Abstract
PRECISE (Predicted and Consensus Interaction Sites in Enzymes) is a database of interactions between the amino acid residues of an enzyme and its ligands (substrate and transition state analogs, cofactors, inhibitors and products). It is available online at http://precise.bu.edu/. In the current version, all information on interactions is extracted from the enzyme–ligand complexes in the Protein Data Bank (PDB) by performing the following steps: (i) clustering homologous enzyme chains such that, in each cluster, the proteins have the same EC number and all sequences are similar; (ii) selecting a representative chain for each cluster; (iii) selecting ligand types; (iv) finding non-bonded interactions and hydrogen bonds; and (v) summing the interactions for all chains within the cluster. The output of the search is the color-coded sequence of the representative. The colors indicate the total number of interactions found at each amino acid position in all chains of the cluster. Clicking on a residue displays a detailed list of interactions for that residue. Optional filters allow restricting the output to selected chains in the cluster, to non-bonded or hydrogen bonding interactions, and to selected ligand types. The binding site information is essential for understanding and altering substrate specificity and for the design of enzyme inhibitors.
INTRODUCTION
One of the most important functions of proteins is serving as enzymes in catalyzing biochemical reactions. Enzyme nomenclature (1) is available through the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB) Enzyme List (2) and the ENZYME database (3). Information on the functions of enzymes is provided by BRENDA (4), which contains an extensive collection of facts related to enzymes, including reaction specificity, functional parameters, substrates, products and inhibitors. IntEnz (5) is a relational database integrating enzyme data from all three of these sources.
The current explosion of structural data has emphasized the need to relate enzyme structures to functions (6). Databases addressing this need are PROCAT (7) and the more recent CSA (8) that provides catalytic residue annotation. However, catalytic sites generally only consist of a few, highly conserved amino acid residues, whereas there are usually 10–20 amino acid positions in an enzyme that directly interact with various ligands, including substrates, products, cofactors and inhibitors. No resource has been developed that permits the systematic study of all residues involved in ligand binding, although the substrate specificity of an enzyme is determined by these rather than only by the catalytic residues. The best tool to obtain information on specific enzyme–ligand interactions is the Sequence Annotated by Structure (SAS) facility of the PDBsum website (9). SAS and PDBsum use the programs HBPLUS (10), to generate lists of hydrogen bonds and non-bonded contacts from the three dimensional (3D) coordinates in a PDB file, and the LIGPLOT program to display the interactions (11). The shortcoming is that each enzyme–ligand complex should be analyzed separately, and then the results be collected to form a summary which provides the consensus characterization of the binding site in a homologous family of enzymes.
Given a query enzyme, the main function of PRECISE is to find all relevant structure files in the PDB (12), align the corresponding sequences, extract all enzyme–ligand interactions and construct the consensus binding site, i.e. identify all residue positions that contribute to ligand binding in any of the structures, ranked on the basis of the frequency of such interactions found for the residues at each position. Future releases of the database will also include interactions predicted by solvent mapping, a novel method for determining protein binding sites based on their structure (13). While this explains the origin of ‘PRE’ in the name of PRECISE, the current version is based exclusively on interactions extracted from the PDB structure files.
BINDING SITES VERSUS CATALYTIC SITES IN ENZYMES
As mentioned above, the broadly defined binding or interaction site of an enzyme means the collection of all amino acid residues that interact with any ligand related to enzyme function, including substrates, products, cofactors and inhibitors. The expressions of ‘functional site’ and ‘recognition site’ are also used in the literature. The binding or interaction site generally includes 10–20 amino acid residues, and should be distinguished from the strict catalytic site that is responsible for the enzyme action and generally comprises only three or four residues. We recall that the PROCAT and CSA databases (7,8) provide catalytic residue annotation. These residues obviously interact with the substrate, and hence are part of the binding site. Since the catalytic mechanism cannot be determined from the structures of complexes alone (although co-crystallized transition state analogs provide useful information), in PRECISE we do not attempt to distinguish the catalytic residues from the rest of the binding site.
ENZYME CLUSTERS WITH HIGH SEQUENCE SIMILARITY
The basis of enzyme classification is the assignment of a specific numerical identifier, the Enzyme Classification (EC) number, which identifies the enzyme in terms of its function. The first digit represents the type of reaction catalyzed. The second digit of the EC number refers to the subclass, which generally contains information about the type of compound or group involved. The third digit, the sub-subclass, further specifies the nature of the reaction and the fourth digit is a serial number that is used to identify the individual enzyme within a sub-subclass. Enzymes with the same EC number, e.g. lysozyme (EC 3.2.1.17) have the same catalytic function, but may include several families (e.g., hen egg-white lysozyme and T4 lysozyme) that significantly differ both in sequence and structure. Collecting consensus information, we group enzymes that have the same EC number and also have very high sequence similarity. Such highly homologous proteins share the same tertiary fold, but can still have localized differences in their binding sites and hence in substrate specificity. However, since the sequences can be reliably aligned, we can assess the role of each amino acid position in forming the binding site.
An important step in the construction of PRECISE is the clustering of enzymes such that members of each cluster have the same EC number and have high sequence similarity to each other. To assure the second property, we have considered the clusters in the Non-Redundant PDB Chain Set (http://www.ncbi.nlm.nih.gov/Structure/VAST/nrpdb.html), also known as the nrPDB database, maintained by the NCBI. The clusters in nrPDB have been constructed by comparing all chains available from PDB (12) with each other using the BLAST algorithm (14). The chains are then clustered into groups of sequence-similar entries using a single-linkage clustering procedure. Chains within a sequence-similar group thus derived are also ranked according to the precision and completeness of their structural data. The following measures of the structural quality are used in this order of priority: (i) lower percentage of residues with unknown amino acid type; (ii) lower percentage of residues with incomplete coordinate data; (iii) lower percentage of residues whose coordinate data are missing; (iv) lower percentage of residues with incomplete side-chain coordinate data; (v) higher resolution; (vi) larger number of chains (subunits) contained in the PDB entry; (vii) larger number of heterogens contained in the PDB entry; (viii) larger number of different types of heterogens; (ix) larger number of residues; and (x) alphanumerical order of their PDB codes.
To form the clusters in PRECISE we consider the enzymes belonging to the same EC number, and group them into clusters of sequence-similar elements that are members of the same nrPDB cluster obtained using the BLAST P-value of 10−40. The top-ranked chain is generally chosen as the representative of the group. In some cases, however, a lower-ranked chain was chosen. For example, if the top-ranked chain was a mutant protein and there was a native protein with reasonably comparable structural quality, then that lower-ranked native protein might replace the mutant as the representative. Representatives from all of the groups together form a non-redundant set of enzymes with different binding sites. The motivation for introducing representatives is 2-fold. First, the interaction frequencies at each amino acid position are projected onto the representative sequence. Second, we will apply computational solvent mapping onto representatives of clusters with no enzyme–ligand complexes available in order to predict putative interaction. While the current version of PRECISE does not include predicted interactions, the availability of consensus interaction sites for a large fraction of enzymes is very useful in a variety of applications.
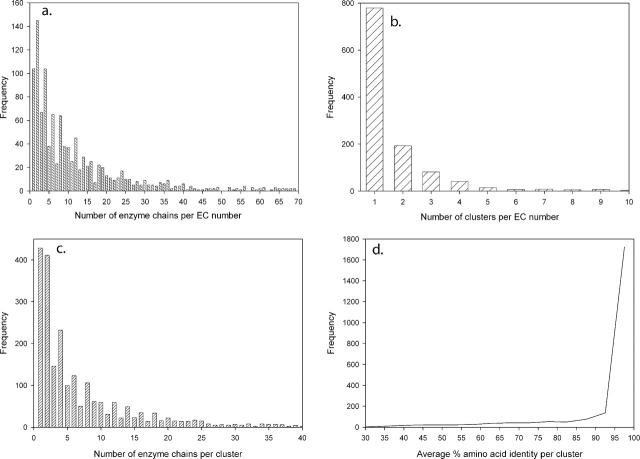
PRECISE version 1.0 is based on the August 31, 2004 version of the nrPDB (12) and the September 17, 2004 version of EC-to-PDB databases, the latter containing 12 532 files. The total number of enzyme chains in these files is 23 872, belonging to 1176 unique EC numbers. Thus, in the PDB the average number of enzyme chains per EC number is 20.29. However, the distribution is very uneven, i.e. the majority of EC numbers is represented with less than 10 chains, whereas a few have very many members (Figure 1a). Clustering chains with the same EC number and pairwise BLAST P-value of 10−40 or less yields 2280 clusters, i.e. on average, 1.93 clusters per enzyme number, with 10.47 chains in each cluster. Figure 1b and c show the distributions for the number of clusters per EC number, and for the number of enzyme chains per cluster, respectively. According to Figure 1b, 779 EC numbers have only chains belonging to a single cluster. More generally, over 90% of EC numbers include only five or fewer clusters of chains that are not homologous to each other. It is interesting to note that the frequencies of EC numbers with even number of chains are always somewhat higher than the frequencies of EC numbers with similar but odd number of chains (Figure 1a). The same observation applies to the distributions of clusters with even and odd number of chains (Figure 1c). While the first appears to be strange, the rule becomes easy to understand when we take into account that many enzymes form homodimers and homotetramers, which increases the fraction of EC numbers and clusters with even number of chains.
Figure 1.
Statistics for enzyme structures in the PDB. (a) Distribution of the 23 872 enzyme chains in the PDB among the 1176 different EC numbers. The graph is truncated at 70 enzyme chains per EC number; there are isolated instances of higher values up to 933 enzyme chains per EC number. (b) Distribution of the 2280 sequence-similar enzyme clusters among the 1176 EC numbers. The graph is truncated at 10 clusters per EC number; there are isolated instances of higher values up to 44 clusters per EC number. (c) Distribution of the 23 872 enzyme chains in the PDB among the 2280 sequence-similar clusters. The graph is truncated at 40 enzyme chains per cluster; there are isolated instances of higher values up to 475 enzyme chains per cluster. (d) Distribution of clusters with given levels of amino acid sequence identity.
Since the selected P-value of 10−40 implies high-sequence identity and similarity within clusters (see Figure 1d), the alignment of sequences within each cluster is very simple. A semi-global dynamic programming algorithm with the GONNET matrix (15), gap penalty of 8, and extension penalty of 0.25 was used to align each sequence to the representative of the cluster. The pairwise alignments within a cluster are stacked upon each other to generate a multiple sequence alignment of all sequences relative to the representative sequence. Because the only concern is that interactions to a residue position in a member of the cluster are related to the correct residue position of the representative, there is no need for a more sophisticated multiple sequence alignment that best aligns every cluster member to each other.
EXTRACTING THE INTERACTIONS
For each chain, the interactions between the protein and the ligand were extracted using the HBPLUS program with the default parameter values to define non-bonded and hydrogen bond interactions. The attributes stored for each interactions are the PDB code and chain identifier of the protein; the name, heteroatom code and type of the ligand; the interacting residue and atom in the protein, the interacting atom in the ligand and the type of the interaction (non-bonded or hydrogen bond). The ligand types we distinguish are peptide, nucleotide, cofactor, metal ion, other inorganic ion and ‘other’, the latter representing the molecules in none of the above categories. The number of interactions for each residue in each chain was obtained by summing the interactions in which the atoms belonging to that residue participate, without any upper bound on the number of interactions. The number of interactions at a particular amino acid position of the aligned sequences was calculated by summing the interactions found at that position for all members of the cluster.
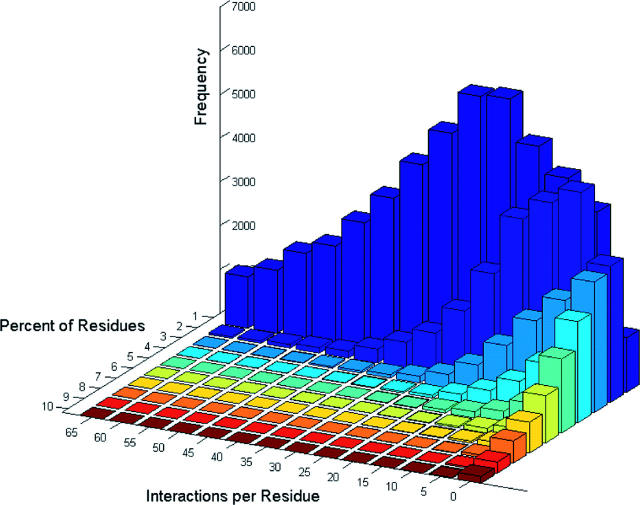
PRECISE currently contains a total of 5 435 107 atom–atom interactions that are distributed among 1533 clusters, i.e. 747 of the 2280 clusters do not have any interactions. The numbers of clusters with different ligand types are as follows: peptides 262, nucleotides 67, cofactors 554, metal ions 733, inorganic ions 673 and ‘others’ 1061. Figure 2 shows the distributions of the percentages of residues with a given number of interactions. According to this figure, the distribution has a robust peak at ∼1–2% of the residues containing 20–30 interactions. This confirms that the information contained in our database is specific enough, i.e. the ligands interact with a relatively small fraction of the residues rather than having interactions broadly distributed over long stretches of the sequence.
Figure 2.
Distribution of the 23 872 enzyme chains among categories defined as having a given percentage of the residues with certain number of interactions, showing that, in most enzymes, 1–2% of the residues have 20–30 interactions which form the peak of the two-dimensional distribution with variables representing the percentage of residues and the number of interactions per residue.
THE PRECISE WEBSERVER
PRECISE is implemented as a relational database using MS SQL Server 2000 as the backend. The database structure can be classified into three sections, Enzyme Information, Cluster Information and Interaction Information. The three sections are related but are independent enough to allow for the update of one without affecting the others. The PRECISE website is hosted on MS IIS 6.0. The web interface is implemented using a combination of ASP.NET and JavaScript. ASP.NET is used for the computationally intensive functions of the site and to provide the database connectivity layer, in the form of ADO.NET. JavaScript is used throughout the site to enhance the usability and responsiveness within a web browser.
The online version of PRECISE is available at http://precise.bu.edu/. Queries can be made using PDB code or EC number. An incomplete EC number will display all clusters that are compatible with the query. If there are non-homologous chains present in structure for the query PDB code, a subsequent page will let users select any of the chains. The main output page (Figure 3a) shows the color-coded sequence of the representative of the cluster. The blue to red color-scheme indicates the residues that belong to the binding site, as well as the total number of interactions found at each amino acid position in all chains of the cluster. This is the most important information provided by the page. Clicking on any ‘colored’ residue (i.e. on a residue that has at least one interaction) displays a separate panel with a detailed list of interactions for the selected residue (Figure 3b). For each interaction, the list shows the PDB code and chain identifier of the protein; the name, heteroatom code and type of the ligand; the interacting residue and atom in the protein, and the type of the interaction (non-bonded or hydrogen bonds). It is important that the list shows both the ‘interaction position’, i.e. the original sequence number of the interacting residue in the PDB file, and the ‘aligned position’, which is the sequence number of the same residue in the alignment of sequences for the entire cluster. A button is provided to open a new window with the alignment. On the right-hand side of the sequence in the main output page, separate panels show all the PDB codes and chain identifiers of the entries that form the cluster. The user may select any subset of these entries and recalculate the list of interactions. Additional panels permit the users to restrict the set of interactions to selected interaction types (i.e. non-bonded or hydrogen bond) and to selected ligand types (i.e. peptides, nucleotides, cofactors, metal ions, other inorganic ions or ‘others’). Again, any subset of these can be selected to produce the list of interactions.
Figure 3.
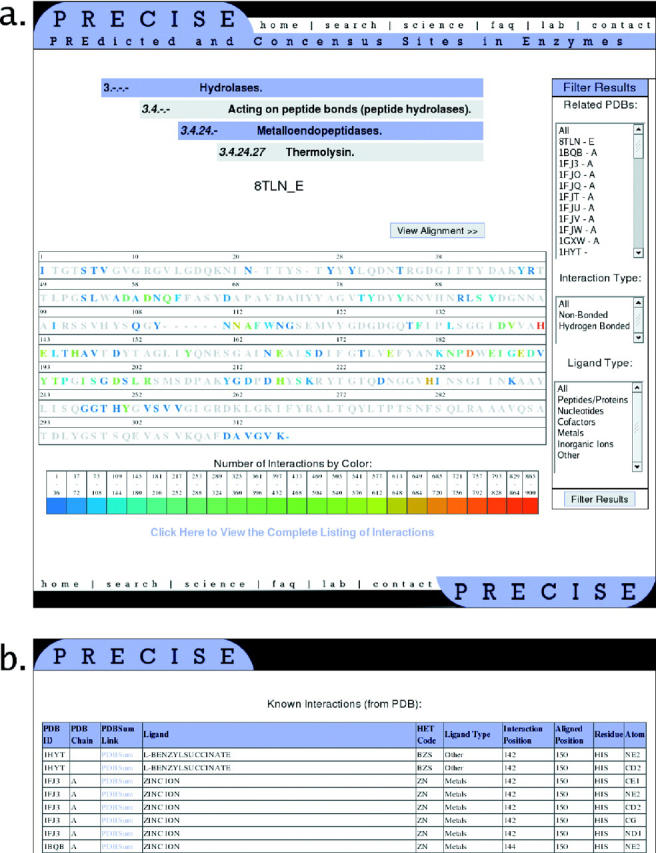
Output of PRECISE for the PDB id 2TLX (thermolysin). (a) Main output page. Chain E of 8TLN is the representative of the cluster containing 2TLX. The color-coded sequence indicates the residues that belong to the binding site, as well as their frequencies of occurrence in enzyme–ligand complexes. The panel on the right shows all the PDB codes and chain identifiers of the entries that form the cluster. The user may select any subset of these entries and recalculate the list of interactions. The two additional panels permit the users to restrict the set of interactions to selected interaction types (i.e. non-bonded or hydrogen bond) and to selected ligand types (i.e. peptides, nucleotides, cofactors, metal ions, other inorganic ions or ‘others’). Any subset of these can be selected to produce the list of interactions. Clicking on any ‘colored’ residue displays the panel shown in (b). (b) Part of the detailed list of interactions for a residue in 2TLX. For each interaction, the list shows the PDB code and chain identifier of the protein; the name, heteroatom code, and type of the ligand; the interacting residue and atom in the protein, and the type of the interaction (non-bonded or hydrogen bond). The list shows both the ‘interaction position’, i.e. the original sequence number of the interacting residue in the PDB file, and the ‘aligned position’, which is the sequence number of the same residue in the alignment of sequences for the entire cluster.
CURRENT WORK AND FUTURE DEVELOPMENT
We are in the process of investigating the origin of some irregularities. In particular, our alignment yields low sequence identity for a limited number of clusters (Figure 1d), in spite of low BLAST P-values, indicating potential problems. The alignment in these clusters is being inspected manually.
As we describe, the 1176 EC numbers in the PDB are subdivided into 2280 sequence-similar clusters. We are in the process of separately annotating these clusters, i.e. identifying the reasons why the same enzymatic action is achieved by non-homologous proteins. The analysis includes the comparison of binding site structures of enzymes that have the same EC number but are in different clusters, and will involve merging some of the clusters.
We will provide a rotatable 3D representation of the binding site in any chain using the Java-based Jmol molecular viewer. The atom set selected for display will include all atoms of the ligand(s), and the side chains of all amino acids that are part of the consensus binding site for the given cluster, including the ones that may not interact with the ligand in the particular chain.
At this point, updating PRECISE to account for new structures in the PDB requires rerunning all our scripts that have been used to generate the database. We will develop scripts that align new PDB entries with the existing representatives and either add them to an existing cluster or create a new cluster, thereby facilitating regular updates without the need for recreating the entire database.
The major addition to PRECISE will be the inclusion of interactions predicted by computational solvent mapping. The latter is a powerful tool for the identification and characterization of binding sites of enzymes (13,16,17). The method moves molecular probes—small organic molecules containing various functional groups—around the protein surface, finds favorable positions using empirical free energy functions, clusters the conformations and ranks the clusters on the basis of the average free energy. The mapping procedure reproduces the available experimental solvent mapping results (18–22). A very important result is that using at least six different solvents as probes, the consensus sites found by the mapping are always in major subsites of the enzyme binding site, and as a result, the amino acid residues that interact with the probes also bind the specific ligands of the enzyme (13). Thus, the method provides detailed and reliable information on the important amino acid residues in the binding site (13). We have already mapped the surface of over 50 enzymes for binding sites. Such predicted interactions will be considered as a separate category in PRECISE. This will permit the comparison of predicted and observed interactions sites if the latter can be determined from the available X-ray structures of complexes. The PRECISE webpage will also contain an email server, and thus users will be able to request the mapping of a specific enzyme by sending an email to the website. Since the mapping runs will be started manually, the response time will be about two days. Once the mapping is finished, the results will be added to the PRECISE database, and an email will be sent to the user.
DISCUSSION
PRECISE provides a summary of interactions between the amino acid residues of an enzyme and its various ligands (substrate and transition state analogs, cofactors, inhibitors and products), thereby complementing other databases that contain enormous wealth of data on enzymes, but do not provide information on the binding site. BRENDA (4) currently has information on 3600 different EC numbers, including nomenclature, isolation and preparation, stability, reaction specificity, functional parameters and references. In particular, BRENDA lists the different ligands (substrates, products, inhibitors, cofactors and metals/ions) but without any type of data on the interactions between these ligands and the protein. The Catalytic Site Atlas (CSA) (8) provides catalytic residue annotation for enzymes in the PDB. Unlike the catalytic residues that are highly conserved, residues that participate in ligand binding but do not directly contribute to the catalytic activity can change through evolution, and are responsible for changes in substrate specificity. Thus, information on the entire ligand binding site is required for understanding the energetic contributions to substrate binding, for developing modified enzymes using methods of protein engineering or directed evolution, and for the design of enzyme inhibitors. Once residues of the binding site are identified and their importance is determined using PRECISE, the subset of catalytic residues can be found by CSA.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Jahnavi Prasad for help with the alignment. This research has been supported by grants DBI-9904834 from the National Science Foundation, GM64700 from the National Institute of Health and P42 ES07381 from the National Institute of Environmental Health Sciences.
REFERENCES
- 1.Tipton K.F. and Boyce,S. (2000) Enzyme classification and nomenclature. In Nature Encyclopedia of Life Sciences. Nature Publishing Group, London. [Google Scholar]
- 2.IUBMB (1992) Enzyme Nomenclature: Recommendations (1992) of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology. Academic Press, San Diego, CA. [Google Scholar]
- 3.Bairoch A. (2000) The ENZYME database in 2000. Nucleic Acids Res., 28, 304–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schomburg I., Chang,A. and Schomburg,D. (2002) BRENDA, enzyme data and metabolic information. Nucleic Acids Res., 30, 47–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fleischmann A., Darsow,M., Degtyarenko,K., Fleischmann,W., Boyce,S., Axelsen,K.B., Bairoch,A., Schomburg,D., Tipton,K.F. and Apweiler,R. (2004) IntEnz, the integrated relational enzyme database. Nucleic Acids Res., 32, 434–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erlandsen H., Abola,E.E. and Stevens,R.C. (2000) Combining structural genomics and enzymology: completing the picture in metabolic pathways and enzyme active sites. Curr. Opin. Struct. Biol., 10, 719–730. [DOI] [PubMed] [Google Scholar]
- 7.Wallace A.C., Laskowski,R.A. and Thornton,J.M. (1996) Derivation of 3D coordinate templates for searching structural databases: application to the Ser-His-Asp catalytic triads of the serine proteinases and lipases. Protein Sci., 5, 1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porter C.T., Bartlett,G.K.J. and Thornton,J.M. (2004) The Catalytic Site Atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res., 32, 129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laskowski R.A. (2001) PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res., 29, 221–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonald I.K. and Thornton,J.M. (1994) Satisfying hydrogen bonding potential in proteins. J. Mol. Biol., 238, 777–793. [DOI] [PubMed] [Google Scholar]
- 11.Wallace A.C., Laskowski,R.A. and Thornton,J.M. (1995). LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng., 8, 127–134. [DOI] [PubMed] [Google Scholar]
- 12.Westbrook J., Feng,Z., Jain,S., Bhat,T.N, Thanki,N., Ravichandran,V., Gilliland,G.L., Bluhm,W., Weissig,H., Greer,D.S., Bourne,P.E. and Berman,H.M. (2002) The Protein Data Bank: unifying the archive. Nucleic Acids Res., 30, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silberstein M., Dennis,S., Brown,L., Kortvelyesi,T., Clodfelter,K. and Vajda,S. (2003) Identification of substrate binding sites in enzymes by computational solvent mapping. J. Mol. Biol., 332, 1095–1113. [DOI] [PubMed] [Google Scholar]
- 14.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 15.Gonnet G.H., Cohen,M.A. and Benner,S.A. (1992) Exhaustive matching of the entire protein sequence database. Science, 256, 1443–1445. [DOI] [PubMed] [Google Scholar]
- 16.Dennis S., Kortvelyesi,T. and Vajda,S. (2003) Computational mapping identifies the binding sites of organic solvents on proteins. Proc. Natl Acad. Sci. USA, 99, 4290–4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kortvelyesi T., Dennis,S., Silberstein,M., Brown,L.,III and Vajda,S. (2003) Algorithms for computational solvent mapping of proteins. Proteins, 51, 340–351. [DOI] [PubMed] [Google Scholar]
- 18.Mattos C. and Ringe,D. (1996) Locating and characterizing binding sites on proteins. Nat. Biotechnol., 14, 595–599. [DOI] [PubMed] [Google Scholar]
- 19.Ringe D. and Mattos,C. (1999) Analysis of the binding surfaces of proteins. Med. Res. Rev., 19, 321–331. [DOI] [PubMed] [Google Scholar]
- 20.Allen K.N., Bellamacina,C.R., Ding,X., Jeffery,C.J., Mattos,C., Petsko,G.A. and Ringe,D. (1996) An experimental approach to mapping the binding surfaces of crystalline proteins. J. Phys. Chem., 100, 2605–2611. [Google Scholar]
- 21.English A.C., Done,S.H., Caves,L.S., Groom,C.R. and Hubbard,R.E. (1999) Locating interaction sites on proteins: the crystal structure of thermolysin soaked in 2% to 100% isopropanol. Proteins, 37, 628–640. [PubMed] [Google Scholar]
- 22.English A.C., Groom,C.R. and Hubbard,R.E. (2001) Experimental and computational mapping of the binding surface of a crystalline protein. Protein Eng., 14, 47–59. [DOI] [PubMed] [Google Scholar]