrWUL-GFP fusion protein, were able to rescue the Arabidopsis wus-1 mutant defects in SAM and floral organ development (fig. 4C). Combined with the sequence analysis (fig. 1D), it seems plausible that inability of the fern CrWUL to maintain the SAM and flower development in Arabidopsis may result from a reduction in its mobility owing to its relatively larger molecular mass. These results suggest that the intercellular mobility of WUS protein may have originated in the common ancestor of gymnosperms and angiosperms after their divergence from the fern lineage.
Fig. 4.
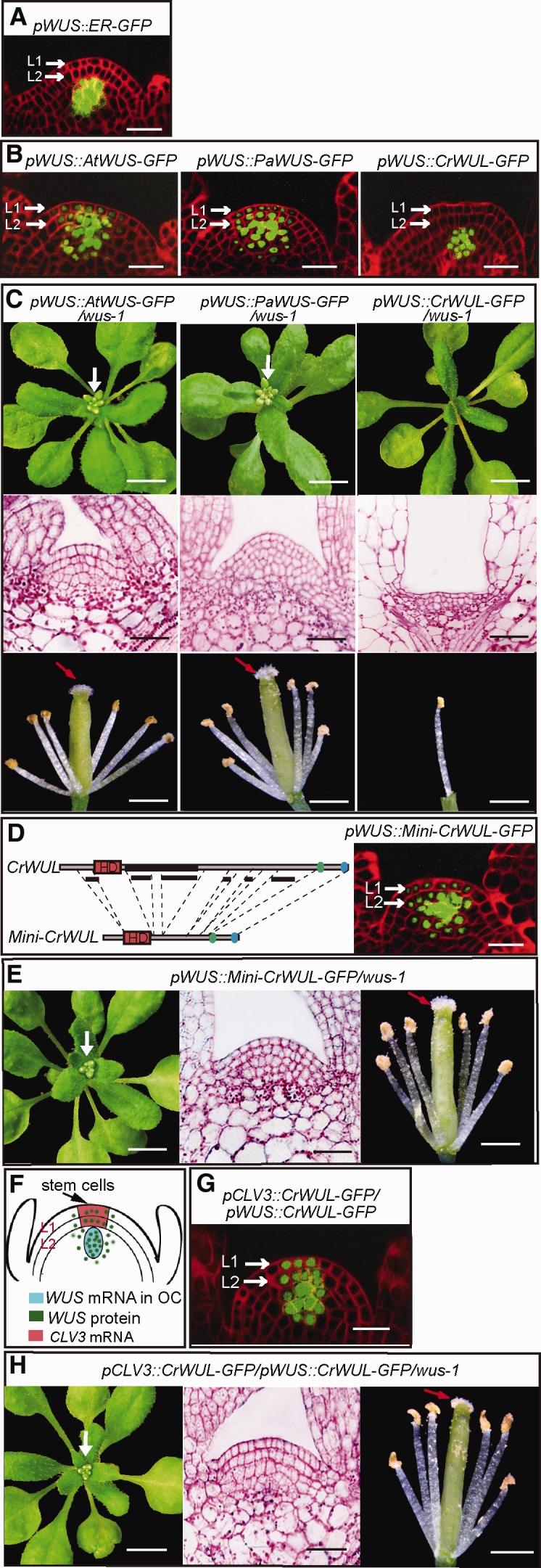
The evolutionary innovations of functional WUS with apical stem-cell maintenance activity and intercellular mobility in Arabidopsis shoots. (A) Lateral view of the SAM with the confocal microscopy showing the specific expression of the transcriptional reporter pWUS::ER-GFP in the OC, but not the L1 or L2 cell layers, in wild-type Arabidopsis shoots. (B) When driven by Arabidopsis WUS promoter, the seed plant WUS-GFP proteins (left and middle panels), but not the fern CrWUL-GFP protein (right panel), migrated into the L1 and L2 layers from the OC in wild-type Arabidopsis shoots. (C) In contrast to the pWUS::AtWUS-GFP and pWUS::PaWUS-GFP constructs, the pWUS::CrWUS-GFP construct failed to rescue the aborted SAM and floral organ defect in wus-1 plants. (D) In pWUS::Mini-CrWUL-GFP plants, the GFP signals were detected both in the OC and in L1 and L2 layers, in Arabidopsis shoots. (E) The pWUS::Mini-CrWUL-GFP construct was able to rescue the wus-1 mutant shoot meristem and flower defects. (F) Schematic representation of Arabidopsis shoot stem-cell niche maintenance. Modified from Laux (Cell 113, 281–283, 2003). (G) In pCLV3::CrWUL-GFP/pWUS::CrWUL-GFP plants, GFP signals were simultaneously detected in the OC and the L1 and L2 layers. (H) The pCLV3::CrWUL-GFP/pWUS::CrWUL-GFP construct was able to rescue the defective shoot meristem and floral organ defects in wus-1 mutant plants. Scale bars: 1 cm (C, E, and H, upper panels), 1 mm (C, E, and H, lower panels), 20 μm (other panels).
Then we generated a truncated version of CrWUL (Mini-CrWUL) by deleting sequences that are not found in gymnosperm PaWUS (supplementary fig. S4, Supplementary Material online). When we expressed the Mini-CrWUL-GFP using the WUS promoter, the protein was observed to migrate into the L1 and L2 layers from the OC (fig. 4D). Moreover, the mobile Mini-CrWUL-GFP driven by Arabidopsis WUS promoter was able to rescue the Arabidopsis wus-1 SAM and floral organ defects (fig. 4E). These results indicate that the WUS predecessor somehow became shorter during the evolution of seed plants after they split from the fern lineages, and thereby gained the intercellular mobility that now appears to be crucial for shoot meristem and floral organ development in flowering plant species.
Although the lack of migration appeared to interfere with the function of the fern CrWUL to complement the Arabidopsis wus-1 mutant, it remained unclear as to whether CrWUL itself possesses the biological activity to maintain shoot apical stem cells and control flower development. The CrWUL-GFP proteins were strictly retained in the OC and were unable to migrate into the L1 and L2 layers to function in stem-cell maintenance activity when driven by the WUS promoter (fig. 4B). It was therefore difficult for us to assess their own biological activity. To address this issue, we took advantage of the fact that CLV3 is known to be specifically expressed in the L1 and L2 layers (fig. 4F) (Ogawa et al. 2008). Thus, to mimic the WUS proteins moving into these layers and to test its shoot apical stem-cell maintenance activity, we simultaneously and ectopically expressed fern CrWUL-GFP in the OC and the L1 and L2 layers by using both the CLV3 and WUS promoters. In pCLV3::CrWUL-GFP/pWUS::CrWUL-GFP plants, GFP signals were detected in both the OC and L1 and L2 layers (fig. 4G). In contrast to the pWUS::CrWUL-GFP construct (fig. 4C), the pCLV3::CrWUL-GFP/pWUS::CrWUL-GFP constructs were able to complement Arabidopsis wus-1 defects (fig. 4H), suggesting that fern CrWUL has acquired the shoot apical stem-cell maintenance activity, and, further, that this activity is independent of intercellular mobility. Combined with the previous interspecies complementation experiments, our results suggest that the shoot apical stem-cell maintenance activity was present in the common ancestor of ferns/seed plants prior to the acquisition of its intercellular mobility.
Intercellular Mobility and Biological Activity of WOX5 in Roots
We used the same approach to examine how the WOX5 protein, a homologue of WUS, evolved its function in regulating the root stem-cell niche. The transcriptional reporter pWOX5::ER-GFP indicated that WOX5 was expressed exclusively in the QC (Chen et al. 2011; Zhang et al. 2015). We generated chimeric WOX5-GFP proteins using coding sequences from three different plant lineages representing the ferns, gymnosperms, and angiosperms. These fusion constructs used the Arabidopsis native WOX5 promoter. We found that seed plant WOX5 proteins—specifically, the gymnosperm PaWOX5 and angiosperm AtWOX5—were able to move into cells surrounding the QC, including the DSC, whereas the fern CrWUL was restricted to the QC and produced no detectable signal in the DSC layer (fig. 5A). These results suggest that the intercellular mobility of WOX5 was an evolutionary innovation of seed plants that has been conserved strongly following the gymnosperm/angiosperm split.
Fig. 5.
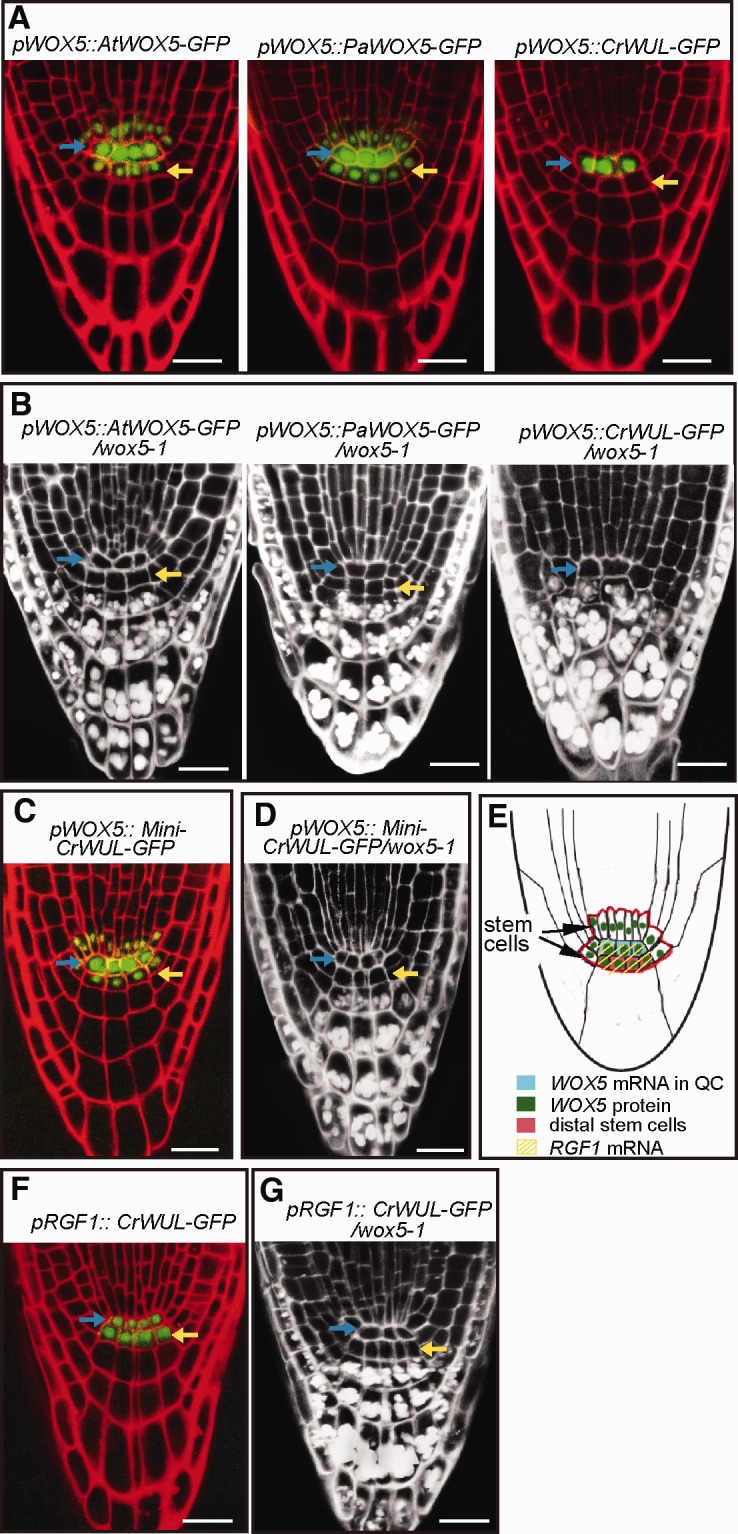
The evolutionary innovations of functional WOX5 with apical stem-cell maintenance activity and intercellular mobility in Arabidopsis roots. (A) Lateral view of the root apical meristem (RAM) showing the seed plant WOX5-GFP proteins (left and middle panels) driven by the native WOX5 promoter, were detected in DSCs. However, the fern CrWUL-GFP protein (right panel) as driven by the native WOX5 promoter, was restricted to the QC. Blue arrow, QC position; Yellow arrow, DSC layer. (B) In contrast to the pWOX5::AtWOX5-GFP and pWOX5::AtWOX5-GFP constructs, the pWOX5::CrWUL-GFP construct was unable to rescue the wox5-1 aborted DSC. (C) In pWOX5::Mini-CrWUL-GFP plants, the GFP signals were detected in the whole root stem-cell niche, including the DSCs. (D) The pWOX5::Mini-CrWUL-GFP construct was able to rescue wox5-1 root defects. (E) Schematic representation of Arabidopsis root stem-cell niche maintenance. The red line outlines the root stem-cell niche. Modified from Sarkar et al. (2007). (F) In pRGF1::CrWUL-GFP plants, the GFP signals were simultaneously detected in the QC and DSCs. (G) The pRGF1::CrWUL-GFP construct was able to rescue the wox5-1 defects. Scale bars, 20 µm.
Interspecies complementation experiments showed that, when using the WOX5 promoter, the expression of the seed plant angiosperm AtWOX5-GFP and gymnosperm PaWOX5-GFP, but not the immobile fern CrWUL-GFP, in Arabidopsis wox5-1 mutants, resulted in the rescue of root meristem defects (fig. 5B), a situation that may partially result from the differences in the molecular masses of WOX5 variants. To test this hypothesis, we expressed the truncated Mini-CrWUL-GFP using the WOX5 promoter. The GFP signals were detected in both the QC and DSC (fig. 5C), similar to the distribution of the gymnosperm PaWOX5-GFP and the angiosperm AtWOX5-GFP (fig. 5A). Finally, the pWOX5::Mini-CrWUL-GFP construct was able to rescue the wox5-1 defect in the DSC (fig. 5D). These results suggest that the intercellular mobility of WOX5 may have been acquired via a decrease in the molecular mass of an ancestral WUS/WOX5 precursor during the evolution of seed plants after they diverged from the fern lineage.
Despite of the immobility of the fern CrWUL protein in root, we want to know whether CrWUL itself possesses root apical stem-cell maintenance regulatory activity. We investigated this by mimicking the movement of the seed plant (gymnosperms and angiosperms) WOX5 proteins in Arabidopsis. In order to mimic the movement, we directly expressed fern CrWUL-GFP in the QC and DSC using the Arabidopsis RGF1 promoter, which is known to induce expression in the QC and DSC (Matsuzaki et al. 2010) (fig. 5E). In pRGF1::CrWUL-GFP plants, GFP signals were observed in both the QC and the DSC (fig. 5F). Furthermore, the pRGF1::CrWUL-GFP construct was able to rescue the aborted DSC in the wox5-1 mutant (fig. 5G). Together, these results indicate that fern CrWUL itself possesses the root apical stem-cell maintenance activity. Notably, only the fern CrWUL and seed plant WUS/WOX5 proteins contain the carboxyl-terminal EAR motif, which is lacked in other WOX proteins (supplementary fig. S1, Supplementary Material online), suggesting that this motif might be responsible for the conserved role of WOX in controlling root stem-cell maintenance activity. Finally, we conclude that this root apical stem-cell maintenance activity may have originated in the common ancestor of ferns and seed plants prior to the capacity for intercellular mobility, and suggesting that capacity has been highly conserved during the course of plant evolution after the fern/seed plant split.
Evolution of WUS/WOX5 Driven by Positive Selection
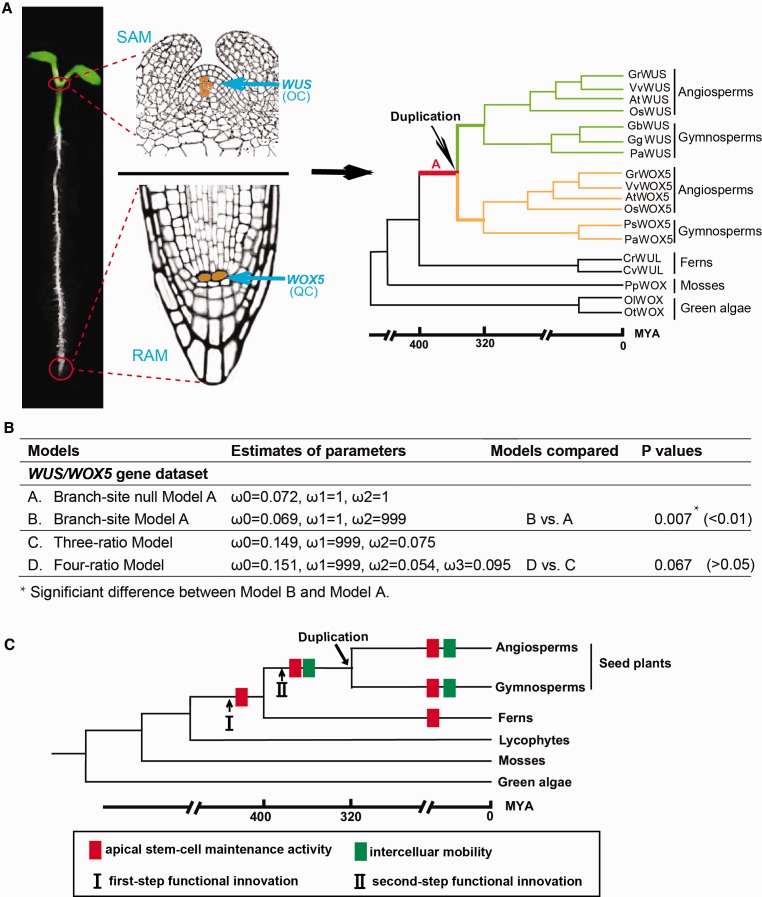
To examine the driving force of WUS/WOX5 evolution, we analyzed the WUS and WOX5 sequences from the green algae to seed plants. Specifically, we used WUS and WOX5 genes from seed plants, WUS-like (i.e. WUL) genes from the ferns, and the WOX genes from the lower plants (moss and green algae) (fig. 6A). We estimated the ratio of nonsynonymous to synonymous substitution rate (termed ω), which is a measure of the driving force behind molecular evolution, with ω < 1 values indicating purifying selection, ω = 1 neutral evolution, and ω > 1 positive selection (Nei and Kumar 2000). Two evolutionary analyses were conducted using codeml in the PAML package (Yang, 2007). First, we constrained the branch (branch A shown in red in fig. 6A) ancestral to all of the WUS/WOX5 genes as the foreground and conducted branch-site models. The branch-site null model A (null hypothesis) fixed the ω for the branch A at 1 (i.e., neutral evolution), whereas the branch-site model A (alternative hypothesis) estimated the same ω allowing it to exceed 1 (i.e., positive selection). A likelihood ratio test between the two models was significant (P = 0.007; fig. 6B), suggesting that positive selection probably had an effect to increase the evolutionary rate of the WUS/WOX5 ancestor after the separation of ferns and seed plants. We also observed that a single copy of WUS-like gene (WUL) was duplicated into two copies (WUS and WOX5) in angiosperms and gymnosperms (fig. 6A). Thus, positive selection may have played an important role in the functional innovations of the WUS/WOX5 genes. Second, we tested the possibility of differential selection on the WUS and WOX5 genes by comparing a three-ratio model with a four-ratio model (fig. 6B). The three-ratio model assumes three ω values: branch A in red has ω1, branches in green have ω2, branches in yellow also have ω2, and other branches have ω0, whereas the four-ratio model assumes one additional ω3 for the branches in yellow (all WOX5 genes) and ω2 for the branches in green (all WUS genes) (fig. 6A). Both ω2 (0.054) and ω3 (0.095) estimated by the four-ratio model are lower than 1 (fig. 6B), suggestive of purifying selection in both WUS and WOX5 genes. A likelihood ratio test between the two models was not significant (P =0.065; fig. 6B), indicating similar levels of purifying selection on the WUS and WOX5 genes. Thus, during the evolution of seed plants, purifying selection of WUS/WOX5 genes may have resulted in the functional conservation of these stem-cell factors in flowering plant shoot/root apical stem-cell maintenance and in flower organ formation. This supposition is concordant with the results of our functional assays of successful inter-species complementation with the seed plant WUS/WOX5 expressed in the Arabidopsis wus-1 and wox5-1 mutants (fig. 2E–J and 3E–J).
Fig. 6.
Selection test and schematic drawing of capabilities acquired for a two-step functional evolution of WUS/WOX5 subfamily. (A) Phylogeny of WUS and WOX5 genes during plant evolution. Left, a one-week-old Arabidopsis seedling with its shoot and root apical tissues highlighted by two red circles. Middle, the enlarged structures of the Arabidopsis shoot and root apical meristems with the WUS expression domain in OC (upper) and WOX5 expression domain in QC (lower). Right, Construction of phylogenetic tree with WUS (upper) and WOX5 (lower) lineages using the NJ, MP, and ML methods. (B) Selection tests suggest that the positive selection had an effect to increase the evolutionary rate of the WUS/WOX5 ancestor after the split of ferns and seed plants, whereas similar levels of purifying selection on the separate WUS and WOX5 genes were observed in seed plants, indicating the functional conservation of these stem-cell factors in the evolution course after duplication. (C) An evolutionary scheme representing the two-step functional innovation of WUS/WOX5 proteins during plant evolution. WUS/WOX5 proteins gained the apical stem-cell maintenance activity in the recent common ancestor of ferns and seed plants; the intercellular mobility was subsequently acquired in the common ancestor of seed plants, after divergence from the fern lineage.
Discussion
Our results showed that the two-step functional innovation of the stem-cell factors WUS/WOX5 plays a pivotal role in flowering plant shoot/root stem cell homeostasis and floral organ development during plant evolution. As depicted in the model (fig. 6C), the first-step functional innovation is the apical stem-cell maintenance activity of WUS/WOX5 originated in the recent common ancestor of ferns and seed plants, which is evidenced by the successful inter-species complementation experiments, with the fern CrWUL ectopically expressed surrounding the OC/QC or exclusively OC-/QC-expressed the seed plants WUS/WOX5 in Arabidopsis WUS or WOX5 knockout mutants. The confocal microscope imaging of GFP fusion proteins revealed that the intercellular mobility of WUS/WOX5, as the second functional innovation, was furtherly acquired in the recent common ancestor of gymnosperms/angiosperms after they diverged from the fern lineage. These two capabilities of WUS/WOX5, originated from two-step evolutionary innovation, have been evolutionarily conserved after the divergence of the gymnosperms/angiosperms. Our findings therefore provide evolutionary insight into the origin of these noncell-autonomous stem-cell factors.
Instead of the single copy of WUS-like gene identified in fern, both the WUS gene and its homologous WOX5 gene is present in seed plants—gymnosperms and angiosperms—suggesting a duplication event occurred after the divergence of ferns/seed plants, but prior to the separation of the angiosperms/gymnosperms (fig. 6C). Moreover, the duplication event may have occurred since the gain of the intercellular mobility function and then both of the discrete WUS and WOX5 proteins have inherited both of the functional capacities developed in the two-step functional innovation history of these genes, which results in the interchangeable function of WUS and WOX5 in Arabidopsis. After the duplication, both the WUS and WOX5 genes underwent similar levels of purifying selection, which was consistent with the conserved functions of gymnosperms/angiosperms WUS/WOX5 in Arabidopsis shoot/root stem-cell maintenance and flower organ formation, as shown by interspecies complementation experiments (figs. 2E–J and 3E–J).
Through plant evolution, one of the most important events was the emergence of the flowering plants. Because the flowering plants contain floral organs, which are conducive to efficient fertilization, they were capable of rapid reproduction, allowing their populations to spread more rapidly than their predecessors. Our results show that the functional WUS, which was acquired by a two-step evolutionary innovation process in plant evolution, is essential for flower organ formation; therefore, suggesting that the two-step functional innovation of WUS/WOX5 should be considered to have played an important role in facilitating the emergence of the flowering plants.
Although neither OC/QC structures nor WOX5/WUS genes are present in the moss P. patens, the WOX13-like genes are known to be involved in the reprogramming of leaf and protoplast cells into stem cells (Barlow 1994; Harrison et al. 2009; Sakakibara et al. 2014). However, the WOX13 lineage seems to have no function in flowering plant apical stem-cell maintenance. In ferns, there is a single large pyramidal cell in the root meristem that is referred to as the apical cell. The apical cell is a QC-like cell that alternates cleavage events along its four faces; it is the ultimate source of all cells in the meristem. However, in contrast to the QC cells in seed plants, the fern apical cell has mitotic activity. The fern C. richardii WUS-like gene is transcribed in the whole root tip instead of in this apical cell (Nardmann and Werr 2012), suggesting the cell-autonomous action of the CrWUL in ferns. Interestingly, it has been reported that in Arabidopsis, when AtWUS was expressed under the control of the CLV3 promoter, transgenic plants showed significantly enlarged shoot meristems with massive increases in their stem cell populations, due to an unbalanced positive-negative feedback loop (Brand et al. 2002; Yadav et al. 2010). The meristem of pCLV3::CrWUL-GFP/pWUS::CrWUL-GFP plants are comparable to wild-type meristem (fig. 4G). These findings indicate that, beyond an inability in intercellular movement, CrWUL may still behave differently from seed plant AtWUS in controlling Arabidopsis shoot stem cell activities. In gymnosperm and angiosperm species, the OC/QC is histologically apparent (Barlow 1994), but studies have shown that the gymnosperm G. gnemon WUS was transcribed in the OC and its surrounding cells, including in the L1 layer (Nardmann et al. 2009), whereas the angiosperm Arabidopsis WUS and WOX5 are exclusively expressed in the OC and QC and then move to the adjacent cells to control the fate of their surrounding stem cells. This information suggests that the noncell-autonomous action of these conserved stem-cell factors might emerge after the divergence of the gymnosperms/angiosperms. It also implies that the coevolution between the OC/QC structural formation and functional WUS/WOX5 acquired by the two-step evolutionary innovation plays a critical role in the origin of the noncell-autonomous regulation mechanism that the flowering plants used to maintain the apical stem-cell and flower organ development. In addition to WUS/WOX5, how the noncell-autonomous activities of other essential plant transcriptional regulators such as SHORTROOT (SHR), KNOTTED1/SHOOT MERISTEMLESS (KN1/STM), and LEAFY (LFY) through plant evolution remain unresolved questions (Sessions et al. 2000; Gallagher et al. 2004; Daum et al., 2014).
Although plants and animals separated very early and evolved independently, they show similar mechanisms in stem cell control. In animals, morphogens are expressed in specific niches and then migrate into adjacent cells to set up concentration gradients that drive the stem cells to differentiate into different cell types, thereby forming all of the tissues and organs of the body. But it remains to be shown how these noncell-autonomous molecules in animals originated during the evolution of animal kingdom.
Materials and Methods
Search for WOX Family Members
The accession numbers or IDs of WOX genes from the following plant species can be found in Nardmann et al. (2009): Ostreococcus tauri, Ostreococcus lucimarinus, Physcomitrella patens, Ginkgo biloba, Gnetum gnemon, Pinus sylvestris, Oryza sativa, and Arabidopsis thaliana. The accession numbers or IDs of the WOX proteins in C. richardii can be found in Nardmann and Werr (2012). WOX genes in other plant species were identified from the following databases: Selaginella moellendorffii and Vitis vinifera from the National Center for Biotechnology Information (NCBI), Gossypium raimondiia from http://www.cottongen.org/(Wang et al. 2012), and P. abies from both NCBI and http://congenie.org/. The accession numbers or IDs of identified WOX genes are given in supplementary table S1, Supplementary Material online. Multiple sequence alignments were performed using ClustalX (Thompson et al. 1997).
Evolutionary Analysis
WUS/WOX5 genes were translated into protein sequences and subsequently aligned with ClustalX (Thompson et al. 1997). Neighbor joining (NJ) and maximum-parsimony (MP) phylogenetic analyses were conducted with MEGA v5.05 (Tamura et al. 2011). Maximum likelihood (ML) phylogenetic analysis was undertaken with PhyML v3.0 (Guindon and Gascuel 2003). NJ analysis was performed using the protein Poisson distances and the pairwise deletion of gap sites. MP analysis used the default parameters. The best-fitting substitution model for the ML analysis was selected with the jModelTest2 program (Darriba et al. 2012). For each of three phylogenetic analyses, 1,000 bootstrap replicates were used to evaluate the reliability of the phylogenetic trees. The estimates of the ratios of nonsynonymous and synonymous nucleotide substitution rates were calculated with the codeml program implemented in PAML (Yang 2007). Likelihood ratio tests were used to compare nested models.
Plant Materials and Growth Conditions
The Arabidopsis mutant and transgenic lines used were as follows: wox5-1 (SALK_038262), wus-1 (NASC ID: N15), J2341 (NASC ID: N9118), and pWUS::ER-GFP (NASC ID: N23897). Transgenic plants with pWUS::AtWUS-GFP in a wild-type background were kindly provided by Dr. J. U. Lohmann (University of Heidelberg, Germany) (Daum et al. 2014). Seeds were surface-sterilized with 0.1% HgCl2, germinated on Murashige and Skoog (MS) medium for 2 weeks, transferred to soil, and grown in an Intellus control system (Percival) with a 16/8-h light/dark cycle at 22 °C and 70% humidity.
Vector Construction, Plant Transformation, and Rescue Analysis
To generate plasmids for genetic complementation analysis, WUS-coding sequences from different plant species were cloned into pQG110 containing either a 4.8-kb WOX5 promoter or a 5.6-kb WUS promoter. To detect the movement of WUS/WOX5 proteins, GFP was fused in-frame to the C terminus of various WUS open reading frames and then driven by the WUS or WOX5 promoter. The primers used in the generation of these constructs are detailed in supplementary table S2, Supplementary Material online. Transgenic Arabidopsis plants were generated by the floral dip method (Han et al. 2008) and selected on solid, half-strength MS medium plates containing 50 mg/ml of the appropriate antibiotics.
For rescue analysis, we extracted the RNA from the pWUS::WOX or pWOX5::WOX rescue/non-rescue transgenic lines, and the cDNA was reverse-transcribed from 5 µg of the total RNA as previously reported (Han et al. 2008). Then we used the forward primer from 5′-untranslated region sequence of WUS mRNA or WOX5 mRNA and the reverse primer from the different WOX CDS for quantitative real-time polymerase chain reaction (QRT-PCR) analysis with the housekeeping gene UBQ5 used as the internal standard. We took care to ensure that the mRNA expression levels of pWUS::WOX or pWOX5::WOX constructs in the nonrescued transgenic lines used for phenotypic analysis were highly expressed and were comparable to the expression level in the rescued transgenic lines to exclude the possibility that the nonrescued phenotype resulted from the insufficient expression of the WOX genes in the wus-1 or wox5-1 mutants. The primers used for analysis are listed in supplementary table S4, Supplementary Material online.
Starch Staining
The starch granules and cell walls in Arabidopsis root tips (7 days old) were stained via the modified pseudo-Schiff propidium iodide staining (mPS-PI) method and imaged with a confocal microscope, as previously described (Truernit et al. 2008). In brief, whole seedlings were fixed in 50% methanol/10% acetic acid at 4 °C for up to 24 h. Tissues were then rinsed briefly with ddH2O and incubated in 1% periodic acid at room temperature for 40 min. The tissue was then rinsed twice with ddH2O and incubated in Schiff reagent with propidium iodide (100 mM sodium metabisulphite, 0.15 N HCl, and 100 mg/ml propidium iodide) for 2 h until the plants were visibly stained. More than three samples were transferred onto microscope slides and covered with chloral hydrate solution (4 g chloral hydrate, 1-ml glycerol, and 2-ml water). The slides were incubated overnight at room temperature, after which excess chloral hydrate was removed. The seedlings were mounted in Hoyer’s solution (30-g gum arabic, 200-g chloral hydrate, 20-g glycerol, and 50-ml water). The slides were left undisturbed for at least 3 d prior to observation (excitation 488 nm, emission 520–720 nm).
Histological Analysis
To prepare semithin sections, shoot tips were stained in 1% (w/v) periodic acid solution containing Schiff’s reagent; these were fixed overnight in 2% (w/v) paraformaldehyde and 2.5% (w/v) glutaraldehyde in phosphate-buffered saline, pH 7.2, at 4 °C. The specimens were then dehydrated in an ethanol series (30%, 50%, 70%, 80%, 90%, 95%, and 100%) and embedded in Spurr’s resin (Spi-Chem). The tissue was mounted in ddH2O and sectioned at a thickness of 2 µm on a Leica RM 2265 microtome. The sections were observed under bright-field optics using a Leica DMRE microscope.
Confocal Microscopy
For confocal microscopy, Arabidopsis shoot tips were stained with 50-µg/ml FM4-64FX (Invitrogen, F34653) (Reddy et al. 2004). Arabidopsis root tips were stained with 10-µg/ml propidium iodide for 5 min (Sarkar et al. 2007), washed briefly in ddH2O, and then visualized at 720–760 nm for FM4-64FX, at 600–640 nm for propidium iodide, and at 500–560 nm for GFP on an LSM 710 NLO confocal microscope with Duoscan functionality.
To confirm the enhancer trap reporter J2341 (a GAL4-GFP enhancer trap reporter) is present in both rescued and nonrescued transgenic lines, we extracted the genomic DNA, utilized the PCR-based genotyping and sequencing of the GFP and GAL4 to confirm that the J2341 reporter was introduced in both the rescued and nonrescued transgenic lines before they were used for confocal microscope imaging. To confirm the pCLV3::CrWUL-GFP/pWUS::CrWUL-GFP lines used for analysis with the strong expression of CrWUL-GFP fusion proteins in both central zone and OC, we utilized the confocal microscopy for selection of these transgenic lines with strong GFP fluorescence under 488 nm of excitation wavelength, and at least three independent transformants were used for phenotypic analysis.
Supplementary Material
Supplementary figures S1–S6 and tables S1–S4 are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank J.U. Lohmann (University of Heidelberg) for kindly providing the pWUS::WUS-GFP and pWUS::WUS-GFP-NLS lines and C.Y. Li (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for kindly providing the WOX5::GFP line. We also acknowledge K. Wang (Wuhan University) for constructive suggestions for this work. This work was supported by a grant from the National Natural Science Foundation of China (90717009).
Footnotes
Associate editor: Hongzhi Kong
References
- Barlow PW. 1994. Structure and function of the root apex–phylogenetic and ontogenetic perspectives on apical cells and quiescent centres. Plant Soil. 167:1–16. [Google Scholar]
- Bennett T, van den Toorn A, Willemsen V, Scheres B. 2014. Precise control of plant stem cell activity through parallel regulatory inputs. Development 141:4055–4064. [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433:39–44. [DOI] [PubMed] [Google Scholar]
- Bond WJ, Scott AC. 2010. Fire and the spread of flowering plants in the Cretaceous. New Phytol. 188:1137–1150. [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. 2000. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289:617–619. [DOI] [PubMed] [Google Scholar]
- Brand U, Grunewald M, Hobe M, Simon R. 2002. Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 129:565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch W, Miotk A, Ariel FD, Zhao Z, Forner J, Daum G, Suzaki T, Schuster C, Schultheiss SJ, Leibfried A, et al. 2010. Transcriptional control of a plant stem cell niche. Dev Cell. 18:841–853. [DOI] [PubMed] [Google Scholar]
- Chen Q, Sun J, Zhai Q, Zhou W, Qi L, Xu L, Wang B, Chen R, Jiang H, Qi J, et al. 2011. The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23:3335–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G, Medzihradszky A, Suzaki T, Lohmann JU. 2014. A mechanistic framework for noncell autonomous stem cell induction in Arabidopsis. Proc Natl Acad Sci U S A. 111:14619–14624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveaux Y, Toffano-Nioche C, Claisse G, Thareau V, Morin H, Laufs P, Moreau H, Kreis M, Lecharny A. 2008. Genes of the most conserved WOX clade in plants affect root and flower development in Arabidopsis. BMC Evol Biol. 8:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Friml J. 2010. Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc Natl Acad Sci U S A. 107:12046–12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Benfey PN. 2008. Plant stem cell niches: standing the test of time. Cell 132:553–557. [DOI] [PubMed] [Google Scholar]
- Forzani C, Aichinger E, Sornay E, Willemsen V, Laux T, Dewitte W, Murray JA. 2014. WOX5 suppresses CYCLIN D activity to establish quiescence at the center of the root stem cell niche. Curr Biol. 24:1939–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillochet C, Daum G, Lohmann JU. 2015. O Cell, Where Art Thou? The mechanisms of shoot meristem patterning. Curr Opin Plant Biol. 23:91–97. [DOI] [PubMed] [Google Scholar]
- Gallagher KL, Paquette AJ, Nakajima K, Benfey PN. 2004. Mechanisms regulating SHORT-ROOT intercellular movement. Curr Biol. 14:1847–1851. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704. [DOI] [PubMed] [Google Scholar]
- Han P, Li Q, Zhu Y-X. 2008. Mutation of Arabidopsis BARD1 causes meristem defects by failing to confine WUSCHEL expression to the organizing center. Plant Cell 20:1482–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P, Zhu Y-X. 2009. BARD1 may be renamed ROW1 because it functions mainly as a REPRESSOR OF WUSCHEL1. Plant Signal Behav. 4:52–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison CJ, Roeder AHK, Meyerowitz EM, Langdale JA. 2009. Local cues and asymmetric cell divisions underpin body plan transitions in the moss Physcomitrella patens. Curr Biol. 19:461–471. [DOI] [PubMed] [Google Scholar]
- Hedman H, Zhu T, von Arnold S, Sohlberg JJ. 2013. Analysis of the WUSCHEL-RELATED HOMEOBOX gene family in the conifer Picea abies reveals extensive conservation as well as dynamic patterns. BMC Plant Biol. 13:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidstra R, Sabatini S. 2014. Plant and animal stem cells: similar yet different. Nat Rev Mol Cell Biol. 15:301–312. [DOI] [PubMed] [Google Scholar]
- Heyman J, Cools T, Vandenbussche F, Heyndrickx KS, Van Leene J, Vercauteren I, Vanderauwera S, Vandepoele K, De Jaeger G, Van Der Straeten D, et al. 2013. ERF115 controls root quiescent center cell division and stem cell replenishment. Science 342:860–863. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M. 2009. Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 21:3493–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473:97–100. [DOI] [PubMed] [Google Scholar]
- Kim I, Cho E, Crawford K, Hempel FD, Zambryski PC. 2005. Cell-to-cell movement of GFP during embryogenesis and early seedling development in Arabidopsis. Proc Natl Acad Sci U S A. 102:2227–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Lu S, Tian H, Ding Z. 2015. WOX5 is shining in root stem cell niche. Trends Plant Sci. 20:601–603. [DOI] [PubMed] [Google Scholar]
- Laux T. 2003. The stem cell concept in plants: a matter of debate. Cell. 113:281–283. [DOI] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J, Jurgens G. 1996. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122:87–96. [DOI] [PubMed] [Google Scholar]
- Lian G, Ding Z, Wang Q, Zhang D, Xu J. 2014. Origins and evolution of WUSCHEL-related homeobox protein family in plant kingdom. Sci World J. doi: 10.1155/2014/534140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Niu L, McHale NA, Ohme-Takagi M, Mysore KS, Tadege M. 2013. Evolutionarily conserved repressive activity of WOX proteins mediates leaf blade outgrowth and floral organ development in plants. Proc Natl Acad Sci U S A. 110:366–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y. 2010. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329:1065–1067. [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. 1998. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95:805–815. [DOI] [PubMed] [Google Scholar]
- Nardmann J, Reisewitz P, Werr W. 2009. Discrete shoot and root stem cell-promoting WUS/WOX5 functions are an evolutionary innovation of angiosperms. Mol Biol Evol. 26:1745–1755. [DOI] [PubMed] [Google Scholar]
- Nardmann J, Werr W. 2012. The invention of WUS-like stem cell-promoting functions in plants predates leptosporangiate ferns. Plant Mol Biol. 78:123–134. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. Oxford: New York Oxford University Press. [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y. 2008. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319:294.. [DOI] [PubMed] [Google Scholar]
- Perilli S, Mambro RD, Sabatini S. 2012. Growth and development of the root apical meristem. Curr. Opin Plant Biol. 15:17–23. [DOI] [PubMed] [Google Scholar]
- Pi L, Aichinger E, van der Graaff E, Llavata-Peris CI, Weijers D, Hennig L, Groot E, Laux T. 2015. Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev Cell. 33:576–588. [DOI] [PubMed] [Google Scholar]
- Reddy GV, Heisler MG, Ehrhardt DW, Meyerowitz EM. 2004. Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development 131:4225–4237. [DOI] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al. 1999. An auxin dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99:463–472. [DOI] [PubMed] [Google Scholar]
- Sakakibara K, Reisewitz P, Aoyama T, Friedrich T, Ando S, Sato Y, Tamada Y, Nishiyama T, Hiwatashi Y, Kurata T, et al. 2014. WOX13-like genes are required for reprogramming of leaf and protoplast cells into stem cells in the moss Physcomitrella patens. Development 141:1660–1670. [DOI] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. 2007. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446:811–814. [DOI] [PubMed] [Google Scholar]
- Scheres B. 2001. Plant cell identity. The role of position and lineage. Plant Physiol. 125:112–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B. 2005. Stem cells: a plant biology perspective. Cell 122:499–504. [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T. 2000. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100:635–644. [DOI] [PubMed] [Google Scholar]
- Sessions A, Yanofsky MF, Weigel D. 2000. Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science 289:779–782. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthélémy J, Palauqui JC. 2008. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20:1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff E, Laux T, Rensing SA. 2009. The WUS homeobox-containing (WOX) protein family. Genome Biol. 10:248.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Wang Z, Li F, Ye W, Wang J, Song G, Yue Z, Cong L, Shang H, Zhu S, et al. 2012. The draft genome of a diploid cotton Gossypium raimondii. Nat Genet. 44:1098–1103. [DOI] [PubMed] [Google Scholar]
- Weigel D, Jürgens G. 2002. Stem cells that make stems. Nature 415:751–754. [DOI] [PubMed] [Google Scholar]
- Yadav RK, Perales M, Gruel J, Girke T, Jönsson H, Reddy GV. 2011. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 25:2025–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Perales M, Gruel J, Ohno C, Heisler M, Girke T, Jönsson H, Reddy GV. 2013. Plant stem cell maintenance involves direct transcriptional repression of differentiation program. Mol Syst Biol. 9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Tavakkoli M, Reddy GV. 2010. WUSCHEL mediates stem cell homeostasis by regulating stem cell number and patterns of cell division and differentiation of stem cell progenitors. Development 137:3581–3589. [DOI] [PubMed] [Google Scholar]
- Yang S, Li C, Zhao L, Gao S, Lu J, Zhao M, Chen CY, Liu X, Luo M, Cui Y, et al. 2015. The Arabidopsis SWI2/SNF2 chromatin remodeling ATPase BRAHMA targets directly to PINs and is required for root stem cell niche maintenance. Plant Cell 27:1670–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang Y, Li G, Tang Y, Kramer EM, Tadege M. 2014. STENOFOLIA recruits TOPLESS to repress ASYMMETRIC LEAVES2 at the leaf margin and promote leaf blade outgrowth in Medicago truncatula. Plant Cell 26:650–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yue J, Liu Z, Zhu Y-X. 2015. ROW1 maintains quiescent centre identity by confining WOX5 expression to specific cells. Nat Commun. doi: 10. 1038/ncomms7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liu X, Engstrom EM, Nimchuk ZL, Pruneda-Paz JL, Tarr PT, Yan A, Kay SA, Meyerowitz EM. 2015. Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 517:377–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.