DNA origami nanotechnology enables selective reconfiguration of plasmonic quasi-enantiomers through simple pH tuning.
Keywords: DNA nanotechnology, Self assembly, active plasmonics, Circular Dichroism, enantioselectivity
Abstract
Selective configuration control of plasmonic nanostructures using either top-down or bottom-up approaches has remained challenging in the field of active plasmonics. We demonstrate the realization of DNA-assembled reconfigurable plasmonic metamolecules, which can respond to a wide range of pH changes in a programmable manner. This programmability allows for selective reconfiguration of different plasmonic metamolecule species coexisting in solution through simple pH tuning. This approach enables discrimination of chiral plasmonic quasi-enantiomers and arbitrary tuning of chiroptical effects with unprecedented degrees of freedom. Our work outlines a new blueprint for implementation of advanced active plasmonic systems, in which individual structural species can be programmed to perform multiple tasks and functions in response to independent external stimuli.
INTRODUCTION
Active control of plasmonic metamolecules is a burgeoning new direction in nanoplasmonics, which is promising for the realization of novel active devices, such as optical switches, transducers, modulators, filters, and phase shifters at different wavelengths (1). In general, there are two major schemes to implement active plasmonic systems. One is based on integration of active media, that is, liquid crystals, phase change materials, and III-V semiconductors, whose material properties can be altered upon application of external stimuli (2–7). The other is based on geometrical reconfiguration (8, 9), that is, structural tuning of plasmonic metamolecules. This latter scheme requires controllable actuation at the nanoscale, which often encounters substantial constraints and technological challenges.
At optical wavelengths, one unique technique for structural tuning of plasmonic nanostructures is DNA self-assembly, taking advantage of the inherent molecular recognition and programmability of DNA chemistry (10–13). This approach allows for large-scale production of plasmonic nanostructures in a highly parallel manner (14–19). It enables reconfigurable plasmonic systems, which are beyond the state of the art of top-down nanofabrication techniques (8, 20, 21). There are various ways to control the active behavior of plasmonic nanostructures through DNA self-assembly. Probably the most versatile and thus widespread approach is based on the so-called “toehold-mediated strand displacement reaction” (22), which uses DNA strands as fuel to regulate spatial configuration (23–25). Photoresponsive molecules, such as azobenzene, can also be used through incorporation with DNA to activate response upon application of light stimuli (20, 26, 27). More intriguing approaches could include reversible reconfiguration based on shape complementarity (28, 29) or structural adaptation of aptamers to the presence of target molecules (30).
To date, only active control of plasmonic metamolecule ensembles that contain single structural species has been investigated. Selective control and active tuning of individual structural species coexisting within one ensemble has not been demonstrated. Therefore, the power of programmable DNA chemistry for nanoplasmonics has not yet been fully explored. These selectively activated systems could be prototypical building blocks for advanced plasmonic sensors, which can carry out multiple tasks and functions following a predesigned “sense-activate” algorithm, because each structural species could be programmed to respond to a specific set of inputs. Here, we experimentally demonstrate selective control of reconfigurable chiral plasmonic metamolecules assembled with DNA origami (31, 32). We use pH-sensitive DNA “locks” as active sites to trigger structural regulation of the chiral plasmonic metamolecules over a wide pH range. These locks exploit triplex DNA secondary structures that display pH-dependent behavior due to the presence of specific protonation sites (33, 34). Crucial advantages of pH control over oligonucleotides lie in simpler implementation, faster modulation, and no generation of waste products, preventing system deteriorations. We further demonstrate discriminative reconfiguration of plasmonic quasi-enantiomers (35), achieving enantioselective tuning of chiroptical effects with unprecedented degrees of freedom.
RESULTS
The working principle of a pH-sensitive DNA triplex formation is shown in Fig. 1A. A DNA triplex can be formed through pH-sensitive sequence-specific parallel Hoogsteen interactions (dots) between a single-stranded DNA (ssDNA) and a duplex DNA, which is formed through pH-insensitive Watson-Crick interactions (dashed). Whereas CGC triplets require protonation of the N3 of cytosine in the ssDNA [average pKa (where Ka is the acid dissociation constant) of protonated cytosines in a triplex structure is ~6.5] and thus are only stable at acidic pH values, TAT triplets are only destabilized at pH values above 10 due to the deprotonation of thymine (pKa ≈ 10) (33, 36–39). By varying the relative content of TAT/CGC triplets, it is thus possible to design DNA locks that can be opened or closed over specific pH windows (33).
Fig. 1. Working principle of the pH-sensitive plasmonic metamolecules.
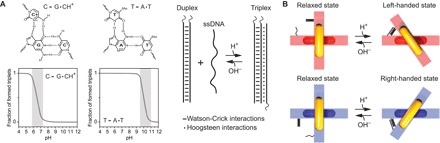
(A) Top left: CGC and TAT triplets can be formed through the combination of Watson-Crick and Hoogsteen interactions. Bottom left: The formation of the CGC triplets requires protonation of cytosines, and they are only stable at acidic pH values. In contrast, the TAT triplets are stable at pH values below 10 and unfold because of deprotonation of thymines. Right: pH-triggered DNA lock. (B) pH regulation of the DNA origami–based chiral metamolecules. The metamolecules can be switched between the relaxed and the LH/RH state by opening or closing the pH-triggered DNA locks.
By implementing these DNA locks on reconfigurable DNA origami, pH-triggered structural tuning of chiral plasmonic metamolecules can be achieved (see Fig. 1B). Each metamolecule comprises two gold nanorods (AuNRs) assembled on a cross-like DNA origami template (see figs. S1 to S3 and tables S1 to S3 in the Supplementary Materials for details of the structural design and assembly process). The DNA lock is composed of a 20–base pair duplex and a 20–base ssDNA positioned on the two respective bundles of the template, such that the pH-regulated triplex formation triggers a conformational change of the metamolecule from a “relaxed” state to a “locked” state (Fig. 1B). Notably, the chiral plasmonic nanostructures can be switched to the left-handed (LH) or the right-handed (RH) state depending on the initial positions of the duplex and the ssDNA on the origami (see Fig. 1B) over different pH windows. The angle between the AuNRs in both the LH and RH configurations is designed to be ~50o.
Figure 2A shows a transmission electron microscopy (TEM) image of the DNA origami/AuNR hybrid nanostructures (see also fig. S4). A high assembly yield has been achieved. Circular dichroism (CD), that is, differential absorption of the LH and RH circularly polarized light by chiral structures, is used as a measure to optically characterize our samples. Representative CD spectra of the metamolecules in the LH and RH states are shown with the red and blue curves in Fig. 2B, respectively. The pH-sensitive DNA lock used in these nanostructures contains equal amounts of TAT and CGC triplets (50% TAT). The CD spectra exhibit characteristic bisignate profiles (40–42).
Fig. 2. DNA assembly of the plasmonic metamolecules and their chiroptical response.
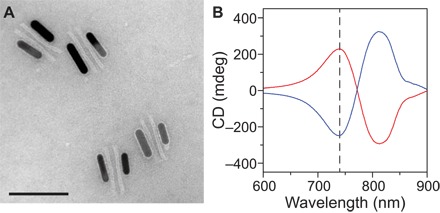
(A) TEM image of the plasmonic nanostructures. The structures tend to lie flat on the TEM grid. Scale bar, 100 nm. (B) Measured CD spectra of the metamolecules in the LH (red line) and the RH (blue line) state from the LH 50% and RH 50% samples at pH 5.5, respectively.
First, we demonstrate that the reconfiguration of our plasmonic metamolecules can be rationally controlled over a wide pH range by simply modulating the relative content of TAT/CGC triplets in the DNA lock (see Fig. 3A). To demonstrate the pH-dependent behavior, we measured the CD responses of the plasmonic metamolecules in buffers of different pH values at 740 nm (Fig. 2B, dashed line) using a CD spectrometer (Jasco J-815). As shown in Fig. 3B, when a DNA lock that contains 50% TAT is used and because of the stabilization of the DNA triplex through the Hoogsteen interactions, the LH or RH plasmonic nanostructures are mostly in the locked state at pH values below 6.5. The triplex-to-duplex transition in the DNA lock occurs upon an increase in pH. For both the LH and RH structures, gradual amplitude decreases in CD response are observed, until it reaches approximately zero at pH value of ~7, indicating that the plasmonic metamolecules are transformed into the relaxed state. The pH of semiprotonation pKaobs (the average pKa due to several interacting protonation cites) for the metamolecules functionalized with 50% TAT locks is ~6.6. When the TAT content is as high as 80% in the DNA lock, the plasmonic nanostructures are opened at a much higher pH (pKaobs = 8.7). The cases of 60 and 70% TAT contents show switching at intermediate pH values with pKaobs of 7.2 and 7.8, respectively (see Fig. 3C). Plasmonic metamolecules with pH-insensitive locks (8) do not exhibit observable CD response changes at different pH values (see fig. S5).
Fig. 3. pH-dependent response of the plasmonic chiral metamolecules.
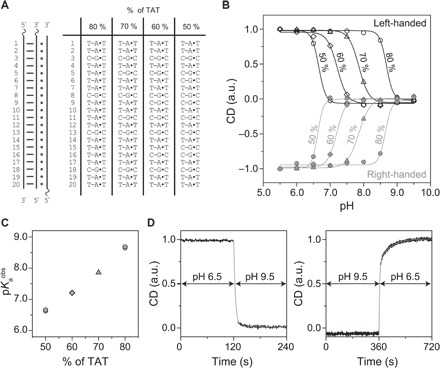
(A) Sequences of the DNA locks with different TAT/CGC contents. (B) Relative CD dependence on pH for the LH and RH metamolecules with DNA locks with different TAT contents. Solid lines are fitting results using Hill function (see Materials and Methods). (C) pH of semiprotonation (pKaobs) dependence on the TAT content for the LH (open black symbols) and RH (solid gray symbols) metamolecules. (D) Kinetic characterization of the metamolecules switching between the LH state and the relaxed state upon pH changes.
Our plasmonic metamolecules exhibit a fast response to pH changes. The time-dependent CD response of an exemplary sample (70% TAT) (Fig. 3D) shows that the reconfiguration upon a dynamic pH change between 9.5 and 6.0 takes place in a few minutes. Reconfiguration is significantly faster than that of the DNA origami–based reconfigurable plasmonic systems previously published (8, 20), which had switching kinetics on the order of several tens of minutes. For the plasmonic systems driven by DNA fuel strands (8, 21) and azobenzene photoisomerization (20), the reconfiguration rates were largely limited by the kinetics of strand displacement reactions and azobenzene-modified DNA hybridization/dehybridization processes, respectively. In our pH-triggered plasmonic system, the high effective concentration of the two triplex-forming domains enables a faster reconfiguration process, which is mainly limited by the rate of protonation/deprotonation of the cytosines/thymines in the ssDNA.
The possibility to engineer plasmonic metamolecules with programmable pH-dependent configurations opens a pathway toward enantioselective control of chiral plasmonic nanostructures. To demonstrate this, we mix LH and RH metamolecules in equimolar amounts (Fig. 4A). These LH and RH nanostructures are plasmonic quasi-enantiomers (35), that is, they are enantiomers as plasmonic objects, but functionalized with DNA locks containing different TAT contents. In the first experiment, only one LH group and one RH group are used. The pH dependence result shown in Fig. 4B (black open circles) is obtained from a plasmonic quasi-racemic solution, in which the TAT contents in the RH and LH nanostructures are 50% (RH 50%; pKaobs = 6.6) and 60% (LH 60%; pKaobs = 7.2), respectively. At pH 5.5, the plasmonic mixture, which is nominally quasi-racemic, exhibits nonzero CD, likely due to structural imperfections. In the pH range of 5.5 to 6.5, where both the LH and RH structures are mainly stable in their locked states, the CD response of the mixture does not experience significant variations upon pH change. When the pH further increases, RH 50% (pKaobs = 6.6) starts to respond and the RH structures in the solution are gradually opened, transitioning the relaxed state. This is reflected by a substantial CD amplitude decrease of RH 50% (see Fig. 3B). In contrast, LH 60% with pKaobs = 7.2 remains unresponsive in the solution. As a result, an abrupt CD peak is observed at pH 7.0 (Fig. 4B, black open circles). This elucidates that discriminative reconfiguration of the plasmonic quasi-enantiomers has been successfully carried out. When the pH continues to increase, LH 60% also becomes responsive and the LH structures in the solution are opened gradually. Therefore, the overall CD response of the mixture decreases until pH 8 is reached. Subsequent pH increases do not introduce drastic CD changes because both the LH and RH structures are in the relaxed state. Similarly, the quasi-racemic solution containing LH 50% and RH 60% exhibits a nearly mirrored pH dependence (Fig. 4B, gray solid circles).
Fig. 4. Enantioselective control of the chiral plasmonic metamolecules.
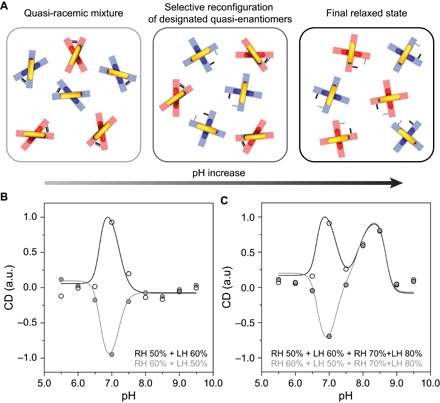
(A) Selective reconfiguration process of the designated quasi-enantiomers. Left: At low pH values, the LH (red) and RH (blue) metamolecules coexist in equimolar amounts. LH and RH metamolecules are quasi-enantiomers, that is, they are enantiomers as plasmonics object but functionalized with DNA locks with different TAT content. The mixture is quasi-racemic. Middle: Upon a pH increase, one type of the quasi-enantiomers (blue) undergoes a locked-to-relaxed transition. Right: Further pH increase results in both quasi-enantiomers being in the relaxed state. (B) Relative CD dependence on pH for a mixture of two quasi-enantiomers; RH 50% and LH 60% (black open circles); RH 60% and LH 50% (gray solid circles). (C) Relative CD dependence on pH for a quasi-racemic mixture composed of four different metamolecules in equimolar amount; RH 50%, LH 60%, RH 70%, and LH 80% (black open circles); RH 60%, LH 50%, RH 70%, and LH 80% (gray solid circles). The predicted CD responses dependent on pH are represented by solid curves (see Materials and Methods).
To demonstrate the unprecedented degrees of freedom of our enantioselective control, we used four groups of metamolecules (two RH and two LH) in the second experiment. The pH-dependent CD result (Fig. 4C, black open circles) is obtained from a racemic solution, containing RH 50% (pKaobs = 6.6), LH 60% (pKaobs = 7.2), RH 70% (pKaobs = 7.8), and LH 80% (pKaobs = 8.6) in equimolar amounts. The experimental results in the pH range of 5.5 to 7.5 resemble those in Fig. 4B. In this range, only RH 50% and LH 60% are pH-triggered, whereas RH 70% and LH 80% are mainly unresponsive to pH change (see Fig. 3B) and remain stable in their respective locked states. When the pH subsequently increases, RH 70% and LH 80% start to undergo structural reconfiguration, transitioning to the relaxed state. In contrast, RH 50% and LH 60% are already in the relaxed state, being pH-unresponsive in the pH range of 7.5 to 9.5. A new CD peak is observed around pH 8.5. This is due to the fact that RH 70% responds to lower pH values than LH 80%. As a result, the RH structures in RH 70% are gradually opened, whereas LH 80% can remain in the locked state until pH 8.5 (see Fig. 3B). When the pH further increases, the LH structures in LH 80% are also gradually opened. Eventually, all four groups are in the relaxed state, resulting in a negligible CD response. The pH-dependent CD result of another sample, which contains RH 60%, LH 50%, RH 70%, and LH 80% in equimolar amounts, is represented by the gray solid circles in Fig. 4C. In the pH range of 5.5 to 7.5, the two samples exhibit a nearly mirrored CD response, whereas in the pH range of 7.5 to 9.5, they show a nearly identical CD response. These results elucidate excellent control selectivity of different plasmonic nanostructure species using simple pH tuning over a wide range. In turn, our strategy outlines an innovative approach to arbitrarily modulate chiroptical response at wish.
DISCUSSION
In conclusion, we have demonstrated selective control of reconfigurable chiral plasmonic metamolecules assembled by DNA origami. The pH-dependent reconfiguration of the plasmonic metamolecules can be tuned in a programmable way by adjusting the relative content of TAT/CGC triplets in the pH-sensitive DNA locks. Excellent enantioselectivity and low cross-activation of the plasmonic enantiomers in quasi-racemic mixtures have been demonstrated upon pH tuning over a wide pH range. Our pH-triggered plasmonic systems could be suitable for optical monitoring of pH changes in biochemical media with higher stability than optical pH sensors based on fluorescence resonance energy transfer because fluorophores generally suffer from low and fluctuating signal intensities as well as photobleaching. In addition, the ability to program DNA structures to perform a specific function only over a certain pH window could find many applications in the field of DNA nanomachines (43) and smart nanomaterials with tailored optical functionalities, such as reversibly modulated optical responses (1, 20). One challenge to achieve excellent performance of reversible modulation is the possible structural degradation upon multiple additions of strong pH agents (either acid or base). This could be circumvented by immobilizing the structures on a surface (for example, a quartz substrate) and then performing buffer exchange. As a future perspective, pH-responsive structures could be applied in the development of new strategies for controlled drug delivery (30). For example, because different cancer types are characterized by an inverted pH gradient between the inside and the outside of cells (44), the possibility to design pH-responsive structures that can release a molecular cargo over a specific pH range could be of value for therapeutic purposes.
MATERIALS AND METHODS
Materials
DNA scaffold strands (p7650) were purchased from tilibit nanosystems. Unmodified staple strands (purification: desalting) were purchased from Eurofins MWG. Capture strands for the AuNRs (purification: desalting) and DNA strands of pH-responsive locks were purchased from Sigma-Aldrich. Thiol-modified strands (purification: high-performance liquid chromatography) were purchased from biomers.net. Agarose for electrophoresis and SYBR Safe nucleic acid stain were purchased from Thermo Fisher. Uranyl formate for negative TEM staining was purchased from Polysciences Inc. AuNRs (10 nm × 38 nm) were purchased from Sigma-Aldrich (catalog no. 716812). Other chemicals were purchased either from Carl Roth or from Sigma-Aldrich.
Design and assembly of DNA origami templates
DNA origami structures were designed using caDNAno 2.0. The strand routing diagram of the origami structures can be found in fig. S1. The sequences of the staple strands and modifications used for pH-sensitive DNA locks are provided in tables S1 and S2. The origami structures were prepared by thermal annealing in a thermal cycling device (Eppendorf Mastercycler pro). For thermal annealing temperatures and times, see table S3. All reaction mixtures contained 10 nM p7650 scaffold and 100 nM of each staple, including those modified with sequences for pH locks. The LH and RH DNA origami templates were assembled separately. More generally, the DNA origami templates with different pH-sensitive locks were assembled in separate polymerase chain reaction tubes. After assembly, the excess staple strands were removed by gel electrophoresis [1.5% agarose gels containing SYBR Safe DNA strain, 0.5× tris–boric acid–EDTA buffer (TBE), and 11 mM MgCl2]. Target bands were cut out, and the DNA origami template structures were extracted with Freeze ’N Squeeze spin columns (Bio-Rad).
Assembly of metamolecules
The AuNR assembly procedure was adopted from previous studies (8, 20). In short, functionalization of the AuNRs with thiolated DNA (SH-5′ T16, biomers.net) was carried out following the low pH route (45, 46). Excess thiolated DNA was removed by centrifugation. The purified AuNRs were added to the purified DNA origami template structures with an excess of 10 AuNRs per DNA origami structure. The mixture was annealed from 40° to 20°C over 15 hours. After thermal annealing, second agarose gel purification step (0.5% agarose gel in 0.5× TBE with 11 mM MgCl2) was performed to remove the excess AuNRs. DNA origami–AuNR structures were extracted with Freeze ’N Squeeze spin columns (Bio-Rad) and further centrifuged at 6000 rcf (relative centrifugal force) for 25 min. The supernatant was carefully removed, and the metamolecules were dispersed in 0.5× TBE with 11 mM MgCl2 and 0.02% SDS. At this stage, different metamolecule samples were diluted to the same concentration, that is, the same absorption (optical density, ~5; 10-mm light path) at the longitudinal plasmon resonance of the AuNRs (780 nm).
TEM characterization
The DNA origami structures (with or without AuNRs) were imaged using a Philips CM-200 TEM operating at 200 kV. For imaging, the DNA origami structures (with or without AuNRs) were deposited on freshly glow-discharged carbon/formvar TEM grids (Science Services). The TEM grids were treated with a uranyl formate solution (0.75%) for negative staining of the DNA structures.
pH regulation of plasmonic metamolecules
Metamolecules functionalized with different pH-sensitive DNA locks were first diluted at a concentration of ~2.5 nM in 0.5× TBE with 11 mM MgCl2 and 0.02% SDS (pH value of ~8.3). These stock solutions were further diluted for CD measurements. The dilution buffers contained 0.5× TBE with 11 mM MgCl2 and 0.02% SDS and varying amounts of acetic acid or sodium hydroxide. The pH of the final metamolecule solutions varied between 5.5 and 9.5 with a step of 0.5 (measurement points in Figs. 3B and 4, B and C). The dilution factors for each type of the metamolecules were 10 (Fig. 3B), 20 (Fig. 4B), and 40 (Fig. 4C). For characterization of the pH change kinetics (Fig. 3D), LH 70% metamolecules were initially dispersed in buffers with pH values of 6.5 and 9.5. Sodium hydroxide or acetic acid was then added to the solution to adjust the pH to 9.5 or 6.5, respectively.
Optical characterizations
The CD and ultraviolet-visible measurements were performed with a J-815 Circular Dichroism Spectrometer (Jasco) using Quartz Suprasil Ultra-Micro cuvettes (105.204-QS, Hellma Analytics) with a path length of 10 mm. Ultraviolet-visible absorption measurements were also performed with a BioSpectrometer (Eppendorf).
The CD response dependent on pH for a single metamolecule group with different TAT contents (Fig. 3B) was analyzed by fitting the measurement results with the Hill function as follows
where CDopen and CDlocked represent the CD intensities of the metamolecules in the relaxed and locked states, respectively. [H+] represents the total concentration of the hydrogen ions ([H+] = 10−pH), Kaobs is the observed acid constant of the metamolecules, and n is the Hill coefficient. pKaobs (Fig. 3C) was calculated using pKaobs = −log (Kaobs).
The kinetics of the metamolecules when undergoing the locked-to-relaxed transition (Fig. 3D, left) was fitted well using a single exponential decay function, suggesting first-order reaction kinetics
where CDopen and CDclosed represent the CD intensities of the metamolecules in the relaxed and locked states, respectively, t0 = 120 s, and k1 = 2.6 × 10−1 s−1.
In contrast, the kinetics of the metamolecules when undergoing the relaxed-to-locked transition (Fig. 3D, right) was fitted well using a double exponential decay function
where CD1 + CD2 = CDopen, t0 = 120 s, k1 = 2.1 × 10−1 s−1, and k2 = 1.4 × 10−2 s−1. The predicted CD responses dependent on pH for the mixed metamolecule groups (Fig. 4, B and C) were obtained by superposition of two (Fig. 4B) and four (Fig. 4C) Hill functions, with parameters obtained from the experimental data analysis for Fig. 3B.
Supplementary Material
Acknowledgments
We thank M. Kelsch for assistance with TEM. TEM images were collected at the Stuttgart Center for Electron Microscopy. Funding: N.L. was supported by the Sofja Kovalevskaja Award from the Alexander von Humboldt Foundation. A.K. was supported by a postdoctoral fellowship from the Alexander von Humboldt Foundation. N.L. was supported by a Marie Curie CIG Fellowship. We also thank the European Research Council (ERC) Starting Grant “Dynamic Nano” for the financial support. F.R. was supported by Associazione Italiana per la Ricerca sul Cancro (project no. 14420), by the ERC (project no.336493), and by Progetti di Ricerca di Interesse Nazionale. Author contributions: A.K., F.R., and N.L. conceived the experiments; A.K. and A.I. designed the DNA origami nanostructures; A.K. and M.J.U. prepared the nanostructures and performed TEM and CD characterization. A.K., F.R., and N.L. wrote the manuscript. All authors discussed the results, analyzed the data, and commented on the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/4/e1602803/DC1
fig. S1. Scaffold/staple layout of the DNA origami template.
fig. S2. Schematics of the DNA origami template.
fig. S3. TEM images of the DNA origami templates after thermal annealing and agarose gel purification.
fig. S4. Additional TEM images of the DNA origami–based metamolecules.
fig. S5. CD response of the plasmonic metamolecules at different pH values with pH-insensitive locks.
table S1. Staple sequences of the DNA origami template.
table S2. Sequences of the pH-sensitive DNA locks.
table S3. Thermal annealing temperatures and times.
REFERENCES AND NOTES
- 1.Zheludev N. I., Plum E., Reconfigurable nanomechanical photonic metamaterials. Nat. Nanotechnol. 11, 16–22 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Driscoll T., Kim H.-T., Chae B.-G., Kim B.-J., Lee Y.-W., Marie Jokerst N., Palit S., Smith D. R., Di Ventra M., Basov D. N., Memory metamaterials. Science 325, 1518–1521 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Liu Q., Yuan Y., Smalyukh I. I., Electrically and optically tunable plasmonic guest–host liquid crystals with long-range ordered nanoparticles. Nano Lett. 14, 4071–4077 (2014). [DOI] [PubMed] [Google Scholar]
- 4.MacDonald K. F., Sámson Z. L., Stockman M. I., Zheludev N. I., Ultrafast active plasmonics. Nat. Photonics 3, 55–58 (2009). [Google Scholar]
- 5.Zhang S., Zhou J., Park Y.-S., Rho J., Singh R., Nam S., Azad A. K., Chen H.-T., Yin X., Taylor A. J., Zhang X., Photoinduced handedness switching in terahertz chiral metamolecules. Nat. Commun. 3, 942 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Zheng Y. B., Yang Y.-W., Jensen L., Fang L., Juluri B. K., Flood A. H., Weiss P. S., Stoddart J. F., Jun Huang T., Active molecular plasmonics: Controlling plasmon resonances with molecular switches. Nano Lett. 9, 819–825 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Wang Q., Rogers E. T. F., Gholipour B., Wang C.-M., Yuan G., Teng J., Zheludev N. I., Optically reconfigurable metasurfaces and photonic devices based on phase change materials. Nat. Photonics 10, 60–65 (2016). [Google Scholar]
- 8.Kuzyk A., Schreiber R., Zhang H., Govorov A. O., Liedl T., Liu N., Reconfigurable 3D plasmonic metamolecules. Nat. Mater. 13, 862–866 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Ou J.-Y., Plum E., Zhang J., Zheludev N. I., An electromechanically reconfigurable plasmonic metamaterial operating in the near-infrared. Nat. Nanotechnol. 8, 252–255 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Seeman N. C., DNA in a material world. Nature 421, 427–431 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Jones M. R., Seeman N. C., Mirkin C. A., Programmable materials and the nature of the DNA bond. Science 347, 1260901 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Rogers W. B., Shih W. M., Manoharan V. N., Using DNA to program the self-assembly of colloidal nanoparticles and microparticles. Nat. Rev. Mater. 1, 16008 (2016). [Google Scholar]
- 13.Krishnan Y., Simmel F. C., Nucleic acid based molecular devices. Angew. Chem. Int. Ed. 50, 3124–3156 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Kuzyk A., Schreiber R., Fan Z., Pardatscher G., Roller E.-M., Högele A., Simmel F. C., Govorov A. O., Liedl T., DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 483, 311–314 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Tian Y., Wang T., Liu W., Xin H. L., Li H., Ke Y., Shih W. M., Gang O., Prescribed nanoparticle cluster architectures and low-dimensional arrays built using octahedral DNA origami frames. Nat. Nanotechnol. 10, 637–644 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chao J., Lin Y., Liu H., Wang L., Fan C., DNA-based plasmonic nanostructures. Mater. Today 18, 326–335 (2015). [Google Scholar]
- 17.Urban M. J., Dutta P. K., Wang P., Duan X., Shen X., Ding B., Ke Y., Liu N., Plasmonic toroidal metamolecules assembled by DNA origami. J. Am. Chem. Soc. 138, 5495–5498 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Schreiber R., Luong N., Fan Z., Kuzyk A., Nickels P. C., Zhang T., Smith D. M., Yurke B., Kuang W., Govorov A. O., Liedl T., Chiral plasmonic DNA nanostructures with switchable circular dichroism. Nat. Commun. 4, 2948 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schreiber R., Do J., Roller E.-M., Zhang T., Schüller V. J., Nickels P. C., Feldmann J., Liedl T., Hierarchical assembly of metal nanoparticles, quantum dots and organic dyes using DNA origami scaffolds. Nat. Nanotechnol. 9, 74–78 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Kuzyk A., Yang Y., Duan X., Stoll S., Govorov A. O., Sugiyama H., Endo M., Liu N., A light-driven three-dimensional plasmonic nanosystem that translates molecular motion into reversible chiroptical function. Nat. Commun. 7, 10591 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou C., Duan X., Liu N., A plasmonic nanorod that walks on DNA origami. Nat. Commun. 6, 8102 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang D. Y., Seelig G., Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 3, 103–113 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Marras A. E., Zhou L., Su H.-J., Castro C. E., Programmable motion of DNA origami mechanisms. Proc. Natl. Acad. Sci. U.S.A. 112, 713–718 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang F., Nangreave J., Liu Y., Yan H., Reconfigurable DNA origami to generate quasifractal patterns. Nano Lett. 12, 3290–3295 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Andersen E. S., Dong M., Nielsen M. M., Jahn K., Subramani R., Mamdouh W., Golas M. M., Sander B., Stark H., Oliveira C. L. P., Pedersen J. S., Birkedal V., Besenbacher F., Gothelf K. V., Kjems J., Self-assembly of a nanoscale DNA box with a controllable lid. Nature 459, 73–76 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Kamiya Y., Asanuma H., Light-driven DNA nanomachine with a photoresponsive molecular engine. Acc. Chem. Res. 47, 1663–1672 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Yang Y., Endo M., Hidaka K., Sugiyama H., Photo-controllable DNA origami nanostructures assembling into predesigned multiorientational patterns. J. Am. Chem. Soc. 134, 20645–20653 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Gerling T., Wagenbauer K. F., Neuner A. M., Dietz H., Dynamic DNA devices and assemblies formed by shape-complementary, non–base pairing 3D components. Science 347, 1446–1452 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Woo S., Rothemund P. W. K., Programmable molecular recognition based on the geometry of DNA nanostructures. Nat. Chem. 3, 620–627 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Douglas S. M., Bachelet I., Church G. M., A logic-gated nanorobot for targeted transport of molecular payloads. Science 335, 831–834 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Rothemund P. W. K., Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Douglas S. M., Dietz H., Liedl T., Högberg B., Graf F., Shih W. M., Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Idili A., Vallée-Bélisle A., Ricci F., Programmable pH-triggered DNA nanoswitches. J. Am. Chem. Soc. 136, 5836–5839 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Hu Y., Ren J., Lu C.-H., Willner I., Programmed pH-driven reversible association and dissociation of interconnected circular DNA dimer nanostructures. Nano Lett. 16, 4590–4594 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Zhang Q., Curran D. P., Quasienantiomers and quasiracemates: New tools for identification, analysis, separation, and synthesis of enantiomers. Chemistry 11, 4866–4880 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Leitner D., Schröder W., Weisz K., Influence of sequence-dependent cytosine protonation and methylation on DNA triplex stability. Biochemistry 39, 5886–5892 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Amodio A., Zhao B., Porchetta A., Idili A., Castronovo M., Fan C., Ricci F., Rational design of pH-controlled DNA strand displacement. J. Am. Chem. Soc. 136, 16469–16472 (2014). [DOI] [PubMed] [Google Scholar]
- 38.M. Egli, W. Saenger, Principles of Nucleic Acid Structure (Springer, 1988). [Google Scholar]
- 39.Ohmichi T., Kawamoto Y., Wu P., Miyoshi D., Karimata H., Sugimoto N., DNA-based biosensor for monitoring pH in vitro and in living cells. Biochemistry 44, 7125–7130 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Auguié B., Alonso-Gómez J. L., Guerrero-Martínez A., Liz-Marzán L. M., Fingers crossed: Optical activity of a chiral dimer of plasmonic nanorods. J. Phys. Chem. Lett. 2, 846–851 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Fan Z., Govorov A. O., Plasmonic circular dichroism of chiral metal nanoparticle assemblies. Nano Lett. 10, 2580–2587 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Shen X., Zhan P., Kuzyk A., Liu Q., Asenjo-Garcia A., Zhang H., de Abajo F. J. G., Govorov A., Ding B., Liu N., 3D plasmonic chiral colloids. Nanoscale 6, 2077–2081 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Bath J., Turberfield A. J., DNA nanomachines. Nat. Nanotechnol. 2, 275–284 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Webb B. A., Chimenti M., Jacobson M. P., Barber D. L., Dysregulated pH: A perfect storm for cancer progression. Nat. Rev. Cancer 11, 671–677 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Zhang X., Servos M. R., Liu J., Instantaneous and quantitative functionalization of gold nanoparticles with thiolated DNA using a pH-assisted and surfactant-free route. J. Am. Chem. Soc. 134, 7266–7269 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Shi D., Song C., Jiang Q., Wang Z.-G., Ding B., A facile and efficient method to modify gold nanorods with thiolated DNA at a low pH value. Chem. Commun. 49, 2533–2535 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/4/e1602803/DC1
fig. S1. Scaffold/staple layout of the DNA origami template.
fig. S2. Schematics of the DNA origami template.
fig. S3. TEM images of the DNA origami templates after thermal annealing and agarose gel purification.
fig. S4. Additional TEM images of the DNA origami–based metamolecules.
fig. S5. CD response of the plasmonic metamolecules at different pH values with pH-insensitive locks.
table S1. Staple sequences of the DNA origami template.
table S2. Sequences of the pH-sensitive DNA locks.
table S3. Thermal annealing temperatures and times.


