Abstract
Working in parallel with the efforts to sequence the chicken (Gallus gallus) genome, the Beijing Genomics Institute led an international team of scientists from China, USA, UK, Sweden, The Netherlands and Germany to map extensive DNA sequence variation throughout the chicken genome by sampling DNA from domestic breeds. Using the Red Jungle Fowl genome sequence as a reference, we identified 3.1 million non-redundant DNA sequence variants. To facilitate the application of our data to avian genetics and to provide a foundation for functional and evolutionary studies, we created the ‘Chicken Variation Database’ (ChickVD). A graphical MapView shows variants mapped onto the chicken genome in the context of gene annotations and other features, including genetic markers, trait loci, cDNAs, chicken orthologs of human disease genes and raw sequence traces. ChickVD also stores information on quantitative trait loci using data from collaborating institutions and public resources. Our data can be queried by search engine and homology-based BLAST searches. ChickVD is publicly accessible at http://chicken.genomics.org.cn.
INTRODUCTION
Chicken (Gallus gallus) is an important model organism for biomedical research, the study of embryology and development (1,2), aging (3), quantitative trait loci (QTL) analysis (4), in addition to being a major food source. Through the comprehensive study of sequence polymorphisms in the chicken, significant progress can be made in understanding the phenotypic differences between individuals/strains and the evolution of populations. Towards this end, the Beijing Genomics Institute (BGI) led an international team of scientists from China, USA, UK, Sweden, The Netherlands and Germany to identify and characterize extensive DNA sequence variation throughout the chicken genome. Sequence variation within the genomes of three different breeds of domesticated chickens [a male broiler (Cornish) from Roslin Institute, a female layer (White Leghorn) from Swedish University of Agricultural Sciences and a female Silkie from the Chinese Agricultural University in Beijing], was identified by direct comparison with the genome sequence of the Red Jungle Fowl (RJF), assembled by the Washington University Genome Sequencing Center (WUGSC), as a reference chicken genome sequence (Chicken Genome Consortium, 2004). From these comparisons 3.1 million unique, high-quality sequence variants were identified, primarily single nucleotide polymorphisms (SNPs). In order to facilitate the use of this information, we created the ‘Chicken Variation Database’ (ChickVD), an integrated information system for the storage, retrieval, visualization and analysis of chicken DNA sequence variation. To enhance the discovery of relationships between sequence variation and genes, we mapped each variant onto the RJF reference genome sequence in the context of gene annotations and other relevant features, such as genetic markers and QTLs. Therefore the ChickVD database, provides both a powerful information resource and an analysis workbench for applications in biological research, medicine and agriculture.
DATA SOURCE
The primary purpose of the chicken SNP project was to discover and map extensive sequence variation in the chicken genome, to increase the density of markers on the genetic map, and to facilitate studies on the genetic basis of phenotypic traits. We compared the sequence reads from the three chicken breeds to the 6.6X RJF reference genome sequence (Chicken Genome Consortium, 2004). For each of the breeds, we sampled ∼25% of the genome (a million sequence reads from each breed) using automated capillary sequencers (Amersham MegaBACE 1000). To minimize sequencing errors, we used the Phred quality score (Q-value) (5,6) to set minimum thresholds. For base substitutions, we used conservative thresholds of >Q25 at the variant site and >Q20 for the two flanking 5 bp regions. Even more stringent thresholds of Q30 and Q25 were used to score insertion–deletions (Indels). Using a genome-wide BlastN search, we were able to determine the sequence position on the RJF genome assembly and eliminate confusion between paralogs. Detailed alignments were performed using CrossMatch (http://www.phrap.org) for increased accuracy. A subset of polymorphisms was confirmed by PCR-based resequencing from the lines in which they were initially detected.
To display each sequence variant in the context of its physical relationship to the nearest gene or other relevant features, we employed a number of methods to identify these target sequences. These included annotations within public databases, homology searches, prediction programs and information on experimentally derived genes. A non-redundant dataset of genes/cDNAs was integrated within ChickVD, including Ensembl gene annotations (7), GenBank genes with ‘complete CDS’ (8), full-length cDNAs (9) (http://pheasant.gsf.de/DEPARTMENT/DT40/dt40Transcript.html) and chicken orthologs of human disease genes (C. Webber and C. P. Ponting, unpublished data). We used BLAT (10) to map these sequences using the RJF reference genome, SIM4 (11) to determine the detailed exon–intron boundaries and then classified coding SNPs into synonymous or non-synonymous substitutions. To associate the location of sequence variants with genetically mapped phenotypic traits, we included information on mapped QTLs, using data provided by a number of collaborating institutions (Roslin Institute, Department of Medical Biochemistry and Microbiology of Uppsala University, USDA-ARS Avian Disease and Oncology Laboratory, Animal Breeding and Genetics Group of Wageningen University) and public resources, such as ChickAce (https://acedb.asg.wur.nl/).
DATA CONTENT
ChickVD contains 3.1 million DNA sequence variants; 2.8 million are categorized as SNPs and 0.3 million are Indels. Follow-up experiments indicate that over 90% of these SNPs are true SNPs, over 70% are common SNPs that segregate in many chicken breeds and the mean nucleotide diversity is estimated to be ∼5 SNP/kb (International Chicken Polymorphism Map Consortium, 2004). For the convenience of data display, all types of DNA sequence variation (substitutions, insertions or deletions) are referred to as ‘SNPs’.
SNP information is detailed in a ‘SNP Report’, including type, allelic differences, flanking sequences and PCR-primer designs, location, associated genes and quality scores of sequence reads, functional site in the chicken genome (e.g. coding region, intron, untranslated regions), and details of any predicted or known functional outcomes (e.g. codon and deduced amino acid changes). For additional information to the potential user, 1.5 million contig-covered (CtgCoV) regions from the broiler, layer and Silkie are aligned to the RJF reference genome and 2.5 million raw sequence traces are available within ChickVD.
For queries about gene-associated polymorphisms, we describe each gene/cDNA in a ‘Gene Report’, including gene structure, functional classification, Gene Ontology (12) and InterPro (13) annotations, gene-associated SNPs, and nucleotide/protein sequences. For chicken orthologs of human disease genes, hypertext links to the given Ensembl files of human disease genes and OMIM entries of disease descriptions (14) are also provided. A collection of 606 QTLs for a wide range of traits are integrated and cross-referenced to markers, genetic maps, genes and SNPs mapped onto the same chromosomal region, and to PubMed (15) for literature sources. ChickVD also hosts a collection of 884 references, which focus on chicken sequence variation, QTL study and basic chicken biology.
A summary of ChickVD content is shown in Table 1 and all data are freely available from our FTP site (http://chicken.genomics.org.cn/chicken/jsp/download.jsp).
Table 1. Data content of ChickVD: August 10, 2004.
| Data type | Data statistics |
|---|---|
| Sequence variations | 3 119 698 |
| SNPs | 2 833 578 |
| Indels | 286 120 |
| Confirmed mRNA transcripts | 3868 |
| GenBank with ‘complete cds’ | 1087 |
| Riken1 full-length cDNAs | 1707 |
| BBSRC full-length cDNAs | 1074 |
| Ensembl gene annotations | 17 909 |
| Chicken orthologs of human disease genes | 995 |
| Percentage length of CtgCoV regions on | |
| RJF genome assembly | |
| Broiler/RJF | 25.6% |
| Layer/RJF | 25.0% |
| Silkie/RJF | 27.3% |
| Raw sequence traces | 2 544 985 |
| SNP-associated traces | 1 205 058 |
| QTLs | 606 |
| References | 884 |
DATABASE USAGE AND ACCESS
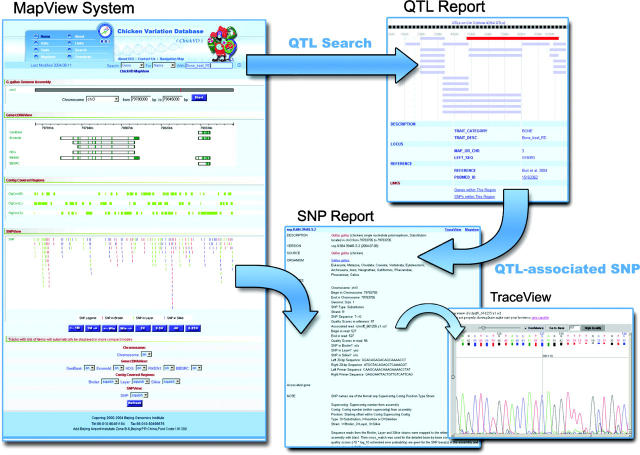
All data housed in ChickVD are uniquely mapped onto the RJF reference genome and graphically represented in MapView, an efficient visualization tool initially developed in our Rice Information System (BGI-RIS) (16) that allows users to browse SNPs in genomic and functional context. MapView is composed of four types of subviewers in hierarchical architecture, namely ChroView, GeneView, SNPView and TraceView (Figure 1). ChroView is based on the reference sequence of RJF with QTLs marked along chromosomes, and displays density and statistics of genes and SNPs. ChroView also allows users to center the map onto a specific chromosome location and make options to expand for more detailed views of genes/cDNAs, SNPs via GeneView and SNPView, respectively. A factual report for each element contained in the visualization system is displayed automatically on demand. TraceView assists users to view the original trace files around the detected SNP. Q-values for each nucleotide and a position-location function facilitate further investigation of the SNPs.
Figure 1.
Screenshots of the MapView system. SNP and QTL reports associated with a SNP are shown, with detailed views on information for SNPs in a selected chromosomal region.
The ChickVD online search tool is the entry point for querying SNPs and other data types in the database. Users can simply query SNPs by identifiers or genomic locations. The results can be further restricted to a specific chicken breed, a certain SNP type, a threshold of quality score or to a specific SNP functional class, such as a coding non-synonymous SNP. BLAST-based SNP search compares a user-supplied sequence against the flanking sequences that immediately surround the sequence variant. Advanced search interfaces for other data types (genes/cDNAs, QTLs, sequence traces and references) are also provided. For sequence searches, a key development is the implementation of SeqGetter, a search tool that allows extraction of all variations residing within genomic domains, as defined by flanking elements that can be other variants, chromosomal positions, genes, or even specific coding sequences, introns or regulatory domains.
SYSTEM DESIGN AND IMPLEMENTATION
ChickVD consists of three hardware components, a World Wide Web server, a database server and a sequence analysis/homology search server. The system is based on an Oracle9i relational database, and the front end consists of a set of JSP scripts running on TomCat web server. The search engine and MapView were developed using Java Servlet and JavaBean. Java Applets are applied for TraceView functions. To handle the large amount of complex data, we developed the standard sets of SNP-centric and QTL-centric XML formats that lay the foundation for our research work and allow ChickVD to accommodate the fast-accumulating data and to integrate new data types when they arise.
FUTURE DEVELOPMENTS
Continued efforts will be made to update SNP data and improve data quality. ChickVD will also be updated for each new chicken genome update, including annotations and more QTLs, markers and references, as soon as they become available. We will also introduce into ChickVD a version system and references around different versions. So in the near future, it will be possible for users to retrieve data from different versions, trace up and locate changes of a given entity between different versions. New versions of ChickVD will integrate allele frequencies of each variation site from ongoing projects with collaborating institutions. To enhance data utility, our development efforts will extend data structures and establish systems to support haplotype information that is expected to become the principal functional unit for chicken genetics. ChickVD continues to make enhancements to user interfaces, improve the functionality of searches and data representation, and evolve the database infrastructure and data model to ensure data quality and consistency. A side-by-side comparative map viewer will make comparative analysis of the SNP map and chicken genes easier. We welcome comments and suggestions from the chicken research community to make ChickVD a user-friendly knowledge resource.
Acknowledgments
ACKNOWLEDGEMENTS
This project was funded by the Chinese Academy of Sciences, Commission for Economy Planning, Ministry of Science and Technology (2002AA104250, 2002AA234011, 2001AA231061, 2001AA231011, 2001AA231101 and 2004AA231050), Danish National Research Foundation (Danish Platform for Integrative Biology) and the China National Grid (2002AA104250).
REFERENCES
- 1.Manner J. and Kluth,D. (2003) A chicken model to study the embryology of cloacal exstrophy. J. Pediatr. Surg., 38, 678–681. [DOI] [PubMed] [Google Scholar]
- 2.Mozdziak P.E. and Petitte,J.N. (2004) Status of transgenic chicken models for developmental biology. Dev. Dyn., 229, 414–421. [DOI] [PubMed] [Google Scholar]
- 3.Holmes D.J. and Ottinger,M.A. (2003) Birds as long-lived animal models for the study of aging. Exp. Gerontol., 38, 1365–1375. [DOI] [PubMed] [Google Scholar]
- 4.Burt D. and Pourquie,O. (2003) Chicken Genome—science nuggets to come soon. Science, 300, 1669. [DOI] [PubMed] [Google Scholar]
- 5.Ewing B., Hillier,L., Wendl,M.C. and Green,P. (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res., 8, 175–185. [DOI] [PubMed] [Google Scholar]
- 6.Ewing B. and Green,P. (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res., 8, 186–194. [PubMed] [Google Scholar]
- 7.Birney E., Andrews,D., Bevan,P., Caccamo,M., Cameron,G., Chen,Y., Clarke,L., Coates,G., Cox,T., Cuff,J. et al. (2004) Ensembl 2004. Nucleic Acids Res., 32, D468–D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J. and Wheeler,D.L. (2004) GenBank: update. Nucleic Acids Res., 32, D23–D26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boardman P.E., Sanz-Ezquerro,J., Overton,I.M., Burt,D.W., Bosch,E., Fong,W.T., Tickle,C., Brown,W.R., Wilson,S.A. and Hubbard,S.J. (2002) A comprehensive collection of chicken cDNAs. Curr. Biol., 12, 1965–1969. [DOI] [PubMed] [Google Scholar]
- 10.Kent W.J. (2002) BLAT—the BLAST-like alignment tool. Genome Res., 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Florea L., Hartzell,G., Zhang,Z., Rubin,G.M. and Miller,W. (1998) A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res., 8, 967–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris M.A., Clark,J., Ireland,A., Lomax,J., Ashburner,M., Foulger,R., Eilbeck,K., Lewis,S., Marshall,B., Mungall,C. et al. (2004) The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res., 32, D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulder N.J., Apweiler,R., Attwood,T.K., Bairoch,A., Barrell,D., Bateman,A., Binns,D., Biswas,M., Bradley,P., Bork,P. et al. (2003) The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res., 31, 315–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamosh A., Scott,A.F., Amberger,J., Bocchini,C., Valle,D. and McKusick,V.A. (2002) Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res., 30, 52–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wheeler D.L., Church,D.M., Edgar,R., Federhen,S., Helmberg,W., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Sequeira,E. et al. (2004) Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res., 32, D35–D40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao W., Wang,J. He,X. Huang,X., Jiao,Y., Dai,M., Wei,S., Fu,J., Chen,Y., Ren,X. et al. (2004) BGI-RIS: an integrated information resource and comparative analysis workbench for rice genomics. Nucleic Acids Res., 32, D377–D382. [DOI] [PMC free article] [PubMed] [Google Scholar]