Abstract
Premise of the study:
Small herbaria represent a significant portion of herbaria in the United States, but many are not digitizing their collections.
Methods:
At the Arkansas State University Herbarium (STAR), we have created a viable workflow to help small herbaria begin the digitization process, including suggestions for publishing data on the Internet. We calculated hourly rates of each phase of the digitization process. We also mapped accessions at the county level to determine geographic strengths in the collection.
Results:
All 17,678 accessioned flowering plant specimens at STAR are imaged, databased in Specify, and available electronically on the herbarium’s website. Students imaged the specimens at a mean rate of 145/h. We found differences in databasing rates between the graduate student leading the project (47/h) and undergraduate assistants (25/h). The majority of specimens at STAR were collected within the counties neighboring the institution.
Discussion:
With this workflow, we estimate that one person can digitize a 20,000-specimen collection in less than 2.5 yr by working only 10 h/wk. Because STAR is a small herbarium with limited resources, the application of the workflow described should assist curators of similar-sized collections as they contemplate and undertake the digitization process.
Keywords: biodiversity informatics, digitization workflow, natural history collection, regional collection, small herbarium
The specimens that comprise many herbaria were collected at a time when plants were collected solely for taxonomic purposes (Chapman, 2005). It is still true that any specimen data used for scientific publications must be traceable back to vouchered sources so that the species’ identities can be verified (Funk et al., 2005). Having access to quality specimens is a requirement for floristic and taxonomic research to advance, and these lines of research are particularly critical for species discovery in our current extinction crisis (Stuessy, 1993). Additionally, herbaria data have emerged as key resources for documenting distributions of biodiversity over time and space (Chapman, 2009; Baird, 2010). The existence of these data, therefore, has important implications for research, education, and public service beyond what was originally envisioned by 19th- and early 20th-century botanists (Funk, 2004; Chapman, 2005). New uses for specimens and their associated data have developed in the past few decades, and technology for linking data virtually through databases has enhanced the utility of these data for answering a variety of scientific questions. For example, changes in land use often result in habitat modification, which can be studied using species occurrences from vouchered herbarium records. Researchers have also used collections data in a variety of other ecological studies. Such studies have used herbarium specimens to track plant viruses over time (Malmstrom et al., 2007), to show that seeds attached to plant specimens are still viable more than 100 yr after collection (Godefroid et al., 2011), and to demonstrate that flowering times are now earlier due to a rise in global temperatures (Hovenden et al., 2008). Effective use of herbarium specimens can assist in detecting and responding quickly to invasive species threats (Baird, 2010). Collections can also be used to study evolutionary change in invasive species as they become established (Marsico et al., 2010).
In the past two decades, there has been a national push to digitize specimen data to make the data more broadly available to the general public and scientific community through Internet access (Owen, 1990; Allen, 1993; Lane, 1996; Network Integrated Biocollections Alliance, 2010; Nelson et al., 2015). The digitizing process consists of predigitization curation, imaging specimens, databasing label and identifying information, and georeferencing locality information (Barkworth and Murrell, 2012; Nelson et al., 2015). Each of these steps can take a considerable amount of time to accomplish, so it is important to note that digitization often happens in stages. Any amount of data coming from a collection is useful, and data can be shared at any stage. Many online platforms publish data that are only images, only label data, only locality information, or a combination of the three.
Having specimen data easily accessible and searchable online increases research efficiency (Chapman, 2005). Time previously spent traveling to collections or waiting for specimen loans could instead be spent gathering data. Taxonomists are often able to use digitized collections to identify and annotate specimens. In addition, digitizing specimens reduces handling and potential damage to specimens (Schmull et al., 2005), which is particularly important for rare and special specimens including type specimens.
An online database of accessioned herbarium specimens promotes sharing of information between institutions and fosters biodiversity information networks. Such resources support the development of professionals in the fields of biodiversity informatics, image services, and geographic information systems (GIS). Online data raise awareness of natural history collections and open these rich resources to education and research. Compiling information for new guides, checklists, and other resources used for understanding botanical information is greatly facilitated by online databases.
In the United States, an estimated 800 herbaria are active and house approximately 90 million specimens (Barkworth and Murrell, 2012), with about 78% of the collections associated with academic institutions. Nearly half of all herbaria in the United States can be considered “small” herbaria; that is, nearly half have fewer than 100,000 specimens (Barkworth and Murrell, 2012). In 2012, about 50% of the small herbaria were databasing, about 25% were imaging, and only 15% had portions of their collections available online. These herbaria are spread out across the United States and occur in many areas where there are no large-sized herbarium collections. With so few of these collections available online, there is a significant portion of plant biodiversity data missing from online databases. To represent the known biodiversity of plants in the United States, online databases must include specimens from small herbaria (Boyd, 2008).
There are many reasons small herbaria are not digitizing their collections and making them available online. Many small herbaria face obstacles such as lack of funding, lack of staff, and curators with many responsibilities in addition to the collection (such as teaching, advising students, and research outside the collections). In many instances, curators at smaller institutions are not given any credit toward promotion for their curatorial duties, making prioritizing these efforts even more difficult. Moreover, curators and collections managers who want to digitize may struggle with knowing where to begin and what the options are (iDigBio, 2013).
The Arkansas State University (STAR) Herbarium houses approximately 20,000 specimens and is run by a curator (T.D.M.) with many other academic responsibilities. The herbarium receives no funding for additional staff and has historically had, and continues to have, no budget for operations. Despite this, the STAR Herbarium now has all of its in-state flowering plant collections reorganized, imaged, databased, and available online. Seedless vascular plants and gymnosperms are imaged and will be databased as part of a recently funded project. This paper describes the protocol we used to digitize more than 16,000 flowering plant specimens of the STAR Herbarium collection in 2.5 yr, which may serve as a model for similar small collections with limited resources. In this publication, we share our experiences as a template for others to apply in similar situations. We provide specific material resources and recommendations for those with a limited budget, and we provide calculations of the time it took us to complete each step of our digitization process.
MATERIALS AND METHODS
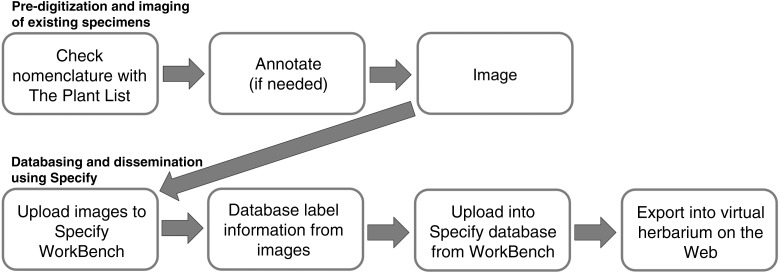
A scholarship from the National Science Foundation (NSF) Scholarships in Science, Technology, Engineering, and Mathematics (S-STEM) program provided funding for one graduate student (K.M.H.) and five undergraduate students to gain practical research experience digitizing the flowering plant collection at the STAR Herbarium. Digitizing the herbarium required six steps: (1) organizing all specimens in the cabinets using updated taxonomy and nomenclature and incorporating as many new collections as possible, (2) purchasing and setting up an imaging station, (3) imaging each specimen individually and developing efficient imaging protocols, (4) choosing the database template that best fit our project, (5) entering all label information into the database, and (6) combining all data to make it accessible to the scientific community and public through an online database (Fig. 1).
Fig. 1.
Arkansas State University (STAR) Herbarium digitization workflow developed and applied in this project.
The equipment we chose was all purchased for US$1500 at the start of the project in 2012. The Dean of the College of Sciences and Mathematics at Arkansas State University (A-State) provided funding for the project. The US$1500 was used to purchase a Nikon D3200 18-megapixel digital single-lens reflex (DSLR) camera (Nikon, Melville, New York, USA) with an 18–55-mm zoom lens (US$760), new lights (Bencher Copymate II fluorescent copy lights; Bencher, Antioch, Illinois, USA; US$670) for an existing copy stand (Bencher Copymate II), a Kodak color separation guide (Kodak, Rochester, New York, USA) and grayscale ruler (US$50), and an extra camera battery (Nikon EN-EL 14; US$20) (Fig. 2). The camera is a crop sensor, but it generates high-quality images with the standard zoom lens that was sold with the camera. The side fluorescent lights are sufficient, but the camera settings have to be carefully manipulated to reduce shadows. The computer used for digitization was already present in the herbarium at the start of the project. We were later able to upgrade to a newer second computer. We used only open-source software for this project. The Nikon View NX2 software was used for remote capture of images. Students recorded their imaging progress in laboratory notebooks by noting species imaged and digitally entered hours worked in a spreadsheet on Google Docs (Google, Mountain View, California, USA). For databasing, we used Specify 6 (Specify Software Project, Biodiversity Institute, University of Kansas, Lawrence, Kansas, USA). This software is free for users, but two people from the project attended a training workshop for Specify. Travel funds to attend this workshop were provided by the Experiential Learning Fellowship Program (NSF DUE-1060209).
Fig. 2.
The imaging station at the Arkansas State University (STAR) Herbarium that was assembled from existing herbarium resources and purchased materials. The figure shows the fluorescent sidelights, the Nikon D3200 camera, the color separation guide and ruler (purchased for the project), and the Bencher Copymate II copy stand (previously existing in the herbarium). The total new cost for the set-up was approximately US$1500.
We began in January 2012 by organizing the specimens according to the most updated nomenclature using The Plant List (2013), an online database with the goal of ensuring each plant species has only a single name that can be traced back to a single nomenclatural source. Each person verifying nomenclature was trained to print and use annotation labels. This ensured updated information was easy to read, and the person responsible for updating the information was clearly documented. Our approach was intended to ensure nomenclature was updated, but it did not account for misidentified specimens. The students verifying nomenclature had little taxonomic training, so specimens that were previously misidentified remained misidentified after the nomenclatural updates.
After the specimens were organized and nomenclatural verifications were nearly complete, we began imaging the collections. Images were captured in JPEG and Nikon raw formats (.NEF). We were able to set our camera settings easily within the software. For optimal lighting, few shadows, and highest quality images, we set camera settings to the following: ISO 100, Shutter speed 1/200, Aperture F/4.0, White Balance Auto, Exposure Comp. 0, Compression Raw + JPEG (Fine), Metering Mode Multi-pattern. To estimate the imaging rate, we asked students to document the number of specimens imaged and the amount of time they worked. With this information, we calculated imaging rates based on 12,108 specimens out of the total 17,678 that were imaged (∼72% of the imaged collection; Table 1).
Table 1.
Digitization rates by task and student. All calculations are in average specimens per hour.
| Task/Individual | No. of recorded work intervals | Specimens/h | SE |
| Nomenclatural verification (Average for all students)a | 150 | ||
| Imaging (Average for all students working alone) | 57 | 145 | 6.3 |
| Imaging (Average for students working in pairs) | 8 | 172 | 16.7 |
| Databasing: | |||
| Undergraduate student 1 | 73 | 26 | 3.2 |
| Undergraduate student 2 | 90 | 27 | 2.9 |
| Undergraduate student 3 | 118 | 21 | 2.0 |
| Undergraduate student 4 | 61 | 26 | 3.3 |
| Undergraduate student 5 | 118 | 26 | 2.4 |
| Undergraduate student average | 460 | 25 | 1.2 |
| Master’s student | 699 | 47 | 1.8 |
| Uploadinga | 331 |
For nomenclatural verification and uploading, we only recorded total time and number of specimens, so we calculated an overall mean without a standard error (SE).
Databasing was divided into two tasks: keystroking label data into the database and uploading the data sets once they were completed. The Specify 6 software allowed for easy updating of nomenclature as well as georeferencing. In addition, support for this software was considered stable, as Specify and its precursors have had continuous federal funding since 1985 (A. C. Bentley, personal communication, 23 September 2016). Specify does, however, require a significant amount of in-house support to set it up and keep it running. For a curator who does not have access to Information Technology (IT) support, it may be simpler to use an online database like Symbiota to maintain a herbarium database and host images, label data, and georeferencing information online. In Specify we used Darwin Core fields (Darwin Core Task Group, 2009) and populated our taxon tree to family level using Plant Systematics: A Phylogenetic Approach (Judd et al., 2008). Data entry was completed in the Specify WorkBench. The WorkBench is a Specify interface that allows several items to be entered at once in a spreadsheet or list format (Fig. 3). The WorkBench is an easy-to-use platform for those new to data entry or accustomed to using Excel spreadsheets. Data records were entered up to family level in the WorkBench so data were able to sync with the taxon tree upon upload. A data set mapping with appropriate Darwin Core fields was created in the WorkBench. In the WorkBench list view, we were able to put the fields in order according to how they were displayed on the specimen label. This set-up allowed students to easily enter data and was less confusing to those who were less familiar with specimen labels. Specimens were databased by species. The images for each species were uploaded into the WorkBench (Fig. 3) as individual data sets and databased by the same individual. After the data were entered, the graduate student project leader (K.M.H.) checked them before they were uploaded into the main database. The rates at which students verified nomenclature, imaged, and databased the specimens were recorded. The amount of time to upload the specimens into the main STAR Herbarium Specify database was also recorded once specimens were databased by the students. The databasing rates were calculated on a per-species work interval with start and end times recorded for all specimens of each species databased. A sample of 11,851 of 16,791 specimens (70%) is used in our database rate calculations.
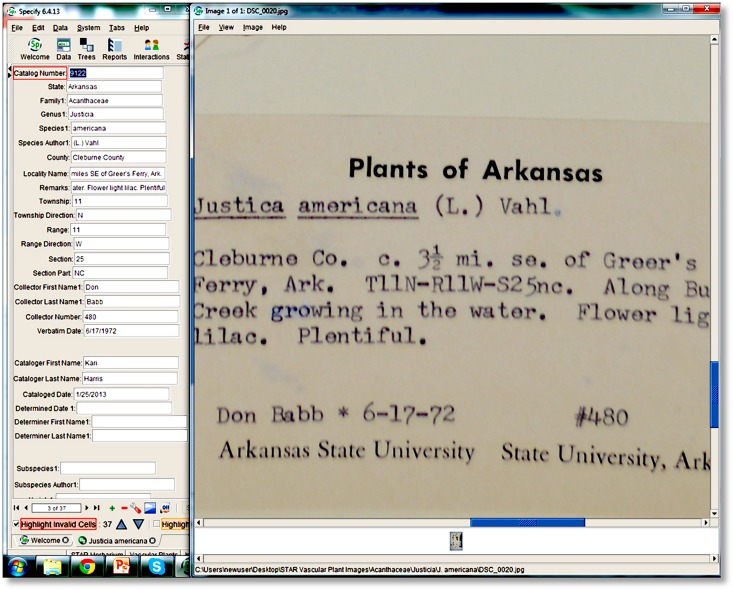
Fig. 3.
Example of a specimen being databased using Specify WorkBench. The form (left) allows for easy entry of the data displayed from the specimen label image (right).
The STAR Herbarium Specify database is backed up on a server maintained by the A-State IT Department. This server must be accessed by Bitvise SSH Client each time users log into the A-State Specify database. Students who use the database are issued log in information for both the A-State server and Specify. This process is somewhat complicated, but it allows the STAR Herbarium records to be securely maintained. In addition, the A-State Specify database and all STAR Herbarium images are backed up on multiple external hard drives. These hard drives are kept at various locations both on and off campus.
To make the records publicly available, we have created a STAR Herbarium website. For this we collaborated with the Department of Computer Science and initiated the website as a project for computer science students. However, rather than create our own online database, we chose to use the Specify Web Portal. We needed the help of an IT expert to create the interface between our database and images on the WorkBench and a web user interface. It is important to note that access to an IT professional is essential in the implementation of a Specify-based workflow. A link to the web portal is available on our website (http://herbarium.astate.edu/). In addition to our own website, we plan to link our data to a larger repository such as the Global Biodiversity Information Facility (GBIF) to make them more widely available. The GBIF Integrated Publishing Toolkit (IPT; http://ipt.gbif.org/) provides directions to export a collection’s database from Specify 6 and upload it into GBIF’s online database (http://www.gbif.org). We will also send our data to the Southeast Regional Network of Expertise and Collections (SERNEC) Portal of Symbiota (http://sernecportal.org/portal/) and iDigBio’s specimen portal (https://www.idigbio.org/portal).
Digitized flowering plant specimen and species data at the STAR Herbarium were examined for their county-level distributions within Arkansas. ArcGIS 10.1 was used to create maps of the accessions in STAR based on five natural breaks determined by the Jenks method (Jenks and Caspall, 1971; de Smith et al., 2015).
RESULTS
During a period of 32 months, a total of 16,791 Arkansas-collected angiosperm (flowering plant) specimens were verified for nomenclature, annotated, imaged, and databased. Nomenclatural verifications were completed at an average of 150 specimens per hour (Table 1). This number included specimens for which only nomenclatural confirmation was required with no corrective label added to the specimens. Imaging had similar rates and was completed at the average rate of 145 images per hour with one student working alone (Table 1). Two students working together resulted in a slightly faster rate of 172 images per hour. This rate increase with two students working together is not nearly as efficient as two students working separately. For example, if two students worked together for one hour, they would image 172 specimens on average, but if these two students worked independently for one hour each they would image 290 specimens total, an increase of 69% for the same person-time worked. At our fastest we had one student imaging 250 specimens per hour. At our slowest we had one student imaging fewer than 100 specimens per hour.
We databased the 16,791 specimens from January 2013 through July 2014. Five undergraduate students databasing specimens were able to do so at an average rate of 25 specimens per hour (Table 1). All had very similar databasing rates with a low of 21 per hour on average and a high of 27 per hour. The graduate student project leader was able to database at a rate of 47 specimens per hour, 81% faster than the undergraduate assistants. The project leader uploaded all data sets. This was completed at a rate of over 300 specimens per hour.
Based on these numbers, we have estimated the time for one person to database a 20,000-specimen collection at 1150 hours. If one person was working on a digitization effort for 10 h/wk, such a collection could be verified for nomenclature and annotated in 13 wk, imaged in 13 wk, databased in 83 wk, and uploaded in 6 wk. This adds up to a total of 115 wk or about 2.4 yr to complete a 20,000-specimen collection with just one person working one quarter of full time.
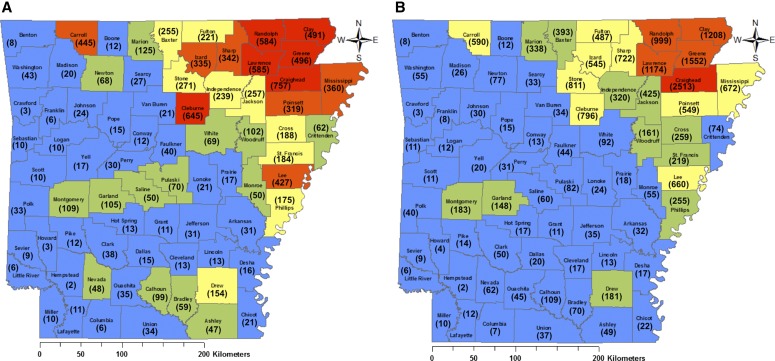
Including additional specimens imaged and databased after August 2014, a total of 17,678 Arkansas angiosperm specimens were digitized at the STAR Herbarium, comprising 154 families, 706 genera, and 1485 species. These specimens represent all 75 Arkansas counties, with Craighead (757), Cleburne (645), Lawrence (585), Randolph (584), and Greene (496) being the most species-rich counties represented at STAR (Fig. 4A). Counties with the most accessioned specimens were Craighead (2513), Greene (1552), Clay (1208), Lawrence (1174), and Randolph (999) (Fig. 4B).
Fig. 4.
Distribution of flowering plant species (A) and vouchers (B) held within the Arkansas State University (STAR) Herbarium. Counties are colored based on five natural breaks determined by the Jenks method in ArcGIS 10.1 (Jenks and Caspall, 1971; de Smith et al., 2015). Colors warm from blue to red with increasing number of species (A) and number of vouchers (B). Numbers of species (A) and vouchers (B) are in parentheses within each county.
DISCUSSION
Our study provides a specific, implemented workflow by which curators can begin digitizing and making their collections available online for minimal monetary investment (Appendix 2). Through the detailed process outlined, we also hope to alleviate any concerns or uncertainty curators may have about beginning the digitization process by providing concrete examples of adequate supplies and personnel time commitments. Through our effort to digitize the flowering plant specimens from Arkansas at the STAR Herbarium, we have developed a viable workflow for those at small institutions with limited support resources and/or competing time commitments. At the beginning of such a project, it is difficult to predict all of the obstacles that will be encountered. Many curators worry that they cannot begin digitizing their collections until the nomenclature has been verified and the collection has been properly organized (iDigBio digitization workshop, personal communication, July 2014). Beginning with nomenclatural verification will give the curator an idea of what is in the collection and how well curated it is. Moreover, examining specimens provides important time for the curator or collection manager to determine the best digitization workflow for the collection. Annotating a collection can also take different forms. In the case of the STAR Herbarium, we only updated nomenclature, without verifying accurate identification of the specimens. Adding the step of verifying identifications would considerably increase the time before digitization can begin, and this step may not be appropriate if the goal is to make collection information more easily available to taxonomic specialists who can verify identifications from online image and database information. Even through our approach of updating nomenclature, we invested time that could have been spent imaging and databasing label information. Some people may decide to eliminate any predigitization curation steps from the workflow. In our case, we used the predigitization curation time to finalize our imaging station purchases and set up.
Obtaining or gaining access to the appropriate imaging and databasing supplies can seem cost prohibitive. There are best practices recommendations available (Nelson et al., 2012, 2015) and specific supply lists from iDigBio (https://www.idigbio.org/wiki/images/8/86/IDigBioImagingGeneralEquipmentRecommendations1_0.pdf) and the New York Botanical Garden (http://sweetgum.nybg.org/science/digitization.php) that suggest a range of adequate to top-of-the-line set-ups for quality imaging and workflow efficiency. These are excellent resources to explore, no matter your budget, but some imaging station or database software options may not be realistic given budgetary constraints. There are advantages, for example, to a full-frame DSLR camera, but if there is no budget for that camera, a cropped sensor camera (for one quarter of the price) will work very well for most herbarium specimen imaging needs. Minimum imaging requirements are (1) a digital camera with at least 18-megapixel capacity, (2) a stable light source that minimizes shadows, (3) an adjustable mount above the specimens where the camera can be affixed, (4) a computer, (5) an external hard drive, and (6) a color separation guide and ruler to be placed alongside the specimens. Barcode labels for naming image files and linking images to database records were not used in this digitization effort, but we strongly recommend them to create unique identifiers (see details in Nelson et al., 2015). The ability to be creative and resourceful in piecing together imaging station components is important for those without funding.
Once the imaging station is assembled and photographs are ready to be taken, access to a reliable workforce is an important consideration. Small collections can complete imaging within several weeks from a single dedicated imager who is spending only 10 h/wk on the imaging effort (see Results). One important consideration in imaging efficiency is the focus of the imager. We suspect that the main cause of variation in imaging rates was the quantity of distractions to the person imaging. For example, the door to the herbarium was often left open to invite visitors and demonstrate to all passers-by that the herbarium is an area of active research. This open-door policy was meant to draw people in and help to raise awareness for the herbarium. However, the adverse effect of the policy seems to have been that visitors distracted imaging personnel. We observed that when few people visited, the student working was able to focus entirely on the task of imaging, and imaging rates were high. When the person imaging was observed to be interrupted, imaging efficiency was lower. In our herbarium, the need to raise awareness and encourage people to use the herbarium outweighed the need to complete the imaging task as quickly as possible.
Some features of the STAR Herbarium made the imaging process efficient. The herbarium consists of a relatively small room (9.75 × 6.7 m), with 21 cabinets of accessioned collections. In this space, it does not take much time for a student to retrieve and replace his/her own specimens. Other models have suggested that it is faster to have two people imaging at once. One person prepares the specimens and another captures the images. At STAR, we found that having two people working together did not increase efficiency. The time required for one person to do all the work was not that much more than the time required for two people to image the same number of specimens. This was a result of the physical size of the herbarium as well as the imaging workflow. We worked through the cabinets one pigeonhole at a time and took care not to disorganize the specimens. The imaging station was made as efficient as possible by moving the computer keyboard close to the imaging station. Pressing the “enter” button on the keyboard captured an image. Then, after the image was taken, the worker quickly switched the imaged specimen out for another specimen. Students could move through folders very quickly this way and still take care not to damage the specimens. If we had verified nomenclature at the same time as imaging, we could have possibly reduced risk of damage to specimens by only removing the specimens from the cabinets once. Yet, we used the preimaging curation step to finalize the imaging station plan and obtain all the necessary supplies. Also, it is possible that combining the nomenclatural review and imaging steps may have actually slowed both of these steps in the process due to combining separate parts of the workflow.
Databasing specimens through the Specify WorkBench allows an easy-to-follow form to be set up. Staff, students, or volunteers can use the WorkBench to transcribe label data directly from specimens or from the images (Fig. 3). A student can be trained to database specimens with this form in about one hour. Another advantage is that utilizing the WorkBench allows specimens to be checked before they are uploaded into the main database.
At the STAR Herbarium, we observed that the undergraduate students databased specimens at a rate about half that of the graduate student project leader. There are many potential reasons for this difference. It is possible that the students were not very invested in the project, but some of the students seemed very interested in it. It is possible that the typing speed of the students played an important factor in the students’ ability to database. However, the students who could not type very well maintained a similar rate to those who were efficient typists. We also considered that better training may have resulted in faster databasing rates if students better understood the herbarium labels. Yet, the five undergraduate students were all well trained, and it did not seem to make a difference whether they had previously taken botany courses or how many years they had been in college. The students came from a variety of backgrounds, and yet all databased at relatively the same rate.
The only obvious difference perceived between the undergraduate students and the graduate student was motivation toward the project. The undergraduates seemed to have been motivated by the time required to spend on databasing, and the graduate student was motivated by the number of specimens that needed to be finished. Based on these observations, one consideration for future digitization projects is to compensate students databasing specimen collections on a per-specimen basis, rather than an hourly basis. For example, if students are to be paid $10/h, they could instead be paid $10 for 30 specimens, no matter how long the databasing actually takes to complete. In a 20-h workweek, the student would be expected to database 600 specimens. Using this approach, a student would database 600 specimens and be paid for a 20-h week. A per-specimen pay rate would motivate the student to finish the number of specimens in a timely manner because the student could possibly complete the work in less time for the same pay. However, the curator or project manager would need to make sure the students were databasing specimens correctly and that increased database rates did not increase error rates. In the Specify 6 WorkBench, there is a button labeled “Show New Records.” If the database is set up to include all possible counties and plant families for a particular project, there should never be new records for geography or family name, as these are already a part of the geography and taxon trees. Entering a name that does not exist will result in a new record being created, and the person uploading the data should double-check any new records in these categories, as they are most likely errors. Agent records represent all individual names (i.e., collectors, annotators, catalogers) entered into the database. After the project is well underway and many of the more common persons have been entered, there should be very few new agent records. This leaves only the locality and habitat data to be checked, which makes error finding and correction efficient. We recommend having a single person (e.g., the curator or collections manager) responsible for uploading data into the database.
Data should be managed so that records are not lost or duplicated. Having a barcoding system and globally unique identifiers (GUIDs) can help prevent duplicated records. Images may represent a significant amount of data and a hard drive with a minimum capacity of 1 terabyte will be needed to store them. We recommend sending data to a portal such as Symbiota, iDigBio, or GBIF as these represent replicated and redundant backups that will ensure the safety of data. If your institution will grant you server space, it is best to back up your images and database on a secure server. At the very least, make sure to store data in multiple locations in both physical and cloud-based systems.
Digitized data are only useful if they are made widely available. In addition to the web portal, we are also determining other portals that are appropriate for disseminating our collections data. Several exist, and they are usually free of charge and with personnel willing to help collections publish their data. Contacting resources such as iDigBio, GBIF, and Symbiota is a good start to making data public and searchable. One important consideration about data availability is the protection of rare species locality records. At the STAR Herbarium, we initially took a very conservative approach by creating a new field and marking all of the species tracked by the Arkansas Natural Heritage Commission as Element Occurrence Records so they were removed from view when the collection went online. In the near future, we plan to restrict from full public view a smaller subset of species in need of protection as agreed upon recently by state botanists.
At the STAR Herbarium, many specimens were collected before Global Positioning System (GPS) data were widely used to provide a reference for the location of a specimen. Instead, specimens have addresses, directions, and township, range, section (TRS) data. These data can be used to approximate GPS coordinates using georeferencing software. Several records can be compiled to provide a map of specimen collection localities. The Specify software is able to georeference specimens in this manner and links them to other collections within the GBIF system. At STAR, steps have not yet been taken to georeference the collection, but we recognize the importance of georeferencing and hope to implement it in the future based on the platform we have established with this project.
Even a small collection like the STAR Herbarium can make a large impact on the known biodiversity of a region. A study using existing specimens from STAR in 2012 found 231 species previously undocumented in Greene County alone (Harris et al., 2012). With collections data from all 75 Arkansas counties now available (Fig. 4), there is much greater access to the state-level biodiversity and distribution knowledge. Through this digitization effort we have discovered that the STAR Herbarium is an important repository for flowering plant species richness and distribution information on Crowley’s Ridge, the northern Mississippi Alluvial Plain, and the eastern Ozark Plateaus in Arkansas (Fig. 4A). Moreover, for those six counties that have the highest species richness in the STAR collection, the number of species represented in this single collection accounts for more than 89% of the total known taxa from Craighead County, 84% for Randolph County, 82% for Lawrence County, 72% for Greene County, 69% for Clay County, and 62% for Cleburne County (Gentry et al., 2013). In terms of the specimens themselves, it is clear that the most specimens have been collected in the counties of northeastern Arkansas, i.e., those geographically closest to the herbarium (Fig. 4B). Arkansas herbaria are scattered throughout different regions of the state, with each likely containing a repository of data for its surrounding counties. Other Arkansas herbaria will soon be digitized under the recently funded SERNEC digitization project. With these data added to data from the STAR Herbarium, the accessible biodiversity data in Arkansas will increase greatly and be readily searchable and analyzable for taxonomic, ecological, and global change biology projects.
Appendix 1.
Detailed acknowledgments of collaborators, student participants, and funding sources.
| Collaborator/Student | Role in project |
| Christopher Burkhart, Crystal Davis, Elizabeth Etzkorn, Eddie Hampton, Ashley Hart, Rebecca Kennedy, Devin Loerch, Starlene Loerch, Angelique Morris, Hunter Nunnally, Emily Panhorst, Mary Rath, Alexander Worm | Arkansas State University students and two student volunteers who contributed by updating nomenclature through specimen annotation, imaging specimens, and databasing label data |
| Drew Prescott | Undergraduate student from University of Arkansas at Monticello who provided advice on imaging set up |
| Alyson Gill, Curtis Steele | Faculty members from Arkansas State University Art Department who assisted with camera settings upon imaging station set up |
| Mary Barkworth, Karen Fawley, Marvin Fawley, John Harris, Tracy Klotz, John Pratte, Thomas Risch, Stan Trauth, iDigBio team, Marsico laboratory students | Individuals who supported and promoted our digitization effort from initiation to publication |
| Joe Burleson | Arkansas State University College of Sciences and Mathematics Project/Program Specialist who led the effort to create the virtual herbarium |
| Theresa Miller and Andrew Bentley | Specify software personnel who provided database training and technical support |
Appendix 2. Workflow for herbarium users considering digitization of their collection.
We recommend the following steps to digitize a herbarium:
1) Organize all specimens in the cabinets using updated taxonomy and nomenclature and incorporate as many new collections as possible.
2) Purchase equipment: The equipment we chose was all purchased for US$1500. This included a Nikon D3200 18-megapixel digital single-lens reflex (DSLR) camera (Nikon, Melville, New York, USA) with an 18–55-mm zoom lens (US$760), new lights (Bencher Copymate II fluorescent copy lights; Bencher, Antioch, Illinois, USA; US$670) for an existing copy stand (Bencher Copymate II), a Kodak color separation guide (Kodak, Rochester, New York, USA) and grayscale ruler (US$50), and an extra camera battery (Nikon EN-EL 14; US$20). The side fluorescent lights are sufficient, but the camera settings must be carefully manipulated to reduce shadows. The computer used for digitization was already present in the herbarium at the start of the project. We used only open-source software for this project. The Nikon View NX2 software was used for remote capture of images.
3) Image each specimen individually and develop efficient imaging protocols. We set our camera settings easily within the software. For optimal lighting, few shadows, and highest quality images, we set camera settings to the following: ISO 100, Shutter speed 1/200, Aperture F/4.0, White Balance Auto, Exposure Comp. 0, Compression Raw + JPEG (Fine), Metering Mode Multi-pattern.
4) Database: We used Specify 6 (Specify Software Project, Biodiversity Institute, University of Kansas, Lawrence, Kansas, USA) and primarily uploaded data though the Specify WorkBench. This allowed us to create an easy-to-use form for students to type label data into. The WorkBench allows data to be transcribed in individual lines similar to an Excel spreadsheet, with corresponding images attached to each line. Collections with little or no IT support may consider Symbiota (http://symbiota.org/docs/) as an alternate database platform.
5) Back up the data: Keep at least two backups in different locations. We back up our data on a university server as well as an external hard drive. As soon as possible, send data to an online portal such as Symbiota, iDigBio (https://www.idigbio.org/), or the Global Biodiversity Information Facility (GBIF; http://www.gbif.org/).
6) Combine all data to make it accessible to the scientific community and public through an online database. We recommend GBIF and iDigBio as potential data repositories.
LITERATURE CITED
- Allen W. H. 1993. The rise of the botanical database: Computerized information offers increased efficiency, access to unpublished records, and new ways of asking questions. BioScience 43: 274–279. [Google Scholar]
- Baird R. 2010. Leveraging the fullest potential of scientific collections through digitization. Biodiversity Informatics 7: 130–136. [Google Scholar]
- Barkworth M. E., Murrell Z. E. 2012. The US Virtual Herbarium: Working with individual herbaria to build a national resource. ZooKeys 209: 55–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A. 2008. Collecting for education: Herbaria at small liberal arts colleges. Plant Science Bulletin 54: 86–91. [Google Scholar]
- Chapman A. D. 2005. Uses of primary species-occurrence data. Report for the Global Biodiversity Information Facility, Copenhagen, Denmark. Website http://www.niobioinformatics.in/books/Uses%20of%20Primary%20Data.pdf [accessed 21 March 2017].
- Chapman A. D. 2009. Numbers of living species in Australia and the world, 2nd ed. Report for the Australian Biological Resources Study, Canberra, Australia. Website https://www.environment.gov.au/system/files/pages/2ee3f4a1-f130-465b-9c7a-79373680a067/files/nlsaw-2nd-complete.pdf [accessed 21 March 2017].
- Darwin Core Task Group. 2009. Biodiversity Information Standards—TDWG. Darwin Core. Website http://rs.tdwg.org/dwc [accessed 13 September 2016].
- de Smith M. J., Goodchild M. F., Longley P. A. 2015. Geospatial analysis: A comprehensive guide to principles, techniques and software tools, 5th ed. Winchelsea Press, Winchelsea, United Kingdom. [Google Scholar]
- Funk V. 2004. 100 uses for an herbarium (well at least 72). U.S. National Herbarium, Smithsonian Institution. Website http://botany.si.edu/documents/2003_Funk_100Uses.pdf [accessed 13 September 2016].
- Funk V. A., Hoch P. C., Prather L. A., Wagner W. L. 2005. The importance of vouchers. Taxon 54: 127–129. [Google Scholar]
- Gentry J. L., Johnson G. P., Baker B. T., Witsell C. T., Ogle J. D. [eds.]. 2013. Atlas of the vascular plants of Arkansas. University of Arkansas, Fayetteville, Arkansas, USA. [Google Scholar]
- Godefroid S., Van De Vyver A., Stoffelen P., Riobbercht E., Vanderborght T. 2011. Testing the viability of seeds from old herbarium specimens for conservation purposes. Taxon 60: 565–569. [Google Scholar]
- Harris K. M., Foard M. B., Marsico T. D. 2012. Understanding floristic diversity through a database of Greene County specimens. Journal of the Arkansas Academy of Science 66: 94–105. [Google Scholar]
- Hovenden M. J., Wills K. E., Vander Schoor J. K., Williams A. L., Newton P. C. D. 2008. Flowering phenology in a species-rich temperate grassland is sensitive to warming but not elevated CO2. New Phytologist 178: 815–822. [DOI] [PubMed] [Google Scholar]
- iDigBio. 2013. Mobilizing Small Herbaria Workshop. 10–11 December 2013. Website https://www.idigbio.org/content/mobilizing-small-herbaria-workshop [accessed 13 September 2016].
- Jenks G. F., Caspall F. C. 1971. Error on choroplethic maps: Definition, measurement, and reduction. Annals of the American Association of Geographers 61: 217–244. [Google Scholar]
- Judd W. S., Campbell C. S., Kellogg E. A., Stevens P. F., Donoghue M. J. 2008. Plant systematics: A phylogenetic approach. Sinauer, Sunderland, Massachusetts, USA. [Google Scholar]
- Lane M. A. 1996. Roles of natural history collections. Annals of the Missouri Botanical Garden 83: 536–545. [Google Scholar]
- Malmstrom C. M., Shu R., Linton E. W., Newton L. A., Cook M. A. 2007. Barley yellow dwarf viruses (BYDVs) preserved in herbarium specimens illuminate historical disease ecology of invasive and native grasses. Journal of Ecology 95: 1153–1166. [Google Scholar]
- Marsico T. D., Burt J. W., Espelend E. K., Gilchrist G. W., Jamieson M. A., Lindström L., Roderick G. K., et al. 2010. Underutilized resources for studying the evolution of invasive species during their introduction, establishment, and lag phases. Evolutionary Applications 3: 203–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson G., Paul D., Riccardi G., Mast A. R. 2012. Five task clusters that enable efficient and effective digitization of biological collections. ZooKeys 209: 19–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson G., Sweeney P., Wallace L. E., Rabeler R. K., Allard D., Brown H., Carter J. R., et al. 2015. Digitization workflows for flat sheets and packets of plants, algae, and fungi. Applications in Plant Sciences 3: 1500065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network Integrated Biocollections Alliance (NIBA). 2010. A strategic plan for establishing a Network Integrated Biocollections Alliance. Website https://digbiocol.files.wordpress.com/2010/08/niba_brochure.pdf [accessed 13 September 2016].
- Owen R. D. 1990. Database computerization and consortium development for vertebrate collections: A collection management perspective. In E. M. Herholdt , Natural history collections: Their management and value, 105–116. Transvaal Museum Special Publication No. 11. Transvaal Museum, Pretoria, South Africa. [Google Scholar]
- Schmull M., Heinrichs J., Baier R., Ullrich D., Wagenitz G., Groth H., Hourticolon S., Gradstein S. R. 2005. The type database at Gottingen (GOET): A virtual herbarium online. Taxon 54: 251–254. [Google Scholar]
- Stuessy T. F. 1993. The role of creative monography in the biodiversity crisis. Taxon 42: 313–321. [Google Scholar]
- The Plant List. 2013. Version 1.1. Website http://www.theplantlist.org/ [accessed 13 September 2016].